Abstract
The GalNAc-type O-glycoproteome is orchestrated by a large family of polypeptide GalNAc-transferase isoenzymes (GalNAc-Ts) with partially overlapping contributions to the O-glycoproteome besides distinct nonredundant functions. Increasing evidence indicates that individual GalNAc-Ts co-regulate and fine-tune specific protein functions in health and disease, and deficiencies in individual GALNT genes underlie congenital diseases with distinct phenotypes. Studies of GalNAc-T specificities have mainly been performed with in vitro enzyme assays using short peptide substrates, but recently quantitative differential O-glycoproteomics of isogenic cells with and without GALNT genes has enabled a more unbiased exploration of the nonredundant contributions of individual GalNAc-Ts. Both approaches suggest that fairly small subsets of O-glycosites are nonredundantly regulated by specific GalNAc-Ts, but how these isoenzymes orchestrate regulation among competing redundant substrates is unclear. To explore this, here we developed isogenic cell model systems with Tet-On inducible expression of two GalNAc-T genes, GALNT2 and GALNT11, in a knockout background in HEK293 cells. Using quantitative O-glycoproteomics with tandem-mass-tag (TMT) labeling, we found that isoform-specific glycosites are glycosylated in a dose-dependent manner and that induction of GalNAc-T2 or -T11 produces discrete glycosylation effects without affecting the major part of the O-glycoproteome. These results support previous findings indicating that individual GalNAc-T isoenzymes can serve in fine-tuned regulation of distinct protein functions.
Keywords: glycosylation, glycobiology, glycosyltransferase, mass spectrometry (MS), cellular regulation, glycomics, GalNAc-transferase, glycoproteomics, inducible expression, O-glycosylation, tandem mass tag, O-GalNAc, protein glycosylation, mucin type, glycan
Introduction
GalNAc-type (mucin-type) O-glycosylation (hereafter simply O-glycosylation) is arguably one of the most abundant and complex types of protein glycosylation, and it is predicted that over 80% of proteins passing through the secretory pathway are O-glycosylated (1). O-Glycosylation is initiated by the transfer of GalNAc moieties to the hydroxyl group of Ser, Thr, and, less frequently, Tyr amino acids, a reaction catalyzed by up to 20 polypeptide GalNAc-transferases (GalNAc-Ts).2 O-Glycans are further sequentially elaborated by a large number of glycosyltransferases to produce diverse structures with various biological functions (2–4). O-Glycans are found in both dense mucin-like regions of proteins or in more isolated positions, and the latter may serve to fine-tune protein functions and, for example, co-regulate limited pro-protein processing, ectodomain shedding, signaling, and receptor dimerization (2, 3, 5–7). No apparent consensus motifs have emerged for O-glycosylation in general or for individual GalNAc-Ts, and apart from features such as glycosites often having adjacent Pro, Ser, or Thr residues, we still have little understanding of how specific sites are selected for glycosylation (1, 8, 9).
The large family of GalNAc-T isoenzymes clearly orchestrate the selection of sites, and there is now substantial evidence demonstrating that distinct GalNAc-Ts initiate O-glycosylation, whereas others follow up by incorporating additional O-glycans at adjacent sites (9, 10). In addition, “long-range” O-glycosylation is directed by the GalNAc-T C-terminal lectin domain that recognizes partially glycosylated substrates and directs glycosylation toward sites ∼10 residues away (8, 11–14). Studies of the initiating type of GalNAc-T enzymes (mainly GalNAc-T1, -T2, -T3, -T6, and -T11) have revealed quite broad substrate specificities with only a small subset of glycosites identified as being uniquely glycosylated by individual isoforms (15–19). The substrate specificities of GalNAc-Ts have mainly been explored by in vitro enzyme assays using short peptide and glycopeptide substrates (12, 20, 21), but more recently, use of isogenic cell lines with knockout (KO) and/or knock-in (KI) of individual GALNT genes coupled with the development of quantitative differential O-glycoproteomics has provided more comprehensive insight into the contributions of individual GalNAc-Ts to the O-glycoproteome (22–25). Perhaps surprisingly, this latter approach has indicated that the nonredundant contributions of GalNAc-T isoenzymes to the O-glycoproteome appear to be minor, and the majority of O-glycosites are redundantly glycosylated among the isoforms present in a cell (22). However, this may be in agreement with the subtle phenotypes found in mice deficient in Galnt genes (26–29) as well as the distinct phenotypes found in individuals with congenital deficiencies in GALNT3 and GALNT2 (25, 30).
Congenital deficiencies caused by structural defects in the coding regions of GALNT genes are very rare (31); however, an increasing number of genome-wide association studies (GWASs) points to GALNT genes for diverse disease traits or dispositions (32). We recently pursued one GWAS candidate, GALNT2, associated with plasma HDL-cholesterol and triglyceride levels (33–35) and demonstrated that two humans with homozygous deleterious mutations in GALNT2 and animal Galnt2 deficiency models exhibited lowered plasma HDL-cholesterol, thus validating the GWAS. These genome-wide association variants are positioned in GALNT2 intron 1, in close proximity to a liver-specific transcriptional regulator, and we and others have demonstrated that these polymorphisms can affect GALNT2 transcription levels (25, 36, 37). Moreover, it appears that most human glycosyltransferase genes identified as GWAS candidates have diagnostic nucleotide variants positioned within intronic regions (32) and thus may also represent gene variants with altered regulation. Surveying the degrees of potential transcriptional regulation of glycosyltransferase genes by reported RNA-Seq transcriptome data indicated indeed that genes with predicted high transcriptional regulation were overrepresented among GWAS candidate genes (32). These findings pose a fundamental question. What is the need for fine-tuned regulation of the glycosylation process, and how may this regulation be achieved? The glycosylation process involves a large number of glycosyltransferases that act sequentially and competitively on diverse substrates, and the kinetic properties, substrate availabilities, and subcellular topologies of these enzymes direct the glycosylation outcome (19, 38). However, many glycosyltransferases appear to be supersaturated in cells because, for example, proficient glycosylation can be achieved for recombinant glycoproteins over a wide range of expression levels (39). Moreover, glycosyltransferase isoenzymes have overlapping redundant functions that may affect regulation of specific functions. The latter is particularly apparent for the large group of GalNAc-T isoenzymes that have a plethora of possible peptide/protein substrates and a high likelihood for redundant activity toward these, but presumably with great variation in kinetic properties. It is therefore of considerable interest to probe the effects of regulating such enzymes.
In this study, we developed isogenic human embryonic kidney HEK293 (HEK) cell lines with inducible expression of GalNAc-T2 or -T11. Two isoenzymes endogenously expressed in these cells and kidney. We constructed isogenic HEK cell lines with KO of GALNT2 or GALNT11 and by KI installed corresponding doxycycline-inducible expression cassettes with the coding region of GALNT2 or GALNT11. We applied a quantitative differential O-glycoproteomics approach to probe effects of gradual induction, up to and beyond the normal endogenous levels of the enzymes, and found impressive selective and gradual induction of glycosylation of specific nonredundant substrates for each isoenzyme in an isoform-specific and dose-dependent manner. The study clearly demonstrates that regulation of glycosyltransferase isoenzymes can fine-tune glycosylation and the functions exerted on proteins.
Results
Establishing isogenic cells with inducible GalNAc-T2 and -T11 expression
HEK cells express a subset of the 20 GALNT genes with the major isoenzymes expressed being GALNT1 and -T2; with lower levels of GALNT3, -T7, -T11, and -T18; and with barely detectable levels of GALNT6 and -T16 (Fig. S1). HEK cells produce mainly sialylated core 1 O-glycans with a minor amount of core 2 structures (40).
We first established HEK WT cells with KO of GALNT2 (HEKΔT2 clone 2A6) or GALNT11 (HEKΔT11 clone 7E5) using zinc-finger nucleases (ZFNs) (the generation of these cell lines will be described elsewhere.3 Loss of expression of GalNAc-T2 and -T11 was verified by direct immunocytology of cells either trypsinized or grown on coverslips with validated mAbs to GalNAc-T2 (4C4) and -T11 (1B2) (15, 41). For GalNAc-T2, flow cytometry of permeabilized cells was also performed. Next, we installed doxycycline-inducible expression cassettes with full coding constructs of human GalNAc-T2 or -T11 in intron 1 of the PPP1R12C gene by KI at a locus known as the adeno-associated virus “safe harbor” (AAVS1) (42) in the corresponding KO cells (Fig. 1A).
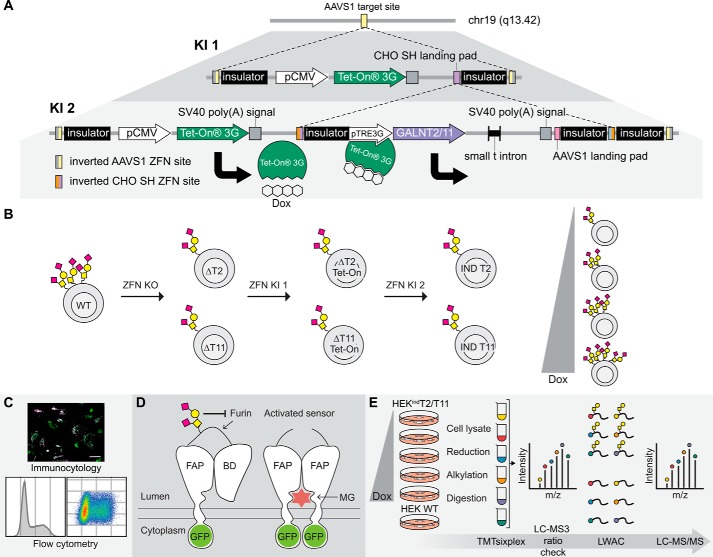
Figure 1.
Generation, validation, and exploration of isogenic HEK cell lines capable of doxycycline-inducible expression of GalNAc-T2 and -T11. A, overview of the tandem KI strategy. The first ZFN-mediated KI directs integration of the pCMV-TETOn3G cassette at the adeno-associated virus “safe harbor” (AAVS1) locus. A second KI targeted to the CHO SH landing pad within the first cassette stacks either GALNT2 or GALNT11 under control of the inducible pTRE3G promotor at the same locus. The inducer, doxycycline, binds to the Tet-On 3G protein, thereby enabling Tet-On 3G to complex with the pTRE3G promotor and drive transcription of the inducible gene. B, HEK ZFN knockouts of GALNT2 (ΔT2) and GALNT11 (ΔT11) were generated and subsequently subjected to the tandem KI strategy to generate HEKindT2 and HEKindT11 monoclonal cell lines. C, the inducible cell lines were validated by immunofluorescence, and HEKindT2 was validated also by flow cytometry. D, a GalNAc-T2 biosensor was used to validate activity of induced GalNAc-T2. The biosensor consists of a fluorogen-activating protein (FAP) and a blocking domain (BD) connected via a linker containing a GalNAc-T2–specific glycosylation site and a furin cleavage site. The sensor traffics via the Golgi en route to the cell membrane, and the absence of GalNAc-T2–mediated glycosylation leads to furin cleavage of the linker, departure of the blocking domain, and dimerization of the fluorogen-activating protein domains, which at the cell surface bind and activate the dye MG. The biosensor is GFP-tagged, allowing ratiometric measurement of activation. E, workflow for generating TMTsixplex-powered quantitative differential O-glycoproteomes for HEKindT2 and HEKindT11. Six parallel cultures, five HEKindT2 or HEKindT11 and one HEK WT, were cultured for 72 h, and the inducible cell lines were differentially induced for the final 48 h. Tryptic peptides from cell lysates of respective cultures were TMT-labeled, the six samples were combined, and a 1% aliquot was analyzed by LC-MS3 (ratio check). Subsequently, O-glycopeptides were enriched from the remaining sample by LWAC and analyzed by LC-MS/MS for O-glycopeptide identification and quantification. Finally, a deep proteome analysis was performed on the LWAC FT.
