Abstract
Alport syndrome (AS) is an inherited disorder characterized by glomerular basement membrane (GBM) abnormality and development of chronic kidney disease at an early age. The cause of AS is a genetic mutation in type IV collagen, and more than 80% of patients have X-linked AS (XLAS) with mutation in COL4A5. Although the causal gene has been identified, mechanisms of progression have not been elucidated, and no effective treatment has been developed. In this study, we generated a Col4a5 mutant mouse harboring a nonsense mutation (R471X) obtained from a patient with XLAS using clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated system. Col4a5 mRNA and protein expressions were not observed in the kidneys of hemizygous R471X male mice. R471X mice showed proteinuria and hematuria. Pathology revealed progression of glomerulosclerosis and interstitial fibrosis by age. Electron microscopy identified irregular thickening in GBM accompanied by irregular lamination. These observations were consistent with the clinical and pathological features of patients with AS and other established models. In addition, our mice models develop end-stage renal disease at the median age of 28 weeks, much later compared to previous models much more consistent with clinical course of human XLAS. Our models have advantages for future experiments in regard with treatment for human XLAS.
Abbreviations: AS, Alport syndrome; GBM, glomerular basement membrane; XLAS, X-linked AS; CKD, chronic kidney disease; ESRD, end-stage renal disease; CRISPR, clustered regularly interspaced short palindromic repeat; sgRNA, single-guide RNA; ssODN, single-stranded oligodeoxynucleotide; PCR, polymerase chain reaction; qPCR, quantitative PCR; ALB, albumin; CRE, urinary creatinine; BUN, blood urea nitrogen
Keywords: Col4a5, Alport syndrome, XLAS, CKD, ESRD, Model mice
Highlights
-
•
Col4a5 R471X mutant mice with a mutation derived from a patient with XLAS were used.
-
•
Hemizygous R471X male mice exhibited proteinuria and hematuria.
-
•
Pathology revealed the progression of glomerulosclerosis and interstitial fibrosis.
-
•
Electron microscopy identified irregular thickening in GBM.
-
•
Pathological features of R471X mice were consistent with that of patients with AS.
1. Introduction
Alport syndrome (AS) is an inherited chronic kidney disease (CKD), characterized by nephritic symptoms that appear during early life and progressive impairment of renal function, leading to end-stage renal disease (ESRD) [1], [2], [3]. One of the symbolic features in the kidney is irregularity of glomerular basement membrane (GBM), which is based on the genetic mutation in type IV collagen [3], [4], [5], [6], [7], [8], [9]. More than 80% of patients with AS have X-linked AS (XLAS), which is caused by a mutation in COL4A5 on the X chromosome [5], [6], the remaining patients have mutations in autosomal COL4A3 or COL4A4 [7]. Because COL4A5 is located on the X chromosome, XLAS occurs more commonly in males, and the condition may progress to ESRD by the age of 40 years [8]. Although the causal genes are known, a detailed mechanism of progression to ESRD has not been elucidated. Therefore, fundamental treatments have not been developed, and dialysis and kidney transplantation at younger age are inevitable [9].
Previous studies have shown that the severity of AS depends on the type of mutations in COL4A3–5 [4], [5], [6], [8], [9], [10]. Various types of mutations in COL4A3–5, such as nonsense, missense, deletion, insertion, and mutation of the splicing site, have been reported; the relationship between these variants and the clinical features has been vigorously investigated [11], [12], [13], [14], [15], [16]. In male patients with XLAS, the disease severity is strongly associated with the position and type of mutation in Col4a5 [11], [12], [14], [15]. Bekheirnia et al. showed that mutations closer to the 5′ end resulted in earlier onset of ESRD, regardless of the type of mutation [14]. Owing to the wide variety of genetic mutations, the genotype–phenotype correlation in patients with AS has not been well characterized. Therefore, it is important to verify this relationship using AS model animals possessing patients’ genetic variations.
Several AS animal models have been developed in dogs or mice [17], [18], [19], [20], [21], [22], [23]. These models are important for pre-clinical studies and elucidation of pathological mechanisms. Using these models, detailed analysis of kidney tissue could be possible in the same genetic background. Especially in the mouse model, differences in the genetic background (e.g., C57BL/6 J or 129/Sv) are associated with different patterns of disease progression, suggesting that animal models are useful to elucidate the underlying mechanisms involved in the development and progression of the disease [24], [25]. Furthermore, pharmacological therapy, such as that with angiotensin-converting enzyme inhibitors, was shown to delay the disease onset as in humans [26], [27]. However, despite the diverse genetic variability observed in patients with AS, only a few animal models have been established. For detailed understanding of AS, development of various animal models that harbor the mutations observed in patients with AS is needed.