The design was built on the PrIITE platform, where the Tet-On 3G (Tet-On) transactivator gene and inducible gene of interest (GOI) are introduced at separate loci (43). We predicted that it would be possible to stably incorporate both the Tet-On 3G gene and the inducible GOI at the same locus without loss of performance, and we used a tandem KI strategy where the first cassette provides a landing pad for the second cassette. This strategy continuously provides a fresh target sequence for the alternating pairs of ZFN used. Paramount to attaining low basal expression or “leakiness” is to prevent transcriptional overlap from the first cassette to the second cassette harboring the inducible GOI, which we expected to achieve by including insulator sequences (500 nucleotides) flanking the cassettes (Fig. 1A).
First the pCMV-TETOn3G cassette was stably integrated at the AAVS1 locus in HEKΔT2 and HEKΔT11 cell lines using ZFN-mediated KI (Fig. 1B). Cells expressing Tet-On, as confirmed by AAVS1 junction PCR and the ability to induce a GFP reporter construct, were isolated either by single cell sorting or FACS. Tet-On–positive cells were subjected to a second round of ZFN-mediated KI for insertion of the coding regions of GALNT2 or GALNT11 under control of the pTRE3G-inducible promoter, at the Chinese hamster “safe harbor” (CHO SH) landing pad within the pCMV-TETOn3G cassette (Fig. 1A and Fig. S2A). Single-sorted cells were screened by immunocytology for the ability to induce expression of the relevant GalNAc-T using mAbs. The AAVS1 KI architecture of the final HEKindT2 clone (clone 385) and HEKindT11 clone (clone 40) (designated HEKindT2 and HEKindT11, respectively) was mapped by AAVS1 junction PCRs spanning the left, internal, and right junction (Fig. S2A). The HEKindT2 was concluded to have a mono-allelic insertion of the Tet-On cassette, likely with a tandem integration of the GALNT2 cassette, based on the finding that the internal junction PCR was negative, whereas the right junction PCR was positive, indicating that the cassette had been inserted with a compromised 5′-end (Fig. S2B). We investigated this further by using different internal junction PCR primer pairs and found that a truncated form of the GALNT2 cassette had been integrated in tandem (Fig. S3A). However, we were still unable to obtain a positive internal junction between the Tet-On and the GALNT2 cassette with this primer pair (G-E). The HEKindT11 clone produced positive PCR products for all junctions and was therefore considered to have biallelic integration of the Tet-On cassette and monoallelic integration of the GALNT11 cassette (Fig. S2C). Proposed structures, based on junction PCRs, of the KI architectures for HEKindT2 and HEKindT11 are shown in Fig. S3 (B and C, respectively). The genetic engineering did not appear to affect growth behavior or gross morphology, and both clones induced reproducibly over more than 20 passages and several freeze/thaw cycles.
The HEKindT2 and HEKindT11 clones were initially characterized by immunocytology, and there was no detectable staining with mAbs to GalNAc-T2 or -T11 when cells were cultured in the absence of doxycycline. Growth in 50 ng/ml doxycycline hyclate (hereafter simply doxycycline) induced strong juxtanuclear Golgi-like staining similar to the subcellular localization found in WT HEK cells, and this staining pattern coincided with the staining produced for the Golgi marker Giantin (Fig. S4). GalNAc-T2 and -T11 were readily induced, and enzyme expression could be detected after overnight doxycycline induction. To conveniently study the effect of induced GalNAc-T2 or -T11, while allowing time for biosynthesis of new glycosylation acceptor proteins, we chose to fix the induction time at 48 h for all experiments.
To further characterize the induction dynamics and determine dose windows of interest, we used indirect antibody staining and flow cytometry to quantify the expression of GalNAc-T2 in fixed and permeabilized HEKindT2 cells with graded induction from 0 to 100 ng/ml doxycycline for 48 h (Fig. 2). In the absence of inducer, there were no cells with detectable expression of GalNAc-T2, in agreement with the immunocytology analysis. The graded induction resulted in a discrete global right shift of the lower-expressing population as the concentration of doxycycline was increased from 0 to 16 ng/ml (Fig. 2B). Importantly, this low to intermediate response corresponds to the expression levels of the endogenous enzyme in HEK WT cells (Fig. 2A, bottom). By increasing the doxycycline concentration further from 50 to 100 ng/ml, we observed a biphasic induction mode with intermediate and high-level expressing cells and a maximum limit of induction ∼10-fold above the endogenous HEK WT levels. At these concentrations of doxycycline, it has been reported that mitochondrial functions may be disturbed (44). Thus, in the following experiments, we chose to study HEKindT2 induced with 0–16 ng/ml doxycycline within the range of WT expression. In parallel, we performed secondary antibody and IgG isotype controls for all samples. The controls showed no change in staining upon GalNAc-T2 induction (Fig. S5). GalNAc-T11 expression in HEK WT is relatively low compared with GalNAc-T2 (Fig. S1), and we were only able to detect weak expression by immunocytology and not by flow cytometry (not shown). However, induction of GalNAc-T11 expression with 0–10 ng/ml doxycycline appeared by immunocytology to reach HEK WT levels (not shown).
Figure 2.
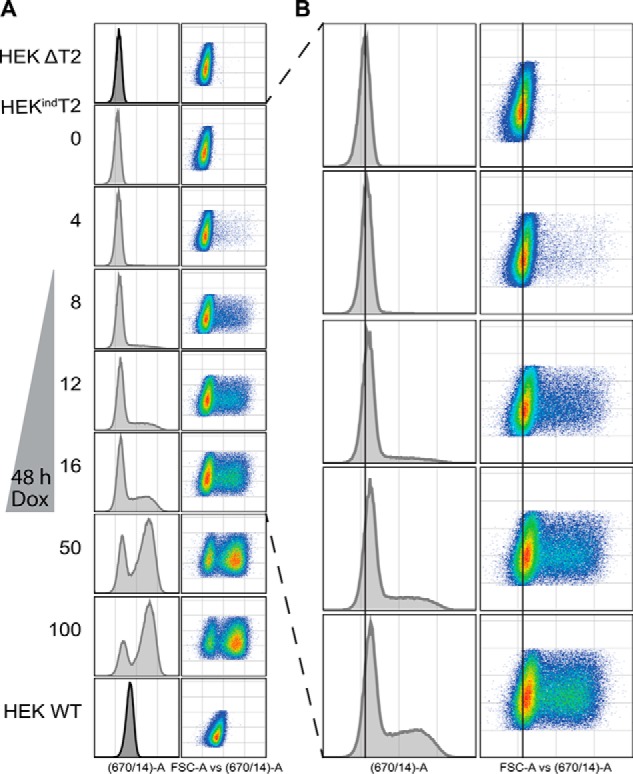
Flow cytometric quantification of induced GalNAc-T2. A, cells were induced for 48 h with doxycycline (Dox) before being fixed, permeabilized, and stained with anti-GalNAc-T2 primary antibody and Alexa Fluor 647 secondary antibody. B, enlargement of HEKindT2 induced from 0 to 16 ng/ml doxycycline with a help line centered at the peak of 0 ng/ml. The induction is biphasic with a distinct global right shift of the lower-expressing population. Data are representative of two biological and two technical replicates.
Functional validation of GalNAc-T2 induction with a biosensor
We took advantage of a recently developed GalNAc-T2–specific cell-based biosensor based on an isoform-specific glycosylation site that regulates proprotein convertase processing of angiopoietin-like 3 (ANGPTL3) (45, 46). The biosensor is composed of a fluorogen-activating protein (FAP) fused to a blocking domain (BD) by a linker sequence containing the O-glycosite and proprotein convertase cleavage site of ANGPTL3 (Fig. 1D). The absence of GalNAc-T2–mediated glycosylation leads to furin cleavage of the linker, departure of the blocking domain, and dimerization of the fluorogen-activating protein domains, which at the cell surface bind and activate the dye malachite green (MG). The biosensor is GFP-tagged, allowing measurement of both sensor expression and activation. Cells stably expressing the T2 biosensor or a control ΔGly biosensor without the T2-specific glycosylation site were generated by puromycin selection and FACS enrichment and subsequently used for sensor activation studies. Live cell images of sensor activation are shown in Fig. S6. Already at 4 ng/ml doxycycline we observed a decrease in MG fluorescence for the T2 biosensor, and further induction resulted in a gradual decrease in MG fluorescence quantitated by flow cytometry (Fig. 3). This is in agreement with induction of GalNAc-T2 leading to an increase in linker glycosylation and blockage of furin cleavage. As expected, activation of the control ΔGly biosensor was not affected by GalNAc-T2 induction.
Figure 3.
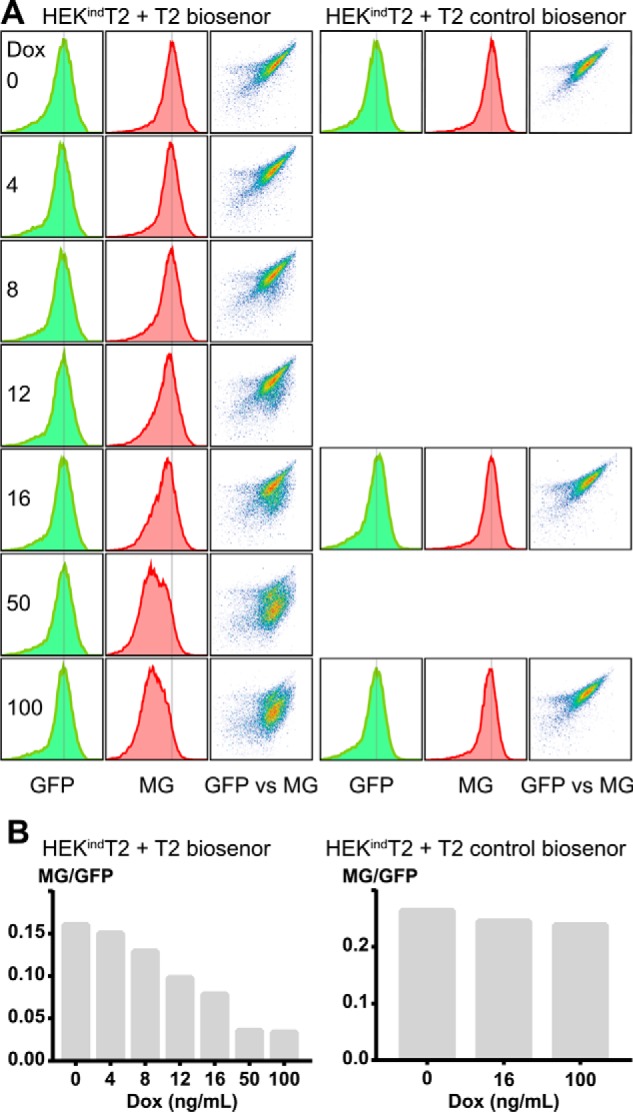
GalNAc-T2 induction inhibits biosensor activation. A, flow cytometric analysis of T2 biosensor and control biosensor activation in HEKindT2. Cells were induced for 48 h, with 0–100 ng/ml doxycycline (Dox), before incubation with MG and analysis by flow cytometry. The sensor expression (GFP) was unaffected by induction, whereas sensor activation (MG) decreased. B, ratios of the geometric mean of the MG and GFP signal from the gated events of a single experiment.
Quantitative analysis of the O-glycoproteome during graded induction of GalNAc-Ts
We modified our quantitative O-glycoproteomic workflow by here using multiplex tandem-mass-tag (TMT) amine-reactive labeling (47) of tryptic peptide digests from cell lysates followed by peanut agglutinin lectin weak-affinity chromatography (PNA LWAC) enrichment of O-glycopeptides (25) (Fig. 1E). The TMTsixplex allowed us to study four levels of induction, the uninduced state, and the original HEK WT cells. Based on the immunocytology, flow cytometry, and biosensor experiments, we chose to induce HEKindT2 or HEKindT11 cells for 48 h at the following concentrations of doxycycline: 0 (uninduced), 4, 8, 12, and 16 ng/ml for HEKindT2 and 0 (uninduced), 2.5, 5, 7.5, and 10 ng/ml for HEKindT11. HEK WT cells were cultured in parallel in the absence of doxycycline. In a separate experiment, we also performed a quantitative differential analysis of cell lysates from HEK WT and HEKΔT2 to identify GalNAc-T2–specific substrates.
We performed a LC-MS3 analysis of the TMT-labeled tryptic digest before O-glycopeptide enrichment (ratio check) and a LC-MS/MS deep proteome analysis of the LWAC flow-through (FT) (Fig. 1E). Each induction level serves as a technical replicate, and in the ratio check (Figs. S7A and S8A and Data S1 (B and F)), we found no substantial variations in the proteome except for increased abundance of peptides from the induced GalNAc-T enzyme in the corresponding HEKindT2 and HEKindT11 cells (Fig. 4). This trend was also seen in the deep proteome analysis of the LWAC FT (Figs. S11 and S12 and Data S1 (D and H)), where both of the induced enzyme protein sequences had ∼40% coverage and peptides showed comparable induction levels. Comparing quantitation of GalNAc-T2– and GalNAc-T11–derived peptides across all levels of induction to HEK WT, we confirmed that the induced GalNAc-T protein levels are comparable with that of WT cells.
Figure 4.
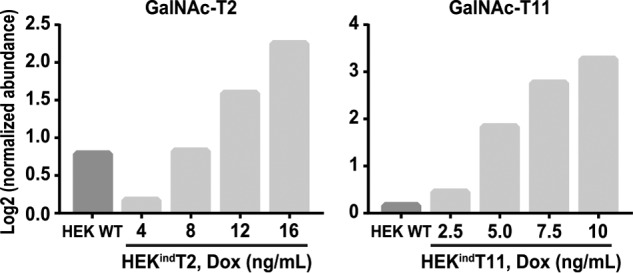
LC-MS quantification of induced GalNAc-T2 and T11 before O-glycopeptide enrichment. LC-MS analysis of TMT-labeled tryptic peptides (ratio check) was performed. Protein abundance was normalized to HEKindT2 and HEKindT11 cultured in the absence of doxycycline (Dox), and log2-transformed. HEKindT2 reaches cumulative HEK WT GalNAc-T2 levels at 8 ng/ml doxycycline and GalNAc-T11 in HEKindT11 at less than 2.5 ng/ml. For GalNAc-T2, the data are representative of analysis of two independent inducible clones, and for GalNAc-T11, data represent analysis of one clone.
The LWAC-enriched O-glycopeptides were analyzed by LC-MS/MS to produce the HEKindT2 and HEKindT11 O-glycoproteome data, which can be found in Data S1 (C and G, respectively) and summarized in Data S1A. Two representative mass spectra showing O-glycopeptide identification and quantification are shown in Fig. S13. The identified O-glycopeptides and glycoproteins were comparable between the two data sets (Fig. 5) and similar to previously obtained data sets derived from human cell lines, plasma, and platelets (Fig. S9) (25, 48). Further, the data included 136 glycoproteins not previously identified in HEK cells, and in agreement with previously reported data, the majority of the identified O-glycopeptides (∼90%) contained one or two O-glycans, and only a minor fraction contained three or more (Data S1A).
Figure 5.
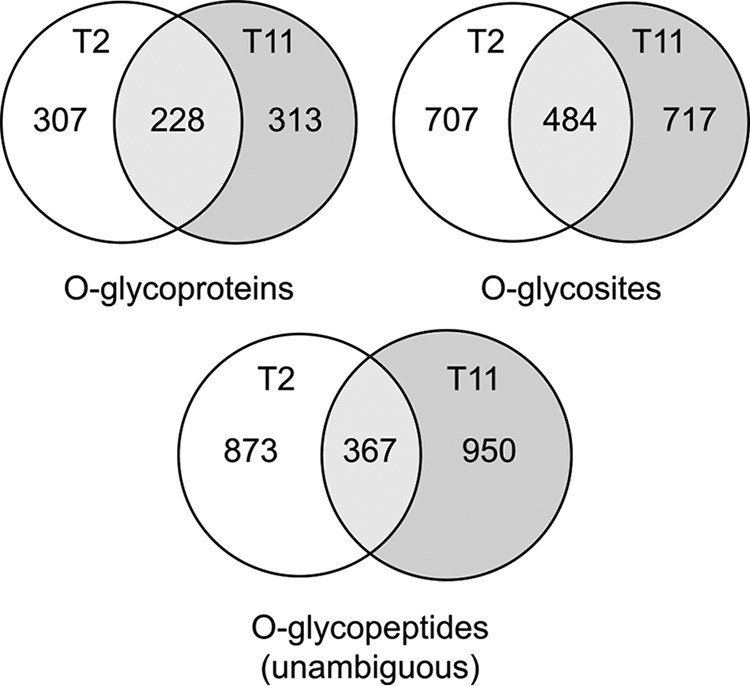
The overall HEKindT2 and HEKindT11 total O-glycoproteomes. Venn diagrams of the total number of identified O-glycoproteins, O-glycosites, and O-glycopeptides with unambiguous glycosite assignment for the two O-glycoproteomes show substantial overlap between the two cell lines.
To identify O-glycopeptides differentially affected by induction of GalNAc-T2 or -T11, we first normalized quantified O-glycopeptides to 0 ng/ml (uninduced), log2-transformed the -fold change values, and collapsed unambiguous O-glycopeptides identified multiple times. We analyzed histograms of the transformed quantitation ratios for each of the two isoforms, and for both GalNAc-T2 and -T11, we observed a rather narrow normal distribution with the appearance of right-skewed positive quant ratios upon induction (Figs. S7B and S8B). Given the highly uniform TMT-labeling and the low intersample variation observed for induced cultures (Figs. S7A and S8A), we selected a log2 -fold change threshold of 0.8 to group differentially induced O-glycopeptides. For the HEKΔT2 versus HEK WT differential O-glycoproteome (Data S1J), we selected a rather conservative cut-off ΔT2/WT log2 -fold change of −3.4 (mean minus 2 S.D. values) (Fig. S10) for selection of candidates for GalNAc-T2–specific glycosylation, similar to what was used in previous studies (22).
GalNAc-T2
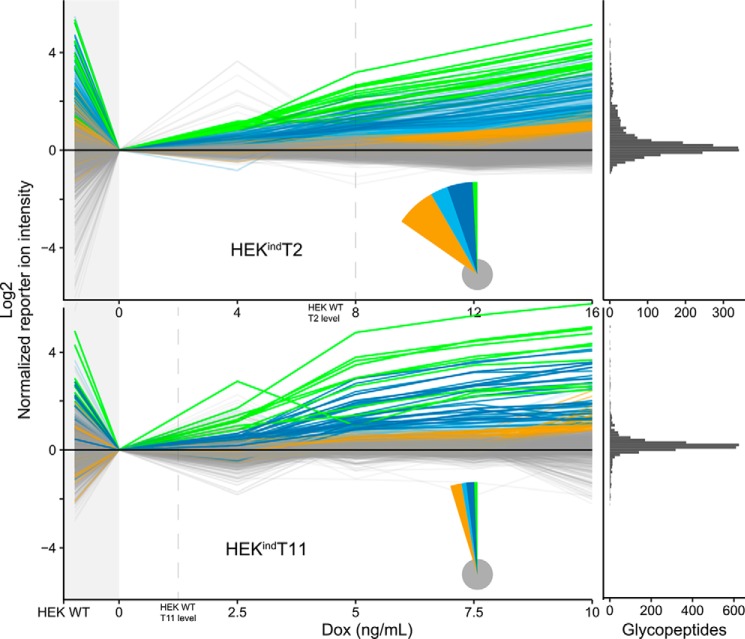
At the lowest concentration of doxycycline (4 ng/ml) 17 O-glycopeptides (hereafter simply glycopeptides) were induced above the threshold, 105 at 8 ng/ml, 76 at 12 ng/ml, and 172 at 16 ng/ml. A line-plot summary of the O-glycoproteome showing the level of induction for each glycopeptide at each doxycycline concentration is shown in Fig. 6. Comparing the differentially induced individual glycopeptides revealed a group of glycopeptides induced already at 4 ng/ml, the lowest concentration of doxycycline (green lines in Fig. 6), and groups of glycopeptides only surpassing the threshold at the higher levels of induction (blue and orange lines in Fig. 6). This made us classify the induced glycopeptides into low-dose or early responders and higher-dose or late responders. The induced glycopeptides from each group partly overlapped with the glycopeptides identified as lost in the differential O-glycoproteome of HEK WT and isogenic KO cells, and interestingly, the overlap with low-dose responders (4 ng/ml, 8 of 12, 66%) was higher compared with higher-dose responders (8 ng/ml, 24 of 73, 32%; 12 ng/ml, 16 of 47, 34%; and 16 ng/ml, 15 of 118, 12%). Comparing the induced glycosylation sites with our previous in vitro enzyme specificity analysis (21, 22, 49) revealed that seven of eight in vitro T2-specific sites were among the low-dose responders and one was among the high-dose responders (Table S1). Nine of nine peptides with an isoform specificity other than T2 or no in vitro activity were among the high-dose responders (Table S2).
Figure 6.
Induced GalNAc-T2 and -T11 O-glycoproteomes. O-Glycopeptides identified and quantified by LC-MS/MS (2,440 for HEKindT2 and 2,743 for HEKindT11) across all six cultures are shown as line graphs. One line represents one O-glycopeptide. The TMT reporter ion intensities were normalized to the uninduced cells (0 ng/ml). HEK WT is shown to the left, followed by uninduced and four increasingly induced HEKindT2 or HEKindT11 cultures. O-Glycopeptides were grouped and colored based on the concentration of doxycycline (Dox) at which their normalized reporter ion intensity is >0.8 (log2 scaled). The green groups surpass at 4 ng/ml (HEKindT2) and 2.5 ng/ml (HEKindT11) respectively, dark blue at 8 and 5 ng/ml, light blue at 12 and 7.5 ng/ml, and orange at 16 and 10 ng/ml. Gray O-glycopeptides are unaffected by induction. The pie chart inset illustrates relative group sizes, and the marginal histogram to the far right shows the distribution of O-glycopeptides at the highest doxycycline concentration. The vertical dashed lines indicate at which concentration of doxycycline the respective cell lines reach cumulative HEK WT levels of the induced GalNAc-T, as quantified in the ratio check (Fig. 4). When inducing GalNAc-T2 or -T11, we observe an increase in glycosylation, selectively of their nonredundant substrate sites, whereas the global O-glycoproteome is unaffected (gray lines). There is a strong agreement between reporter ion intensities in HEK WT and the response of the intensities upon induction. O-Glycopeptides selectively lost when comparing uninduced cells to HEK WT are rescued by enzyme induction in HEKindT2 or HEKindT11. For GalNAc-T2, the data are representative of analysis of two independent inducible clones, and for GalNAc-T11, data represent analysis of one clone.