In this study, we generated novel Col4a5 mutant mice harboring a nonsense mutation of exon 21 (c. 1411 C>T, p. R471X) of Col4a5, which was derived from a patient with XLAS, as reported by Yamamura et al.[16]. We developed the model possessing this variant because the male members in the family having this variant showed typical clinical and pathological findings for XLAS and expected to have XLAS symptoms in mice. Mutant mice were generated using clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) system [28], [29]. Their phenotypes were investigated by the measurement of urinary and blood biochemical parameters and light and electron microscopy to investigate whether hemizygous R471X male mice developed AS-like pathology.
2. Materials and methods
2.1. Animals
All mice were provided ad libitum access to a normal chow diet (CE-2; CLEA Japan) and water and were housed in a temperature- and humidity-controlled room. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Shonan Research Centre (IACUC of SRC), Takeda Pharmaceutical Company Limited, which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International. All procedures were conducted in accordance with the guidelines of the IACUC of SRC.
2.2. Production of Col4a5 knock-in mice
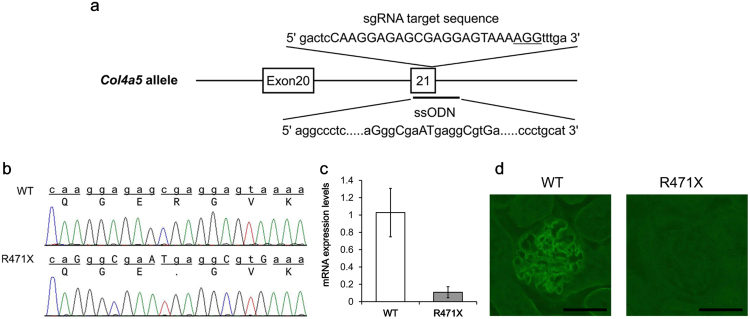
A single-guide RNA (sgRNA) and single-stranded oligodeoxynucleotide (ssODN) were designed to produce mutant mice harboring a mutation, which causes a stop codon in exon 21 (c. 1411C>T, p. R471X) of Col4a5 (Fig. 1a) [16]. For in vitro transcription, T7 promoter was added to the sgRNA sequence using polymerase chain reaction (PCR). The amplified PCR fragment was transcribed to sgRNA using MEGA shortscript T7 Kit (Life Technologies, California, USA), and sgRNA was purified using RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. ssODN, which has a target mutation and several silent mutations to prevent re-cleavage by Cas9 nuclease, was purchased from Eurofins Genomics (Tokyo, Japan). The Cas9 mRNA (5meC, ψ) was purchased from TriLink Biotechnologies (California, USA).
Fig. 1.
Targeting strategy, sequencing analysis, and expression assay for Col4a5 mutant mice. (a) Target sequence of sgRNA is capitalized and NGG protospacer-adjacent motif sequence is underlined. Exons are indicated by closed boxes. ssODN contains 120 bases, which have the target mutation (c. 1411 C>T), several silent mutations, and homology regions. (b) Sequencing results of wild-type (WT) and mutant (R471X) alleles are shown. The upper row shows the nucleotide sequence and lower row shows the amino acid sequence. Substituted bases are capitalized. (c) Col4a5 mRNA level was measured in WT and R471X male mice at 6 weeks of age (n = 5). Values are normalized by the expression level of WT and presented as the mean ± standard deviation (SD). (d) Representative images of immunostaining for COL4A5 in the kidneys of WT and R471X male mice at 6 weeks of age. Scale bar: 50 µm.
Cas9 mRNA (final, 100 ng/μL), sgRNA (50 ng/μL), and ssODN (50 ng/μL) were mixed in 0.1 × TE buffer [10 mM Tris-HCl, 0.1 mM EDTA (pH 8.0)]. The mixture was microinjected into the cytoplasm of C57BL/6 J fertilized eggs (CLEA Japan, Tokyo, Japan). The injected eggs were incubated at 37 °C until the 2-cell stage. The 2-cell-stage embryos were transferred into the ampulla of the oviduct of pseudopregnant ICR females (Charles River Laboratories Japan, Yokohama, Japan). Newborn mice were genotyped, and KI (R471X) mice were crossed with C57BL/6 J mice for two generations.
2.3. Genotyping
Genomic DNA in the ear was extracted from each offspring using a DNeasy Kit (Qiagen) or Puregene Kit (Qiagen) according to the manufacturer's protocol. The copy number of wild-type Col4a5 was analyzed using quantitative PCR (qPCR) with the genomic DNA as a template. qPCR was performed using TaqMan Fast Universal PCR MasterMix (Life Technologies), and TaqMan MGB probe kit was used to design the sgRNA target sequence (Supplementary Table S1). All samples were analyzed using 7900HT Fast Real Time PCR system (Life Technologies) and normalized against the nerve growth factor (Ngf). To confirm the insertion of R471X mutation, PCR was performed using primers flanking exon 21 of Col4a5 (5′-AGTTTGTGGTTTTGGTCATTCGTCT-3′, 5′-GGAGGTGTGCTGGAGTCAAGTTATT-3′, product size: 464 bp). The following cycle conditions were used: 98 °C for 10 s, 60 °C for 15 s, and 68 °C for 15 s for 32 cycles. The PCR fragments were treated with ExoSAP-IT (Affymetrix, California, USA) and sequenced using the same primers as above and BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies) with 3130xl Genetic Analyzer (Life Technologies).
2.4. Survival test
Mice that died suddenly or were euthanized were counted as dead. Mice that exhibited > 20% weight loss since the preceding week were euthanized, and their blood and kidneys were harvested for investigation of end-stage pathology. The survival test was terminated when the survival rate of hemizygous R471X male mice fell below 30%.
2.5. mRNA expression analysis
Total RNA was extracted from the kidney cortex using ISOGEN II (Nippon Gene), followed by DNase treatment and total RNA purification using RNeasy mini kit (Qiagen). Reverse transcription reaction was performed with SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, Massachusetts, USA) using Oligo (dT) primer. Expression levels of Col4a5 mRNA were measured by quantitative PCR (qPCR) with TaqMan Gene Expression Assays (Mm00801606_m1) (Life Technologies). For the endogenous control, TaqMan Rodent GAPDH Control Reagent (Life Technologies) was used.
2.6. Urine parameters
To collect urine samples, mice were placed individually in metabolic cages for 16 h, and urine was collected under conditions of free access to water and diet. Urine was collected from the age of 6–30 weeks at 4-week intervals. Urinary albumin (ALB) level was measured using ELISA with Albuwell M (Exocell, Philadelphia, USA). Urinary creatinine (CRE) level was measured by Hitachi 7170 type automatic analyzer (Hitachi, Tokyo, Japan) with L-Type Wako CREM (Wako Pure Chemicals Industries, Osaka, Japan). Hematuria was also examined using Uriace (Kc) (Terumo, Tokyo, Japan). These measurements were performed according to the manufacturers’ protocols.
2.7. Serum biochemical parameters
Mice were anesthetized under triple mixed anesthesia (medetomidine, midazolam and butorphanol: 0.15, 3.0, and 3.75 mg/kg), then abdominalized and blood was collected from the heart. Serum biochemical parameters [total protein (TP), ALB, blood urea nitrogen (BUN) and CRE] were measured using Hitachi Automatic Biochemical Analyzer 7180 (Hitachi), along with Total Protein-HR2 (Wako), Albumin 2-HA (Wako), UN-S (Denka Seiken, Tokyo, Japan), and L-type Creatinine M (Wako). All measurements were performed according to the manufacturers’ protocols.
2.8. Immunohistochemistry
The kidneys were removed and fixed in 4% paraformaldehyde for 4 h and then placed in 18% sucrose solution for over 16 h. Fixed kidneys were frozen and sectioned. The sections were stained with FITC-conjugated anti-COL4A5 antibody (H53; Shigei Medical Research Institute, Okayama, Japan) according to the manufacturer's protocol.
2.9. Histology
Kidneys were fixed in 4% paraformaldehyde before being embedded in paraffin for the preparation of sections. Fixed sections were stained with periodic acid methenamine silver (PAM), hematoxylin and eosin (HE) and Masson trichrome (MT).