Glycopeptides from nine glycoproteins were induced at 4 ng/ml (green in Fig. 6 and Data S1C), and among these was a glycopeptide from GalNAc-T2 itself 55DLHHSNGEEK64 (unambiguous glycosites in boldface type). This glycosite in GalNAc-T2 is not a specific substrate for the enzyme, and we interpret that this finding reflects induction of the enzyme protein and subsequent glycosylation by the other endogenous GalNAc-Ts expressed in HEK cells (Fig. S1). The other low-dose responders were GRP-78/HSPA5/BiP (634LYGSAGPPPTGEEDTAEKDEL654), PAPS transporter 1/SLC35B2 (137SYGATATSPGER148), β4Gal-T3/B4GALT3 (219SFPDQFSTGEPPALDEVPEVR239) (ambiguous glycosites italicized), calnexin/CANX (62APVPTGEVYFADSFDR77), GP73/GOLM1 (233SQTPAPSSEVVLDSK247 and 233SQTPAPSSE241), LRP11 (299AAYSTGGCLHTCSR312), TGN46/TGOLN2 (346MSGSASSENR355), and carboxypeptidase D/CPD (219SFPDQFSTGEPPALDEVPEVR239). Glycosylation of 36 other proteins was induced at 8 ng/ml doxycycline (dark blue in Fig. 6A and Data S1C). In addition to the glycopeptides induced at the lowest level, we found specifically induced glycosylation on, for example, neuroligin-1/NLGN1 (677QQPSPFSVDQR687), ATF6-α/ATF6 (405MNPSVSPANQR415), G protein–coupled receptor 64/GPR64 (252GPPFSSSQSIPVVPR266), and solute carrier family 38 member 10/SLC38A10 (787PGGRPAPSQDLNQR801). Further increasing the level of induction (above 8 ng/ml) resulted in broader glycosylation effects with more general induction of a larger number of glycopeptides.
GalNAc-T11
Approximately the same number of glycopeptides (2,500 glycopeptides) were quantified in the HEKindT11 and HEKindT2 analysis; however, substantially fewer glycopeptides were found to be induced above the threshold (Fig. 6 and Data S1G). Classifying induced glycopeptides similarly into low-dose, early responders and high-dose, late responders revealed that the early responders (2.5 ng/ml doxycycline) were essentially limited to the linker regions in LDLR class A repeats (LA module) in LRP2 (megalin) LA18-19 (2816NCPDRTCQSGYTK2828) and LRP1 LA8-9 (1096SCEGVTHVCDPSVK1109) (cysteines underlined and unambiguous glycosites in boldface type). Similar to the finding for GalNAc-T2, we also found an early increase in quantification of several glycopeptides from GalNAc-T11 (44GSGPHGPSPK53, 59FTRGPSRVLEPQFK72, 313AEGATAPIK321, and 384RPYGSPEGQDTMTHN398). At 5 ng/ml doxycycline, an additional 10 glycoproteins were found, including the LA-2 module in VLDLR (70KTCAESDFVCNNGQCVPSR88) (ambiguous glycosites italicized), a glycosite just C-terminal to the single LA module in SPINT1 (328HPVCSGTCQPTQFR341) not previously identified situated between two Kunitz inhibitor domains, and another class A repeat in LRP1 (873CDGDNDCLDNSDEAPALCHQHTCPSDR899) (Data S1G). The remaining induced glycopeptides were from BCR-associated protein 29/BCAP29, cathepsin D/CTSD, neuferricin/CYB5D2, HLA-A, GRP-78/HSPA5/BiP, podocalyxin/PODXL, xyloside xylosyltransferase 1/XXYLT1, and golgin 1/GLG1, and four of these sites were located in cysteine-rich structured domains: CYB5D2 (PRCVCVR222TTGPPSGQMPDNPPHR237NRGDLDH) (cysteines and identified glycopeptide underlined), HLA-A (CVEWLRRYLENGK201ETLQRTDAPK210THMTHHAVSDHEATLRC), XXYLT1 (CEGHVK381IYHGNCNTPIPED393), and PODXL (PSP334TVAHESNWAK343CEDLETQTQSEKQLVLNLTGNTLCAGGASDEKLISLICRAVKATFNPAQDKC).
Importantly, among the low-dose responders in the HEKindT2 and HEKindT11 data set, there was no overlap (Fig. 7). Direct comparison of the 367 overlapping unambiguous glycopeptides from the two data sets showed that glycopeptides induced in HEKindT2 were not induced in HEKindT11 and vice versa, with the exception of a single glycopeptide from GRP-78/HSPA5/BiP (634LYGSAGPPPTGEEDTAEKDEL654). We also aligned the unambiguous single-site glycopeptides of each group for both the HEKindT2 and HEKindT11 data set (Fig. S14 and S15). The number of aligned sequences is small, especially for the low-dose responders, because few peptides are both unambiguous and single-site. Still, we could validate GalNAc-T2's preference for Pro at position −1 and identified an enrichment of Pro at position +3 for medium to high-dose responders. For GalNAc-T11, the LA module linker sequence XXC6XXXTC1XX is highly enriched for low- to medium-dose responders, and a more general GalNAc-T motif with Pro at position −1 appears for high-dose responders.
Figure 7.
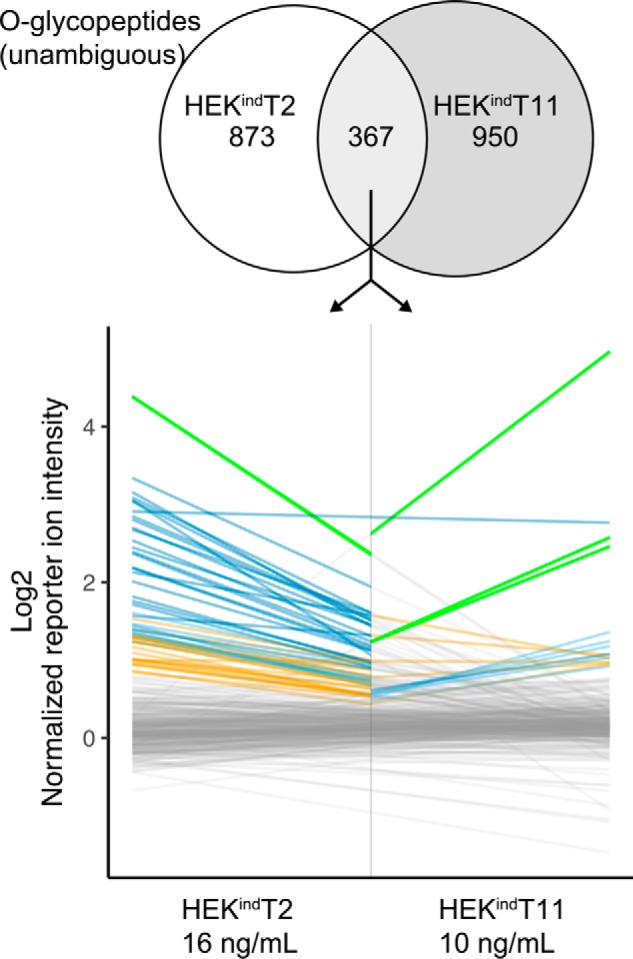
Induced GalNAc-T2 and -T11 glycosylate distinct O-glycosites. The 367 overlapping unambiguous O-glycopeptides are joined as lines for the highest doxycycline concentrations. Coloring represents respective grouping in the two data sets. Only one dark blue peptide, 34LYGSAGPPPTGEEDTAEKDEL654 (glycosite in boldface type), from GRP-78/HSPA5/BiP is shared between the two data sets (horizontal blue line).
Taken together, we interpret the data to demonstrate that induction of GalNAc-T2 and -T11 in HEK cells produces discrete glycosylation of distinct O-glycosites in a tight and dose-dependent manner without affecting the major part of the O-glycoproteome, which is covered by functional redundancy among GalNAc-T isoforms, even though many of the unaffected sites serve as substrates for the induced enzymes in in vitro assays (21).
Discussion
Whereas it seems logical that an increase in the amount of a glycosyltransferase enzyme in a cell should lead to an increase in glycosylation efficiency, this may not be the case given the complex and intertwined pathways of glycan assembly, where multiple enzymes utilize the same substrates, and isoenzymes have overlapping functions but potentially different subcellular localizations. Moreover, it is conceivable that many glycosyltransferases are present at supersaturated levels given that, for example, high expression levels of recombinant therapeutic glycoproteins in mammalian cells do not appear to exhaust the glycosylation capacity (39). We chose to study the effects of enzyme dose in the context of the large family of GalNAc-T isoenzymes, especially because several of these enzymes appear to be involved in human disorders through dysregulation of transcription rather than through structural defects in the enzyme proteins (32). We chose to study two isoenzymes, GalNAc-T2, a major contributor to the O-glycoproteome of all cells and co-regulator of lipid metabolism, and GalNAc-T11, which selectively glycosylates the LDLR-related receptors (24, 25, 34). Although both enzymes have been shown to have broad substrate specificities by in vitro analyses, we found that gradual induction of either enzyme in isogenic cell lines resulted in exquisite concomitant increase in glycosylation, selectively of their nonredundant substrate sites. The global O-glycoproteome was unaffected by the increase in GalNAc-T2 or -T11 except at induction levels 2–3-fold above the endogenous levels of these enzymes, where a more general but minor increase in the O-glycoproteome was found (Fig. 6).
Our insights into the functions of GalNAc-Ts and their isoform-specific contributions to the O-glycoproteome have so far been explored using in vitro assays with recombinant soluble enzymes and short peptide substrate (10, 20, 21) and isogenic cell lines with complete loss or gain of individual GALNT genes (22, 50). Additional information has been obtained from studies of deficiencies in model organisms (5, 15, 27, 51–53) and, for GALNT3 and GALNT2, also in humans (25, 30). Whereas in vitro studies with short synthetic peptides demonstrate quite broad substrate specificities with considerable overlap among isoforms, the available analyses of deficiencies in GALNTs in cells or organisms suggest more restricted and specific functions. Perhaps the most illustrative example is the isoform GalNAc-T3, which in vitro glycosylates a wide range of peptide substrates (54) but, by differential O-glycoproteomics of cell lines with KO of GALNT3, was identified to have only a small subset of nonredundant substrates (22, 23), whereas in vivo studies suggest that GalNAc-T3 primarily co-regulates the proprotein processing of FGF23 and hence phosphate homeostasis (29, 55). Another important example is GalNAc-T11, originally shown to have substrate specificities very similar to and overlapping with those of GalNAc-T1-T3 in vitro (15) but later identified by us as the only isoform glycosylating a highly conserved O-glycosite in the short linker regions XC6XXXTC1X present between the ligand-binding LDL class A repeats (LA modules) of all LDLR-related receptors (56). Moreover, these O-glycans and, in particular, their sialic acids potentiate receptor/ligand binding affinity and endocytosis, at least for LDLR and VLDLR (24). Interestingly, the Drosophila ortholog, dGalNAcT1 or l(2)35a, is essential for embryonal development (15) and appears to have an analogous function, glycosylating LA modules of the fly lipophorin receptors (24). Human GALNT11 is a GWAS candidate gene associated with chronic kidney decline, and GalNAc-T11 is critical for glycosylation of the linker regions between the LA modules of the endocytic receptor LRP2. Whereas the functions of GalNAc-T11–mediated O-glycosylation of LRP2 remain to be established, deficiency of Lrp2 in mice causes proteinuria and kidney disease (57), suggesting that O-glycans on LRP2 are important for function (58).