2.10. Transmission electron microscopy (TEM)
For electron microscopy, 2- to 3-mm pieces of external renal cortex were pre-fixed with 2% glutaraldehyde and 2% paraformaldehyde in phosphate buffer and then fixed in 2% osmium tetroxide solution for 3 h in ice bath. Fixed sections were embedded in Epon 812 and were then ultrasliced. Ultrathin sections were mounted on 200 mesh copper grids and stained with 2% uranyl acetate for 15 min at 60 °C and subsequently with lead citrate solution (Sigma-Aldrich, St. Louis, MO) for 5 min at room temperature. The stained ultrathin sections were examined using H-7600 TEM (Hitachi).
3. Results
3.1. Establishment of Col4a5 knock-in mice
Using the CRISPR/Cas9 system, we created Col4a5 mutant mice harboring R471X nonsense mutation based on a mutation described in a patient with AS [16]. As a result of microinjection of Cas9 mixture into C57BL/6 J fertilized eggs, the target mutation was detected in six mice (two males and four females) out of the 21 weaning offsprings (10 males and 11 females) (Fig. 1a, b). Because they were fertile, we bred them for two generations on the C57BL/6 J background for phenotypic analysis. The offspring were genotyped as per the Mendelian ratio. The body weight of hemizygous R471X male mice increased normally until 14 weeks of age and did not increase as much as that in the wild-type after 16 weeks of age (Supplementary Fig. S1). They started dying from 26 weeks of age, and 72.2% died by 30 weeks of age (n = 18) (Supplementary Fig. S2). Female mutants were not examined in this study.
For the analysis of Col4a5 mRNA expression level, qPCR was performed using total RNA extracted from the renal cortex of 6-week-old male mice (Fig. 1c). The expression level of Col4a5 mRNA was remarkably decreased in mutant mice compared with that in wild-type mice. The decrease in Col4a5 mRNA levels in R471X mice was maintained at 14 and 22 weeks of age (data not shown). As illustrated in Fig. 1d, GBM of R471X mice was not stained, suggesting that COL4A5 was not expressed because of the nonsense mutation.
3.2. Urine and serum parameters
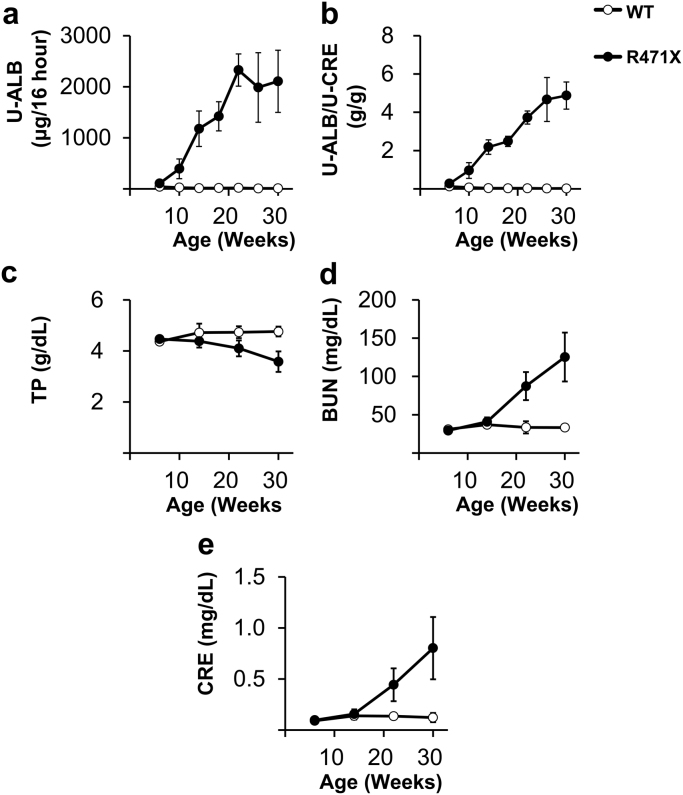
Urinary ALB levels showed an increasing tendency at 6 weeks of age in R471X mice, which increased with aging and remained high after 22 weeks of age [mean ± standard deviation (SD), 2108 ± 608 μg/16 h; n = 5–6] (Fig. 2a). Urinary ALB corrected by CRE also increased with aging (Fig. 2b), and hematuria was observed after 22 weeks of age (data not shown). With regard to serum parameters, R471X mice showed a decrease in TP and an increase in BUN and CRE levels with aging (Fig. 2c–e).
Fig. 2.
Urine and serum biochemical parameters. (a) Urine albumin (U-ALB) levels in 16-h urine collection; (b) U-ALB to urine creatinine (U-CRE) ratio (U-ALB/U-CRE); (c) blood total protein (TP); (d) blood urea nitrogen (BUN); and (e) blood CRE. Data are presented as the mean ± standard deviation (n = 5–6).
3.3. Pathological findings
Kidney histology was inspected using light and TEM in R471X mice and controls at 6 and 22 weeks of age.
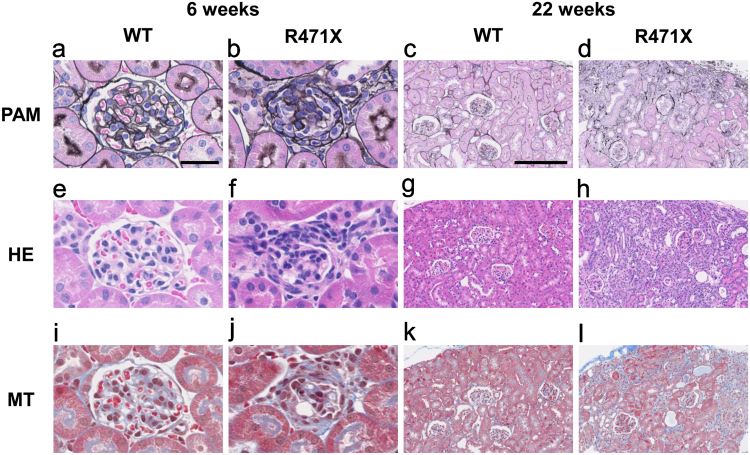
In light microscopy (Fig. 3), few glomeruli in R471X mice exhibited collapsed glomerular tuft accompanied by parietal cell hyperplasia mimicking crescent formation. Tubulointerstitial changes were limited to the peri-glomerular area of involved glomeruli. Such lesions are entirely absent in controls of same age. Marked glomerular changes are recognized in R471X mice of 22 weeks, including glomerular tuft collapse with thickened Bowman's capsule. Parietal cell hyperplasia was occasionally associated. Tubulointerstitial changes are generally associated with accumulated glomerulosclerosis. Again, such lesions were not observed in controls of the same age.
Fig. 3.
Histological changes in kidney sections of mutant male mice. Representative images of PAM-, HE- and MT-stained kidney sections of wild-type (WT) and mutant (R471X) male mice. In 6 weeks, R471X mice revealed tuft collapse with crescentic formation and tubulointerstitial fibrosis (b, f, j). In 22 weeks, R471X mice showed glomerular collapse with extraglomerular hypercellularity similar to that already found in 6 weeks (d, h, l). In both 6 and 22 weeks, control mice showed no apparent abnormalities (a, e, i, c, g, k). Scale bars: 50 µm in a, applies to a, b, e, f, i and j; 200 µm in c, applies to c, d, g, h, k, and l.
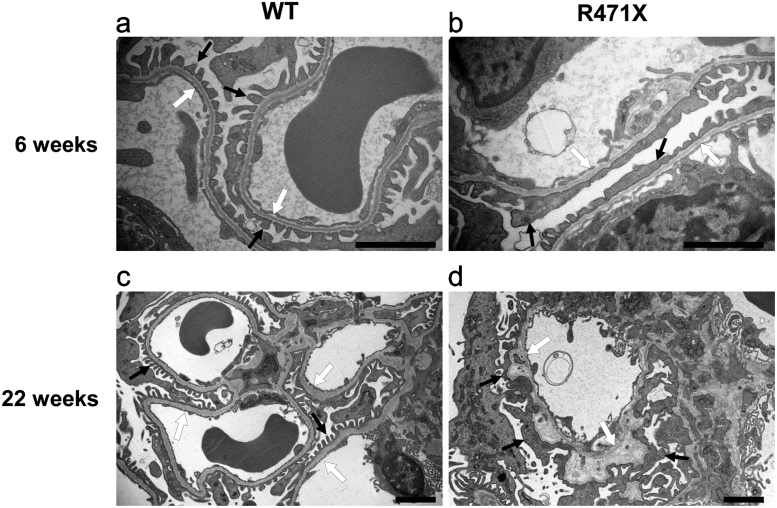
Ultrastructure of the glomeruli at 6 and 22 weeks of age was observed using TEM (Fig. 4). Six-week-old R471X mice showed focal irregularity of GBM and occasional foot process effacement; however, the average width of GBM in R471X mice was not different from that in wild-type mouse. At 22 weeks of age, mutant mice revealed marked thickening with matrix lamination in GBM. Mesangial matrices were increased.