Recent studies have demonstrated that common genetic variants in a regulatory locus of GALNT2 are likely drivers of the low-HDL and high-triglyceride genome-wide association traits (25, 36, 37), pointing for the first time to an important role of tight regulation and enzyme dose of a GalNAc-T isoenzyme. Further, the respective roles of GalNAc-T3 and -T11 in regulating the function of FGF23- and LDLR-related receptors suggest that expression of these isoenzymes could also be regulated as a means to fine-tune important and disease-relevant functions (3). In the present report, we produced isogenic cell models, which enabled us to study gradually increased levels of GalNAc-T2 or -T11 expression up to and beyond the endogenous levels of a HEK cell and to evaluate the global effect of enzyme dose on the O-glycoproteome. We demonstrate that a small subset of glycopeptides were induced at the lowest level of enzyme dose (early responders), whereas the majority of detectable glycopeptides were unaffected (Fig. 6). Previously, we identified a number of GalNAc-T2–specific candidates, including apolipoprotein C-III, angiopoietin-like 3, and phospholipid transfer protein, but none of these are expressed in HEK cells. Instead, for GalNAc-T2, the early responders were mainly localized to the endoplasmic reticulum (calnexin and GRP-78) or Golgi (GP73/GOLM1, TGN46, β4Gal-T3/B4GALT3, and PAPS transporter 1/SLC35B2). One explanation for these substrates being found among the early responders may be that they represent the limited exposure of the induced biosynthetic pool of GalNAc-T2 that is moving from the endoplasmic reticulum to the Golgi. Interestingly, GalNAc-T2 was recently associated with PAPS transporter 1/SLC35B2, GP73/GOLM1, GOLIM4, and GLG1 as proteins depleted from endocytic vesicles in response to KO of AP-5 adaptor protein complex, suggesting involvement in the late endosome-to-Golgi retrieval process (59). For GalNAc-T11, the early responders identified were LRP1 and LRP2, which is in agreement with our previous findings and supports our hypothesis that O-glycosylation of these receptors serves regulatory functions (24). We recently characterized O-glycosylation of LDLR-related proteins in cell lines and rat organs, identifying LA linker glycosylation in LDLR, VLDLR, LRP1, LRP1B, SorLA, LRP8, and LRP2 (24), but in HEK cells, most of these genes are expressed at very low levels. In addition to the LRPs, we also identified a novel O-glycosite in the single LA linker of SPINT1, the matriptase inhibitor also known as HAI-1. In SPINT1, the LA domain affects the Kunitz domain 1 inhibition of matriptase (60), and it is possible that an O-glycan in the LA linker may modulate this effect. Besides glycosylation of LA linkers, increasing GalNAc-T11 expression induced a number of O-glycosites located in cysteine-rich globular domains. For example, an O-glycosite in the C terminus of XXYLT1 is located just 2 residues from one of four cysteine residues involved in disulfide bridge formation, providing structure to the catalytic domain (61). Similarly, the induced O-glycosite in podocalyxin is located in the most C-terminal part of the mucin domain, just N-terminal to a globular domain with two disulfide bridges. O-Glycosylation of podocalyxin affects ezrin/NHERF2 interaction and is essential to protect the integrity of the glomerulus (62).
The molecular basis for the observed tight regulation of highly select substrates is still unclear. The large group of common substrates that are not affected by the induction levels of a particular isoenzyme should in principle serve as a reservoir of competing substrates. The relative kinetic properties for individual O-glycosites are believed to drive selectivity and order of incorporation in peptides (19), but the isoform-specific glycosites utilized by individual GalNAc-Ts do not appear to be kinetically preferred, at least as evaluated by in vitro assays in the past (21, 55). In particular, we were unable to show activity of GalNAc-T11 with peptides derived from LDLR linker regions, and only very weak activity was demonstrated with a folded construct containing multiple LA domains in vitro (56). We cannot rule out the possibility that higher-order organization of GalNAc-Ts in the Golgi and trans-Golgi network may drive the selectivity. Hassinen et al. (63) have suggested that heteromeric glycosyltransferase complex formation and Golgi stack pH drive specificity and velocity of the respective enzymes, and the GalNAc-Ts are believed to be differentially distributed throughout the Golgi, although this has only been studied in detail for GalNAc-T1-T3 (38). Further studies are definitely needed to resolve the mechanism, but it is clear that altering the level of expression of a single GalNAc-T drives specific and tight changes in the O-glycoproteome.
In conclusion, our study demonstrates that the expression levels of GalNAc-T2 and -T11 in HEK cells are not supersaturated at or below the endogenous expression levels of these enzymes. The presented data indicate that within reasonable expression levels, there is a tight correlation between enzyme dose and glycosylation efficiency and hence stoichiometry of O-glycans on highly select glycoproteins. Thus, it is plausible that tight regulation of GalNAc-Ts directly co-regulates specific functions, such as proprotein processing and ectodomain shedding. The results further suggest that even slight dysregulation of GalNAc-T expression levels can have adverse effects on glycosylation and resulting biological functions, and this may explain the many GWAS traits associated with the GALNT gene family.
Experimental procedures
ZFN gene-targeting plasmids and construction of donor plasmids
AAVS1 CompoZr ZFN pair, targeting intron 1 of the human PPP1R12C gene at the AAV integration safe harbor site (ACCCCACAGTGGggccacTAGGGACAGGAT) and a CHO SH CompoZr ZFN pair targeting the landing pad (TCTTCCCCGACCCAGGTCACTTCTGGgttataGCTGAGACTCCGGACAGCATGCAACC) of EPB71 were obtained from Sigma. ZFN binding sites are shown in capital letters, and the linker cut site is shown in lowercase. To enable FACS enrichment and improve editing efficiency, ZFNs are tagged with a 2A peptide fused to GFP or Crimson (64).
Knock-in donor plasmids were based on Obligate ligation-gated recombination (ObLiGaRe) and designed as in Maresca et al. (65), the only modification being that inverted ZFN binding sites were positioned flanking the donor cassette (42). Synthetic ObLiGaRe donor vector frame works possessing a multiple cloning site, a universal single-stranded donor oligonucleotide (ssODN) site, and a ZFN landing pad flanked by insulators and inverted ZFN binding sites were synthesized by Genewiz USA. EPB71 (Addgene ID 90018) was used for AAVS1 targeting and EPB69 for CHO SH targeting.
pCDX2-pCMV-TETOn3G-ObLiGaRe (42) (Tet-On®3G, Clontech/Takara) was linearized with XhoI and modified by annealed oligonucleotide cloning introducing a PacI site 5′ of pCMV. The resulting plasmid was subsequently linearized with HindIII and modified by a second round of annealed oligonucleotide cloning introducing a PmeI site 3′ of SV40UTR. pCMV-TETOn3G-SV40UTR was then PacI/PmeI-excised and inserted into the PacI/PmeI site of EPB71, generating pAAVS1-EPB71-TETOn3G-ObLiGaRe donor plasmid.
The NdeI site of EPB71 was removed by NdeI digestion, overhang-blunting by T4 DNA polymerase, and religation. A PacI/PmeI fragment encoding pTRE3G-T2EYFP was excised from pAAVS1-pTRE3G-T2EYFP-ObLiGaRe (43) and inserted into NdeI-free EPB71. This plasmid was subsequently digested with BglII/NdeI, and a BglII/NdeI oligonucleotide containing internal BamHI and NotI sites was inserted, generating pAAVS1-EPB71-pTRE3G-BamHINotI-ObLiGaRe.
pTRE3G-BamHINotI was PacI/PmeI-subcloned into EPB69, generating pCHO_SH-EPB69-pTRE3G-BBamHINotI-ObLiGaRe.A BamHI/BamHI fragment encoding GALNT2 was inserted into BamHI/BamHI sites of pCHO_SH-EPB69-pTRE3G-BamHINotI-ObLiGaRe, generating pCHO_SH-EPB69-pTRE3G-GALNT2-ObLiGaRe. A BamHI/BamHI fragment encoding codon-optimized human GALNT11 was inserted into BamHI/BamHI sites of pCHO_SH-EPB69-pTRE3G-BamHINotI-ObLiGaRe, generating pCHO_SH-EPB69-pTRE3G-GALNT11-ObLiGaRe.
pTRE3G (Clontech/Takara) was digested with ApaI/BamHI and modified by annealed oligonucleotide cloning introducing a KpnI and a NotI site in the MCS. A KpnI/NotI fragment encoding secreted EYFP was inserted into KpnI/NotI-digested pTRE3G, generating pTRE3G-interferonγ-EYFP.
Cell culture and tandem knock-in
HEK cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 2 mm l-glutamine. ObLiGaRe-mediated KI was performed as described previously (43). In brief, ∼60% confluent HEKΔT2 (2A6) and HEKΔT11 (7E5) cells in 6-well plates were transfected with GFP/E2 Crimson-tagged AAVS1 ZFN plasmids and pAAVS1-EPB71-TETOn3G-ObLiGaRe donor construct in a ratio of 1:1:3 μg, using Lipofectamine 3000 according to the manufacturer's instructions. 24 h after transfection, cells expressing both GFP and E2 Crimson were sorted out in a FACS ARIA III (BD Biosciences) as described previously (64). HEKΔT2 stably expressing the Tet-On transactivator were selected after eight passages by the ability to express EYFP when transfected with pTRE3G-EYFP and cultured in medium supplemented with 50 ng/ml doxycycline (D9891, Sigma). EYFP-positive cells were sorted, expanded, and subjected to a second round of ObLiGaRe-mediated KI by transfection with GFP/E2 Crimson-tagged CHO SH ZFN plasmids and pCHO_SH-EPB69-pTRE3G-GALNT2-ObLiGaRe donor construct in a ratio of 1:1:3 μg. Bulk-sorted cells were expanded and single cell–sorted.
HEKΔT11 pAAVS1-EPB71-TETOn3G-ObLiGaRe bulk-sorted cells were expanded and single cell–sorted. A clone stably expressing the Tet-On transactivator was identified by AAVS1 junction PCR. This clone (HEKΔT11 7E5 Tet3G clone 58) was subjected to a second round of ObLiGaRe-mediated KI by transfection with GFP/E2 Crimson-tagged CHO_SH ZFN plasmids and pCHO_SH-EPB69-pTRE3G-GALNT11-ObLiGaRe donor construct in a ratio of 1:1:3 μg. Bulk-sorted cells were expanded and single cell–sorted.
Initial immunocytology screen
Expanded single-cell clones were cultured in 50 ng/ml doxycycline (D9891, Sigma) for 24 h before being harvested and seeded on Teflon-coated glass slides for microscopy. Slides with dried cells were fixed in ice-cold acetone for 7 min, dried, and incubated with anti-T2 hybridoma supernatant (4C4) overnight at 4 °C. After washing, a rabbit anti-mouse IgG FITC-labeled secondary antibody (F0261, Dako A/S, Glostrup, Denmark) diluted 1:500 in PBS, 1% BSA was added for 1 h at room temperature (RT), protected from light. After a final wash, the cells were mounted with Fluoromount-GTM (00-4958-02, Thermo Scientific).
Junction PCR
Integration of the cassettes at the AAVS1 locus was verified by junction PCR using primer pairs shown in Fig. S2 and Fig. S3 and listed in Table S2. We used TEMPase Hot Start DNA Polymerase (Ampliqon) in combination with an ammonium buffer system and a final DMSO concentration of 2.5%. The touch-down PCR program is shown in Table S3. PCR products were visualized on 2% agarose gels with GelStar stain (Lonza).