Fig. 4.
Transmission electron microscopy (TEM) images of the glomerular basement membrane. Representative TEM images of glomerular capillary loops from wild-type (WT) and mutant (R471X) male mice are shown. In 6 weeks, R471X mice showed focal thinning and mild irregularity in GBM and focal foot process effacement in podocytes (b). In mutant mice of 22 weeks of age, marked irregularity in GBM, such as thickening and lamination, were noted (d). White arrows indicate glomerular basement membrane. Black arrows indicate podocyte foot processes. Scale bar: 2 µm.
4. Discussion
Our mice models have particular merit harboring the actual genetic abnormality of nonsense mutation in human XLAS (R471X). As a consequences, the model revealed more consistent clinical and histological features of XLAS in humans than any other animal models of AS. In this study, we demonstrated AS-like renal pathology in Col4a5 hemizygous, mutant male mice harboring nonsense mutation from a patient with XLAS (R471X). R471X mice showed loss of renal expression of Col4a5 mRNA and protein and increased levels of urinary ALB and serum BUN. These results indicated impaired renal function. Histological investigation revealed glomerular abnormalities in the mutant mice as early as 6 weeks, i. e., glomerular collapse with parietal cell hyperplasia and occasional abnormalities in GBM as revealed by TEM. These findings are mounted in 22 weeks of mutant mice. These pathological features correspond to those of previously reported Col4a3 KO [21], [22] and Col4a5 KO (G5X) mice [23], and the clinical features mimicked those observed in humans [2], [8].
R471X mice showed slower disease onset than that in other AS model mice [21], [22], [23]. The median survival time of Col4a3 KO, G5X and R471X mice were 14, 23, and 28 weeks, respectively [21], [23]. Col4a3 KO mice had a mixed genetic background of the 129/Sv and C57BL/6, and G5X mice were backcrossed from the 129/Sv to C57BL/6 for nine generations. G5X mice also showed the highest levels of urinary ALB before 20 weeks of age [23], whereas the highest values in R471X mice were seen after 22 weeks of age. The slower disease progression in R471X mice may suggest that the genetic background of C57BL/6 J or the mutation site in Col4a5 affected the progression of the disease. This hypothesis was supported by previous reports [14], [24], [25]. Andrews et al. described that Col4a3 KO mice with C57BL/6 J background showed slower disease onset than that in mice with 129/Sv background [24]. Bekheirnia et al. described that the mutation closer to the 5'-end resulted in the early onset of ESRD [14]. The C57BL/6 J background and a nonsense mutation almost 1400 bp downstream from the 5'end of Col4a5 were possibly responsible for the delayed disease onset in R471X mice.
U-ALB and U-ALB/U-CRE are primary contributors of renal dysfunction. These markers started to increase from the age of 10 weeks in R471X mice. The elevation of these markers preceded changes in serum parameters after the age of 14 weeks and the suppression of weight gain after the age of 16 weeks. The precedence of proteinuria is assumed to be due to the collapse of the glomerular filtration barrier at a young age (≤6–10 weeks). As shown in the histological analysis of R471X mice, the gradual spreading of partial glomerular collapses was noted throughout the kidney with aging. Furthermore, R471X mice developed ESRD with the progression of the disease, resulting in death. This process is consistent with the clinical course of XLAS in humans, except for hematuria [8].
Alport syndrome is also characterized by hearing loss, ocular abnormalities and hematuria [8]. As these features are measurable by non-invasive methods without renal biopsy, they are important for the diagnosis of AS. In this study, hearing loss and ocular abnormalities were not investigated, but hematuria was detected after 22 weeks of age in R471X mice. In most patients with AS, hematuria is detected at a very young age (<10 years) [8]. In Col4a3 KO mice, micro-hematuria was detected very early (at 2 weeks of age) [21]. R471X mice exhibited slow onset of hematuria. The difference in the timing of hematuria may be affected by the mutation sites or genetic background. Investigation of the genotype–phenotype correlation of these non-invasive features in AS mice models may lead to more accurate prediction of disease progression without performing renal biopsy.