Immunocytology
2.5 × 104 cells were seeded on glass coverslips in 24-well plates. Cells were induced with doxycycline after 24 h by complete replacement of the culture medium and again after 24 h of induction. After 72 h of total culture time, cells were fixed in 4% paraformaldehyde for 15 min and washed three times in PBS. Fixed cells were permeabilized in block buffer (PBS + 5% FBS, 0.5% Triton X-100, and 50 mm glycine) for 30 min at RT, stained with anti-GalNAc-T2 (4C4) or anti-GalNAc-T11 (1B2) mouse hybridoma supernatant and polyclonal rabbit anti-giantin (1:1000) (66) for 1 h at RT, washed with PBS, and incubated for 1 h at RT with Alexa Fluor 647–conjugated goat anti-mouse and Alexa Fluor 488–conjugated goat anti-rabbit diluted 1:500 in block buffer. After a final wash in PBS, coverslips were mounted with Fluoromount-GTM (00-4958-02, Thermo Scientific). Images were captured by spinning disk confocal microscopy using an FEI CorrSight system utilizing 488- and 640-nm lasers and a ×63/1.4 numerical aperture oil objective. Captured z-stacks were collapsed for maximum intensity projection in ImageJ.
Characterization of induction dynamics by indirect flow cytometric measurement of GalNAc-T2 expression
1 × 106 of HEKindT2 cells were seeded in T25 flasks and cultured for a total of 72 h before analysis. Cells were induced with 0, 4, 8, 12, 16, 50, and 100 ng/ml doxycycline after 24 h by complete replacement of the culture medium and again after 24 h of induction. HEK WT and HEKΔT2 cells were cultured in parallel in the absence of doxycycline. Cells were stained as described (67). In brief, trypsinized cells were washed in PBS; fixed with 4% paraformaldehyde for 10 min; permeabilized in PBS, 0.1% Triton X-100 for 4 min; washed in PBS; and blocked in PBS, 5% BSA for 30 min. 0.5–1 × 106 cells were stained in a 200-μl volume with GalNAc-T2 antibody (4C4), at a concentration of 10 μg/ml in PBS, 1% BSA, for 1 h at RT. After washing in PBS, cells were incubated with Alexa Fluor 647–conjugated goat anti-mouse IgG (A-21235, Thermo Scientific) (1:1000 in PBS, 1% BSA) for 1 h at RT, washed, and stored in PBS, 1% BSA at 4 °C until analysis. Secondary-only and IgG isotype control was performed in parallel by replacing 4C4 with negative control mouse IgG1 (X0931, Dako A/S) diluted 1:100 in PBS, 1% BSA. A Fortessa flow cytometer (BD Biosciences) using a 540-nm laser and a 570/10-nm bandpass filter was used to analyze cells. A population of 1 × 105 events was recorded for each analysis. Gates were set using FSC-A versus SSC-H for exclusion of debris and FCS-H versus FSC-W to gate for single cells. Events were plotted using FlowJo. Two independent clones, clone 385 (HEKindT2) and clone 468, were analyzed.
Functional validation of GalNAc-T2 induction with an established biosensor
HEKindT2 cells stably expressing biosensors were generated and imaged as described previously (46). In brief, HEKindT2 cells were transfected with T2 biosensor (linker sequence: -KPRAPRGTPF-) or ΔGly T2 biosensor (linker sequence: -KPRAPRGGPF-) and selected with puromycin for 2 weeks. GFP-positive cells were sorted on a FACSVantage fluorescence cell sorter (BD Biosciences), expanded, and used for induction experiments. Sensor activation response to doxycycline was first investigated by live cell imaging. Cells were passed onto coverslips, induced for 48 h with 50 ng/ml doxycycline, and right before imaging incubated with 110 nm MG dye. Images were captured using an LSM 510 Meta DuoScan spectral confocal microscope equipped with a ×40 objective (Zeiss).
For flow experiments, 5 × 104 HEKindT2 sensor-expressing cells were seeded in 24-well plates and cultured for a total of 72 h before analysis. Cells were induced with 0, 4, 8, 12, 16, 50, and 100 ng/ml doxycycline after 24 h by complete replacement of the culture medium and again after 24 h of induction. Cells were released with 300 ml of PBS containing 110 nm MG dye and 5 mm EDTA for 5 min at 37 °C. Up to 50,000 events were recorded on a FACSVantage flow cytometer (BD Biosciences). GFP was detected using a 488-nm laser and a 530/30-nm filter. MG was detected using a 635-nm laser and a 685/35-nm filter. Events were gated through FSC-A versus SSC-H to select viable cells before producing histograms and dot plots. Geometric means of the gated populations (Fig. 3A) were used to calculate the MG/GFP ratio (Fig. 3B). All analysis was performed in FlowJo.
Differential O-glycoproteomics
2.5 × 106 HEKindT2 or HEKindT11 cells were seeded in T75 flasks and cultured for a total of 72 h and induced for 48 h before harvest. Cells were induced with doxycycline at 24 and 48 h by medium replacement. HEKindT2 was induced at 0, 4, 8, 12, and 16 ng/ml and HEKindT11 at 0, 2.5, 5, 7.5, and 10 ng/ml of doxycycline. 2.5 × 106 HEK WT cells were cultured in parallel in the absence of doxycycline. Total cell lysates were prepared as described previously (22). In brief, packed cell pellets were lysed in 50 mm ammonium bicarbonate, 0.2% RapiGest SF Surfactant (Waters Corp.), and the lysate was homogenized by sonication. Cleared lysates were diluted in 50 mm ammonium bicarbonate to bring the final concentration of RapiGest below 0.2% before being subjected to reduction with DTT, alkylation with iodoacetamide, and digestion with trypsin (Roche Applied Science). Each tryptic digest was purified using a 1-ml Sep-Pak C18 column (Waters Corp.), and peptide concentration was measured on a NanoDrop. Equal amounts of each digest were analyzed by LC-MS to ensure sample uniformity before labeling; 200 μg of each digest was labeled by TMTsixplexTM isobaric labeling (Thermo Scientific) following the manufacturer's instructions, providing one channel for HEK WT and five channels for uninduced and induced HEKindT2 or HEKindT11. 1% of each labeling reaction was combined, and labeling efficiency was verified in a LC-MS ratio check. Labeled peptides were pooled and treated with neuraminidase (N3001, Sigma), and O-glycopeptides were enriched by PNA LWAC. Selected elution fractions were StageTip-purified, pooled, and fractionated using the PierceTM high pH reversed-phase peptide fractionation kit (Thermo Scientific) following the manufacturer's instructions. PNA LWAC flow-through was purified on a 1-ml Sep-Pak C18 column, and 100 μg of FT peptides were fractionated using the PierceTM high-pH reversed-phase peptide fractionation kit. For GalNAc-T2, two independent clones were analyzed, HEKindT2 (clone 385) and clone 468. One inducible GalNAc-T11 clone HEKindT11 (clone 40) was analyzed.
Mass spectrometry
EASY-nLC 1000 UHPLC (Thermo Scientific) interfaced via a PicoView nanoSpray ion source (New Objectives) to an Orbitrap Fusion mass spectrometer (Thermo Scientific) was used for glycoproteomic and proteomic studies. Nano-LC was operated in a single analytical column setup using PicoFrit Emitters (New Objectives, 75-μm inner diameter) packed in-house with Reprosil-Pure-AQ C18 phase (Dr. Maisch, 1.9-μm particle size, ∼19-cm column length), with a flow rate of 200 nl min−1. All samples dissolved in 0.1% formic acid were injected onto the column and eluted in a gradient from 2 to 25% acetonitrile in either 95 (for glycoproteomic samples) or 155 min (for proteomic samples), from 25 to 80% acetonitrile in 10 min, followed by isocratic elution at 80% acetonitrile for 15 min (total elution time 120 or 180 min, respectively). The nanoSpray ion source was operated at 2.1-kV spray voltage and 300 °C heated capillary temperature. A precursor MS1 scan (m/z 350–1,700) of intact peptides was acquired in the Orbitrap at a nominal resolution setting of 120,000. For glycoproteomic samples, the five most abundant multiply charged precursor ions in the MS1 spectrum at a minimum MS1 signal threshold of 50,000 were triggered for sequential Orbitrap HCD MS2 and ETD MS2 (m/z of 100–2,000). MS2 spectra were acquired at a resolution of 50,000 for HCD MS2 and 50,000 for ETD MS2. Activation times were 30 and 200 ms for HCD and ETD fragmentation, respectively; isolation width was 4 mass units, and 1 microscan was collected for each spectrum. Automatic gain control targets were 1,000,000 ions for Orbitrap MS1 and 100,000 for MS2 scans, and the automatic gain control for the fluoranthene ion used for ETD was 300,000. Supplemental activation (20%) of the charge-reduced species was used in the ETD analysis to improve fragmentation. Dynamic exclusion for 60 s was used to prevent repeated analysis of the same components.
For proteomic samples, the 10 most abundant multiply charged precursor ions in the MS1 spectrum at a minimum MS1 signal threshold of 100,000 were triggered for sequential Orbitrap HCD MS2 at a resolution of 60,000. In addition, for some proteomic samples, a synchronous-precursor selection MS3 method was used for quantitative analysis (68, 69). Polysiloxane ions at m/z 445.12003 were used as a lock mass in all runs. Raw data have been deposited to the ProteomeXchange Consortium (70) via the PRIDE partner repository with the data set identifier PXD010155.
Mass spectrometry data analysis
Data processing was performed using Proteome Discoverer version 1.4 software (Thermo Scientific) using Sequest HT Node as described previously (1) with minor changes. Briefly, all spectra were initially searched with full cleavage specificity, filtered according to the confidence level (medium, low, and unassigned), and further searched with the semi-specific enzymatic cleavage. In all cases, the precursor mass tolerance was set to 6 ppm and fragment ion mass tolerance to 20 milli-mass units. Carbamidomethylation on cysteine residues was used as a fixed modification. Methionine oxidation and HexNAc attachment to serine, threonine, and tyrosine were used as variable modifications for ETD MS2. All HCD MS2 data were preprocessed as described (1) and searched under the same conditions mentioned above using only methionine oxidation as a variable modification.
For the quantitative analysis, only HCD MS2 spectra were used. In the case of ETD MS2 spectra, the group of TMT reporter fragment ions (m/z range of 126–132) was extracted from the adjacent HCD MS2 spectrum (the same precursor ions), concatenated with the corresponding ETD MS2 spectrum, and later used for quantification. Processing of the TMT MS3 data was performed using Proteome Discoverer version 2.1 software (Thermo Scientific).
All spectra were searched against a concatenated forward/reverse human-specific database (UniProt, January 2013, containing 20,232 canonical entries and another 251 common contaminants) using a target false discovery rate of 1%. False discovery rate was calculated using the target decoy peptide–spectrum match validator node. The resulting list was filtered to include only peptides with glycosylation as a modification.
Visualization of MS data
Raw lists of quantified O-glycopeptides were imported into R. O-Glycopeptides with missing quantification(s) were dropped. Glycopeptides with HCD misassigned glycans were dropped. Next, data were normalized to HEKindT2 or HEKindT11 cultured in 0 ng/ml doxycycline, and the ratios were log2-transformed. Subsequently, unambiguous HCD and ETD O-glycopeptides were independently collapsed to reduce redundancy. The two collapsed lists were rejoined with the list of ambiguous HCD glycopeptides, generating Data S1C (HEKindT2), Data S1G (HEKindT11), and Data S1J (HEKΔT2). A similar pipeline was applied to proteins quantified in the ratio check (Data S1, B and F) and LWAC FT (Data S1, E and I).
Author contributions
J. H., E. P. B., S. Y. V., and K. T. S. designed and performed experiments, analyzed data, and wrote the manuscript. Z. Y. and C. B. designed and performed experiments. Y. N., T. D. M., H. J., and U. M. provided materials and contributed to data interpretation and manuscript preparations. A. L., C. K. G., and Y. N. contributed to the design of experiments.
Supplementary Material
Acknowledgment
We acknowledge the Core Facility for Integrated Microscopy, Faculty of Health and Medical Sciences, University of Copenhagen for assisting with confocal microscopy.
Note added in proof
Christoffer K. Goth was inadvertently omitted from the Papers in Press version of this article published October 16, 2018.
This work was supported by Læge Sofus Carl Emil Friis og hustru Olga Doris Friis' Legat, Kirsten og Freddy Johansen Fonden, the Lundbeck Foundation, A. P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til Almene Formaal, the Mizutani Foundation, the Novo Nordisk Foundation, the Danish Research Council Sapere Aude Research Talent Grant (to K. T. S.), and Danish National Research Foundation Grant DNRF107. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Data S1, Tables S1–S3, and Figs. S1–S15.
The mass spectrometry raw data have been deposited to the ProteomeXchange Consortium (70) via the PRIDE partner repository with the data set identifier PXD010155.
Y. Narimatsu, H. J. Joshi, K. T. Schjoldager, A. Halim, J. Hintze, C. Steentoft, U. Mandel, E. P. Bennett, H. Clausen, and S. Y. Vakhrushev, manuscript in preparation.
- GalNAc-T
- GalNAc-transferase isoenzyme
- KO
- knockout
- KI
- knock-in
- GWAS
- genome-wide association study
- HEK
- HEK293
- GOI
- gene of interest
- CHO SH
- Chinese hamster “safe harbor”
- MG
- malachite green
- TMT
- tandem-mass-tag
- PNA
- peanut agglutinin
- LWAC
- lectin weak-affinity chromatography
- FT
- flow-through
- HDL
- high-density lipoprotein
- RT
- room temperature
- HCD
- higher-energy c-trap dissociation
- ETD
- electron transfer dissociation
- ZFN
- zinc finger nuclease
- AAVS1
- adeno-associated virus “safe harbor”.
References
- 1. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T. B. G., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., et al. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schjoldager K. T., and Clausen H. (2012) Site-specific protein O-glycosylation modulates proprotein processing: deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim. Biophys. Acta 1820, 2079–2094 10.1016/j.bbagen.2012.09.014 [DOI] [PubMed] [Google Scholar]
- 3. Goth C. K., Vakhrushev S. Y., Joshi H. J., Clausen H., and Schjoldager K. T. (2018) Fine-tuning limited proteolysis: a major role for regulated site-specific O-glycosylation. Trends Biochem. Sci. 43, 269–284 10.1016/j.tibs.2018.02.005 [DOI] [PubMed] [Google Scholar]
- 4. Varki A. (2017) Biological roles of glycans. Glycobiology 27, 3–49 10.1093/glycob/cww086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tian E., Stevens S. R., Guan Y., Springer D. A., Anderson S. A., Starost M. F., Patel V., Ten Hagen K. G., and Tabak L. A. (2015) Galnt1 is required for normal heart valve development and cardiac function. PLoS One 10, e0115861 10.1371/journal.pone.0115861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boskovski M. T., Yuan S., Pedersen N. B., Goth C. K., Makova S., Clausen H., Brueckner M., and Khokha M. K. (2013) The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature 504, 456–459 10.1038/nature12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maxson J. E., Luty S. B., MacManiman J. D., Abel M. L., Druker B. J., and Tyner J. W. (2014) Ligand independence of the T618I mutation in the colony-stimulating factor 3 receptor (CSF3R) protein results from loss of O-linked glycosylation and increased receptor dimerization. J. Biol. Chem. 289, 5820–5827 10.1074/jbc.M113.508440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Revoredo L., Wang S., Bennett E. P., Clausen H., Moremen K. W., Jarvis D. L., Ten Hagen K. G., Tabak L. A., and Gerken T. A. (2016) Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology 26, 360–376 10.1093/glycob/cwv108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., and Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 10.1093/glycob/cwr182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gerken T. A., Jamison O., Perrine C. L., Collette J. C., Moinova H., Ravi L., Markowitz S. D., Shen W., Patel H., and Tabak L. A. (2011) Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J. Biol. Chem. 286, 14493–14507 10.1074/jbc.M111.218701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hassan H., Reis C. A., Bennett E. P., Mirgorodskaya E., Roepstorff P., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., and Clausen H. (2000) The lectin domain of UDP-N-acetyl-d-galactosamine: polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities. J. Biol. Chem. 275, 38197–38205 10.1074/jbc.M005783200 [DOI] [PubMed] [Google Scholar]
- 12. Gerken T. A., Revoredo L., Thome J. J., Tabak L. A., Vester-Christensen M. B., Clausen H., Gahlay G. K., Jarvis D. L., Johnson R. W., Moniz H. A., and Moremen K. (2013) The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J. Biol. Chem. 288, 19900–19914 10.1074/jbc.M113.477877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Las Rivas M., Coelho H., Diniz A., Lira-Navarrete E., Compañón I., Jiménez-Barbero J., Schjoldager K. T., Bennett E. P., Vakhrushev S. Y., Clausen H., Corzana F., Marcelo F., and Hurtado-Guerrero R. (2018) Structural analysis of a GalNAc-T2 mutant reveals an induced-fit catalytic mechanism for GalNAc-Ts. Chemistry 24, 8382–8392 10.1002/chem.201800701 [DOI] [PubMed] [Google Scholar]
- 14. Lira-Navarrete E., Iglesias-Fernández J., Zandberg W. F., Compañón I., Kong Y., Corzana F., Pinto B. M., Clausen H., Peregrina J. M., Vocadlo D. J., Rovira C., and Hurtado-Guerrero R. (2014) Substrate-guided front-face reaction revealed by combined structural snapshots and metadynamics for the polypeptide N-acetylgalactosaminyltransferase 2. Angew. Chem. Int. Ed. Engl. 53, 8206–8210 10.1002/anie.201402781 [DOI] [PubMed] [Google Scholar]
- 15. Schwientek T., Bennett E. P., Flores C., Thacker J., Hollmann M., Reis C. A., Behrens J., Mandel U., Keck B., Schäfer M. A., Haselmann K., Zubarev R., Roepstorff P., Burchell J. M., Taylor-Papadimitriou J., et al. (2002) Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J. Biol. Chem. 277, 22623–22638 10.1074/jbc.M202684200 [DOI] [PubMed] [Google Scholar]
- 16. Bennett E. P., Chen Y. W., Schwientek T., Mandel U., Schjoldager K., Cohen S. M., and Clausen H. (2010) Rescue of Drosophila melanogaster l(2)35Aa lethality is only mediated by polypeptide GalNAc-transferase pgant35A, but not by the evolutionary conserved human ortholog GalNAc-transferase-T11. Glycoconj. J. 27, 435–444 10.1007/s10719-010-9290-5 [DOI] [PubMed] [Google Scholar]
- 17. Bennett E. P., Hassan H., Mandel U., Mirgorodskaya E., Roepstorff P., Burchell J., Taylor-Papadimitriou J., Hollingsworth M. A., Merkx G., van Kessel A. G., Eiberg H., Steffensen R., and Clausen H. (1998) Cloning of a human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J. Biol. Chem. 273, 30472–30481 10.1074/jbc.273.46.30472 [DOI] [PubMed] [Google Scholar]
- 18. Nehrke K., Hagen F. K., and Tabak L. A. (1998) Isoform-specific O-glycosylation by murine UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-T3, in vivo. Glycobiology 8, 367–371 10.1093/glycob/8.4.367 [DOI] [PubMed] [Google Scholar]
- 19. Wandall H. H., Hassan H., Mirgorodskaya E., Kristensen A. K., Roepstorff P., Bennett E. P., Nielsen P. A., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., and Clausen H. (1997) Substrate specificities of three members of the human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J. Biol. Chem. 272, 23503–23514 10.1074/jbc.272.38.23503 [DOI] [PubMed] [Google Scholar]
- 20. Gerken T. A., Raman J., Fritz T. A., and Jamison O. (2006) Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J. Biol. Chem. 281, 32403–32416 10.1074/jbc.M605149200 [DOI] [PubMed] [Google Scholar]
- 21. Kong Y., Joshi H. J., Schjoldager K. T.-B. G., Madsen T. D., Gerken T. A., Vester-Christensen M. B., Wandall H. H., Bennett E. P., Levery S. B., Vakhrushev S. Y., and Clausen H. (2015) Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. Glycobiology 25, 55–65 10.1093/glycob/cwu089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schjoldager K. T., Joshi H. J., Kong Y., Goth C. K., King S. L., Wandall H. H., Bennett E. P., Vakhrushev S. Y., and Clausen H. (2015) Deconstruction of O-glycosylation–GalNAc-T isoforms direct distinct subsets of the O-glycoproteome. EMBO Rep. 16, 1713–1722 10.15252/embr.201540796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lavrsen K., Dabelsteen S., Vakhrushev S. Y., Levann A. M. R., Haue A. D., Dylander A., Mandel U., Hansen L., Frödin M., Bennett E. P., and Wandall H. H. (2018) De novo expression of human polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) in colon adenocarcinoma inhibits the differentiation of colonic epithelium. J. Biol. Chem. 293, 1298–1314 10.1074/jbc.M117.812826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang S., Mao Y., Narimatsu Y., Ye Z., Tian W., Goth C. K., Lira-Navarrete E., Pedersen N. B., Benito-Vicente A., Martin C., Uribe K. B., Hurtado-Guerrero R., Christoffersen C., Seidah N. G., Nielsen R., et al. (2018) Site-specific O-glycosylation of members of the low-density lipoprotein receptor superfamily enhances ligand interactions. J. Biol. Chem. 293, 7408–7422 10.1074/jbc.M117.817981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khetarpal S. A., Schjoldager K. T., Christoffersen C., Raghavan A., Edmondson A. C., Reutter H. M., Ahmed B., Ouazzani R., Peloso G. M., Vitali C., Zhao W., Somasundara A. V., Millar J. S., Park Y., Fernando G., et al. (2016) Loss of function of GALNT2 lowers high-density lipoproteins in humans, nonhuman primates, and rodents. Cell Metab. 24, 234–245 10.1016/j.cmet.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lowe J. B., and Marth J. D. (2003) A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 72, 643–691 10.1146/annurev.biochem.72.121801.161809 [DOI] [PubMed] [Google Scholar]
- 27. Tenno M., Ohtsubo K., Hagen F. K., Ditto D., Zarbock A., Schaerli P., von Andrian U. H., Ley K., Le D., Tabak L. A., and Marth J. D. (2007) Initiation of protein O-glycosylation by the polypeptide GalNAcT-1 in vascular biology and humoral immunity. Mol. Cell Biol. 27, 8783–8796 10.1128/MCB.01204-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ten Hagen K. G., Fritz T. A., and Tabak L. A. (2003) All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 13, 1R–16R 10.1093/glycob/cwg007 [DOI] [PubMed] [Google Scholar]
- 29. Ichikawa S., Sorenson A. H., Austin A. M., Mackenzie D. S., Fritz T. A., Moh A., Hui S. L., and Econs M. J. (2009) Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyperphosphatemia despite increased Fgf23 expression. Endocrinology 150, 2543–2550 10.1210/en.2008-0877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., and Sprecher E. (2004) Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 36, 579–581 10.1038/ng1358 [DOI] [PubMed] [Google Scholar]