In conclusion, we generated a novel mouse model harboring a nonsense mutation in exon 21 (R471X) of Col4a5 and confirmed the presence of AS-like pathology. R471X mice could help to provide new insights into the mechanisms involved in the development of AS and could be useful models for the pre-clinical study of pharmacological therapies for patients with AS and other various kidney diseases including CKD leading to ESRD.
Acknowledgments
The authors would like to thank Takashi Yano, Takeshi Yamamura, Naoya Nishimura, and Tadahiro Nambu for their technical supports. The authors acknowledge Keiji Yamamoto and Hiroyuki Kobayashi for their contributions.
Acknowledgments
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.12.003.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.12.003.
Appendix A. Transparency document
Transparency document
Appendix A. Supplementary material
Supplementary material
References
- 1.Alport A.C. Hereditary familial congenital haemorrhagic nephritis. Br. Med. J. 1927;1:504–506. doi: 10.1136/bmj.1.3454.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spear G.S., Slusser R.J. Alport's syndrome. Emphasizing electron microscopic studies of the glomerulus. Am. J. Pathol. 1972;69:213–224. [PMC free article] [PubMed] [Google Scholar]
- 3.Barker D.F., Hostikka S.L., Zhou J. Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science. 1990;248:1224–1227. doi: 10.1126/science.2349482. [DOI] [PubMed] [Google Scholar]
- 4.Harvey S.J., Zheng K., Sado Y. Role of distinct type IV collagen networks in glomerular development and function. Kidney Int. 1998;54:1857–1866. doi: 10.1046/j.1523-1755.1998.00188.x. [DOI] [PubMed] [Google Scholar]
- 5.Hostikka S.L., Eddy R.L., Byers M.G. Identification of a distinct type IV collagen alpha chain with restricted kidney distribution and assignment of its gene to the locus of X chromosome-linked Alport syndrome. Proc. Natl. Acad. Sci. USA. 1990;87:1606–1610. doi: 10.1073/pnas.87.4.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin P., Heiskari N.I.I.N.A., Zhou J.I.N.G. High mutation detection rate in the COL4A5 collagen gene in suspected Alport syndrome using PCR and direct DNA sequencing. J. Am. Soc. Nephrol. 1998;9:2291–2301. doi: 10.1681/ASN.V9122291. [DOI] [PubMed] [Google Scholar]
- 7.Mochizuki T., Lemmink H.H., Mariyama M. Identification of mutations in the alpha 3(IV) and alpha 4(IV) collagen genes in autosomal recessive Alport syndrome. Nat. Genet. 1994;8:77–81. doi: 10.1038/ng0994-77. [DOI] [PubMed] [Google Scholar]
- 8.Kashtan C.E., Michael A.F. Alport syndrome. Kidney Int. 1996;50:1445–1463. doi: 10.1038/ki.1996.459. [DOI] [PubMed] [Google Scholar]
- 9.Kashtan C.E., Ding J., Gregory M. Clinical practice recommendations for the treatment of Alport syndrome: a statement of the Alport Syndrome Research Collaborative. Pediatr. Nephrol. 2013;28:5–11. doi: 10.1007/s00467-012-2138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu X.J., Nozu K., Eguchi A. X-linked Alport syndrome associated with a synonymous p.Gly292Gly mutation alters the splicing donor site of the type IV collagen alpha chain 5 gene. Clin. Exp. Nephrol. 2016;20:699–702. doi: 10.1007/s10157-015-1197-9. [DOI] [PubMed] [Google Scholar]
- 11.Jais J.P., Knebelmann B., Giatras I. X-linked Alport syndrome: natural history in 195 families and genotype-phenotype correlations in males. J. Am. Soc. Nephrol. 2000;11:649–657. doi: 10.1681/ASN.V114649. [DOI] [PubMed] [Google Scholar]
- 12.Gross O., Netzer K.O., Lambrecht R. Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: impact on clinical counselling. Nephrol. Dial. Transplant. 2002;17:1218–1227. doi: 10.1093/ndt/17.7.1218. [DOI] [PubMed] [Google Scholar]