- 31. Hansen L., Lind-Thomsen A., Joshi H. J., Pedersen N. B., Have C. T., Kong Y., Wang S., Sparso T., Grarup N., Vester-Christensen M. B., Schjoldager K., Freeze H. H., Hansen T., Pedersen O., Henrissat B., et al. (2015) A glycogene mutation map for discovery of diseases of glycosylation. Glycobiology 25, 211–224 10.1093/glycob/cwu104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joshi H. J., Hansen L., Narimatsu Y., Freeze H. H., Henrissat B., Bennett E., Wandall H. H., Clausen H., and Schjoldager K. T. (2018) Glycosyltransferase genes that cause monogenic congenital disorders of glycosylation are distinct from glycosyltransferase genes associated with complex diseases. Glycobiology 28, 284–294 10.1093/glycob/cwy015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., Wahlstrand B., Hedner T., Corella D., Tai E. S., Ordovas J. M., et al. (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40, 189–197 10.1038/ng.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., Strait J., Duren W. L., Maschio A., Busonero F., Mulas A., et al. (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40, 161–169 10.1038/ng.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cavalli M., Pan G., Nord H., and Wadelius C. (2016) Looking beyond GWAS: allele-specific transcription factor binding drives the association of GALNT2 to HDL-C plasma levels. Lipids Health Dis. 15, 18 10.1186/s12944-016-0183-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roman T. S., Marvelle A. F., Fogarty M. P., Vadlamudi S., Gonzalez A. J., Buchkovich M. L., Huyghe J. R., Fuchsberger C., Jackson A. U., Wu Y., Civelek M., Lusis A. J., Gaulton K. J., Sethupathy P., Kangas A. J., et al. (2015) Multiple hepatic regulatory variants at the GALNT2 GWAS locus associated with high-density lipoprotein cholesterol. Am. J. Hum. Genet. 97, 801–815 10.1016/j.ajhg.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Röttger S., White J., Wandall H. H., Olivo J. C., Stark A., Bennett E. P., Whitehouse C., Berger E. G., Clausen H., and Nilsson T. (1998) Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J. Cell Sci. 111, 45–60 [DOI] [PubMed] [Google Scholar]
- 39. Walsh G. (2010) Post-translational modifications of protein biopharmaceuticals. Drug Discov. Today 15, 773–780 10.1016/j.drudis.2010.06.009 [DOI] [PubMed] [Google Scholar]
- 40. Fujitani N., Furukawa J., Araki K., Fujioka T., Takegawa Y., Piao J., Nishioka T., Tamura T., Nikaido T., Ito M., Nakamura Y., and Shinohara Y. (2013) Total cellular glycomics allows characterizing cells and streamlining the discovery process for cellular biomarkers. Proc. Natl. Acad. Sci. U.S.A. 110, 2105–2110 10.1073/pnas.1214233110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mandel U., Hassan H., Therkildsen M. H., Rygaard J., Jakobsen M. H., Juhl B. R., Dabelsteen E., and Clausen H. (1999) Expression of polypeptide GalNAc-transferases in stratified epithelia and squamous cell carcinomas: immunohistological evaluation using monoclonal antibodies to three members of the GalNAc-transferase family. Glycobiology 9, 43–52 10.1093/glycob/9.1.43 [DOI] [PubMed] [Google Scholar]
- 42. DeKelver R. C., Choi V. M., Moehle E. A., Paschon D. E., Hockemeyer D., Meijsing S. H., Sancak Y., Cui X., Steine E. J., Miller J. C., Tam P., Bartsevich V. V., Meng X., Rupniewski I., Gopalan S. M., et al. (2010) Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 20, 1133–1142 10.1101/gr.106773.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pinto R., Hansen L., Hintze J., Almeida R., Larsen S., Coskun M., Davidsen J., Mitchelmore C., David L., Troelsen J. T., and Bennett E. P. (2017) Precise integration of inducible transcriptional elements (PrIITE) enables absolute control of gene expression. Nucleic Acids Res. 45, e123 10.1093/nar/gkx371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moullan N., Mouchiroud L., Wang X., Ryu D., Williams E. G., Mottis A., Jovaisaite V., Frochaux M. V., Quiros P. M., Deplancke B., Houtkooper R. H., and Auwerx J. (2015) Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 10, 1681–1691 10.1016/j.celrep.2015.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schjoldager K. T., Vester-Christensen M. B., Bennett E. P., Levery S. B., Schwientek T., Yin W., Blixt O., and Clausen H. (2010) O-Glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J. Biol. Chem. 285, 36293–36303 10.1074/jbc.M110.156950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song L., Bachert C., Schjoldager K. T., Clausen H., and Linstedt A. D. (2014) Development of isoform-specific sensors of polypeptide GalNAc-transferase activity. J. Biol. Chem. 289, 30556–30566 10.1074/jbc.M114.599563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., and Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 10.1021/ac0262560 [DOI] [PubMed] [Google Scholar]
- 48. King S. L., Joshi H. J., Schjoldager K. T., Halim A., Madsen T. D., Dziegiel M. H., Woetmann A., Vakhrushev S. Y., and Wandall H. H. (2017) Characterizing the O-glycosylation landscape of human plasma, platelets, and endothelial cells. Blood Adv. 1, 429–442 10.1182/bloodadvances.2016002121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T. B. G., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., and Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 10.1038/nmeth.1731 [DOI] [PubMed] [Google Scholar]
- 50. Schjoldager K. T., Vakhrushev S. Y., Kong Y., Steentoft C., Nudelman A. S., Pedersen N. B., Wandall H. H., Mandel U., Bennett E. P., Levery S. B., and Clausen H. (2012) Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc. Natl. Acad. Sci. U.S.A. 109, 9893–9898 10.1073/pnas.1203563109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tran D. T., Zhang L., Zhang Y., Tian E., Earl L. A., and Ten Hagen K. G. (2012) Multiple members of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family are essential for viability in Drosophila. J. Biol. Chem. 287, 5243–5252 10.1074/jbc.M111.306159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakayama Y., Nakamura N., Kawai T., Kaneda E., Takahashi Y., Miyake A., Itoh N., and Kurosaka A. (2014) Identification and expression analysis of zebrafish polypeptide α-N-acetylgalactosaminyltransferase Y-subfamily genes during embryonic development. Gene Expr. Patterns 16, 1–7 10.1016/j.gep.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 53. Miwa H. E., Gerken T. A., Jamison O., and Tabak L. A. (2010) Isoform-specific O-glycosylation of osteopontin and bone sialoprotein by polypeptide N-acetylgalactosaminyltransferase-1. J. Biol. Chem. 285, 1208–1219 10.1074/jbc.M109.035436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bennett E. P., Hassan H., and Clausen H. (1996) cDNA cloning and expression of a novel human UDP-N-acetyl-α-d-galactosamine: polypeptide N-acetylgalactosaminyltransferase, GalNAc-t3. J. Biol. Chem. 271, 17006–17012 10.1074/jbc.271.29.17006 [DOI] [PubMed] [Google Scholar]
- 55. Kato K., Jeanneau C., Tarp M. A., Benet-Pagès A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., and Clausen H. (2006) Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J. Biol. Chem. 281, 18370–18377 10.1074/jbc.M602469200 [DOI] [PubMed] [Google Scholar]
- 56. Pedersen N. B., Wang S., Narimatsu Y., Yang Z., Halim A., Schjoldager K. T.-B. G., Madsen T. D., Seidah N. G., Bennett E. P., Levery S. B., and Clausen H. (2014) Low density lipoprotein receptor class A repeats are O-glycosylated in linker regions. J. Biol. Chem. 289, 17312–17324 10.1074/jbc.M113.545053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nykjaer A., Dragun D., Walther D., Vorum H., Jacobsen C., Herz J., Melsen F., Christensen E. I., and Willnow T. E. (1999) An endocytic pathway essential for renal uptake and activation of the steroid 25-(OH) vitamin D3. Cell 96, 507–515 10.1016/S0092-8674(00)80655-8 [DOI] [PubMed] [Google Scholar]
- 58. Gorski M., Tin A., Garnaas M., McMahon G. M., Chu A. Y., Tayo B. O., Pattaro C., Teumer A., Chasman D. I., Chalmers J., Hamet P., Tremblay J., Woodward M., Aspelund T., Eiriksdottir G., et al. (2015) Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int. 87, 1017–1029 10.1038/ki.2014.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hirst J., Itzhak D. N., Antrobus R., Borner G. H. H., and Robinson M. S. (2018) Role of the AP-5 adaptor protein complex in late endosome-to-Golgi retrieval. PLoS Biol. 16, e2004411 10.1371/journal.pbio.2004411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu M., Yuan C., Jensen J. K., Zhao B., Jiang Y., Jiang L., and Huang M. (2017) The crystal structure of a multidomain protease inhibitor (HAI-1) reveals the mechanism of its auto-inhibition. J. Biol. Chem. 292, 8412–8423 10.1074/jbc.M117.779256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu H., Takeuchi M., LeBarron J., Kantharia J., London E., Bakker H., Haltiwanger R. S., Li H., and Takeuchi H. (2015) Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat. Chem. Biol. 11, 847–854 10.1038/nchembio.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Song K., Fu J., Song J., Herzog B. H., Bergstrom K., Kondo Y., McDaniel J. M., McGee S., Silasi-Mansat R., Lupu F., Chen H., Bagavant H., and Xia L. (2017) Loss of mucin-type O-glycans impairs the integrity of the glomerular filtration barrier in the mouse kidney. J. Biol. Chem. 292, 16491–16497 10.1074/jbc.M117.798512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hassinen A., Pujol F. M., Kokkonen N., Pieters C., Kihlström M., Korhonen K., and Kellokumpu S. (2011) Functional organization of Golgi N- and O-glycosylation pathways involves pH-dependent complex formation that is impaired in cancer cells. J. Biol. Chem. 286, 38329–38340 10.1074/jbc.M111.277681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Duda K., Lonowski L. A., Kofoed-Nielsen M., Ibarra A., Delay C. M., Kang Q., Yang Z., Pruett-Miller S. M., Bennett E. P., Wandall H. H., Davis G. D., Hansen S. H., and Frödin M. (2014) High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res. 42, e84 10.1093/nar/gku251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Maresca M., Lin V. G., Guo N., and Yang Y. (2013) Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 23, 539–546 10.1101/gr.145441.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Puthenveedu M. A., and Linstedt A. D. (2001) Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J. Cell Biol. 155, 227–238 10.1083/jcb.200105005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chia P. Z. C., Ramdzan Y. M., Houghton F. J., Hatters D. M., and Gleeson P. A. (2014) High-throughput quantitation of intracellular trafficking and organelle disruption by flow cytometry. Traffic 15, 572–582 10.1111/tra.12161 [DOI] [PubMed] [Google Scholar]
- 68. McAlister G. C., Huttlin E. L., Haas W., Ting L., Jedrychowski M. P., Rogers J. C., Kuhn K., Pike I., Grothe R. A., Blethrow J. D., and Gygi S. P. (2012) Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 84, 7469–7478 10.1021/ac301572t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Quirós P. M., Prado M. A., Zamboni N., D'Amico D., Williams R. W., Finley D., Gygi S. P., and Auwerx J. (2017) Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 10.1083/jcb.201702058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 10.1038/nbt.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.