- 13.Pescucci C., Mari F., Longo I. Autosomal-dominant Alport syndrome: natural history of a disease due to COL4A3 or COL4A4 gene. Kidney Int. 2004;65:1598–1603. doi: 10.1111/j.1523-1755.2004.00560.x. [DOI] [PubMed] [Google Scholar]
- 14.Bekheirnia M.R., Reed B., Gregory M.C. Genotype-phenotype correlation in X-linked Alport syndrome. J. Am. Soc. Nephrol. 2010;21:876–883. doi: 10.1681/ASN.2009070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savige J., Storey H., Cheong H.I. X-linked and autosomal recessive Alport Syndrome: pathogenic variant features and further genotype-phenotype correlations. PLoS One. 2016;14:e0161802. doi: 10.1371/journal.pone.0161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamura T., Nozu K., Fu X.J. Natural history and genotype-phenotype correlation in female X-linked Alport syndrome. Kidney Int. Rep. 2017;4:850–855. doi: 10.1016/j.ekir.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kashtan C.E. Animal models of Alport syndrome. Nephrol. Dial. Transplant. 2002;17:1359–1362. doi: 10.1093/ndt/17.8.1359. [DOI] [PubMed] [Google Scholar]
- 18.Zheng K., Thorner P.S., Marrano P. Canine X chromosome-linked hereditary nephritis: a genetic model for human X-linked hereditary nephritis resulting from a single base mutation in the gene encoding the alpha 5 chain of collagen type IV. Proc. Natl. Acad. Sci. USA. 1994;26:3989–3993. doi: 10.1073/pnas.91.9.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox M.L., Lees G.E., Kashtan C.E. Genetic cause of X-linked Alport syndrome in a family of domestic dogs. Mamm. Genome. 2003;14:396–403. doi: 10.1007/s00335-002-2253-9. [DOI] [PubMed] [Google Scholar]
- 20.Lu W., Phillips C.L., Killen P.D. Insertional mutation of the collagen genes Col4a3 and Col4a4 in a mouse model of Alport syndrome. Genomics. 1999;15:113–124. doi: 10.1006/geno.1999.5943. [DOI] [PubMed] [Google Scholar]
- 21.Cosgrove D., Meehan D.T., Grunkemeyer J.A. COL4A3 knockout: a mouse model for autosomal Alport syndrome. Genes Dev. 1996;10:2981–2992. doi: 10.1101/gad.10.23.2981. [DOI] [PubMed] [Google Scholar]
- 22.Miner J.H., Sanes J.R. Molecular and functional defects in kidneys of mice lacking collagen alpha 3(IV): implications for Alport syndrome. J. Cell Biol. 1996;135:1403–1413. doi: 10.1083/jcb.135.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rheault M.N., Kren S.M., Thielen B.K. Mouse model of X-linked Alport syndrome. J. Am. Soc. Nephrol. 2004;15:1466–1474. doi: 10.1097/01.asn.0000130562.90255.8f. [DOI] [PubMed] [Google Scholar]
- 24.Andrews K.L., Mudd J.L., Li C. Quantitative trait loci influence renal disease progression in a mouse model of Alport syndrome. Am. J. Pathol. 2002;160:721–730. doi: 10.1016/S0002-9440(10)64892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosgrove D., Kalluri R., Miner J.H. Choosing a mouse model to study the molecular pathobiology of Alport glomerulonephritis. Kidney Int. 2007;71:615–618. doi: 10.1038/sj.ki.5002115. [DOI] [PubMed] [Google Scholar]
- 26.Grodecki K.M., Gains M.J., Baumal R. Treatment of X-linked hereditary nephritis in Samoyed dogs with angiotensin converting enzyme (ACE) inhibitor. J. Comp. Pathol. 1997;117:209–225. doi: 10.1016/s0021-9975(97)80016-3. [DOI] [PubMed] [Google Scholar]
- 27.Gross O., Schulze-Lohoff E., Koepke M.L. Antifibrotic, nephroprotective potential of ACE inhibitor vs AT1 antagonist in a murine model of renal fibrosis. Nephrol. Dial. Transplant. 2004;19:1716–1723. doi: 10.1093/ndt/gfh219. [DOI] [PubMed] [Google Scholar]
- 28.Shen B., Zhang J., Wu H. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Yang H., Shivalila C.S. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document
Supplementary material