Abstract
Ezh2 is a histone methyltransferase that suppresses osteoblast maturation and skeletal development. We evaluated the role of Ezh2 in chondrocyte lineage differentiation and endochondral ossification. Ezh2 was genetically inactivated in the mesenchymal, osteoblastic, and chondrocytic lineages in mice using the Prrx1-Cre, Osx1-Cre, and Col2a1-Cre drivers, respectively. WT and conditional knockout mice were phenotypically assessed by gross morphology, histology, and micro-CT imaging. Ezh2-deficient chondrocytes in micromass culture models were evaluated using RNA-Seq, histologic evaluation, and Western blotting. Aged mice with Ezh2 deficiency were also evaluated for premature development of osteoarthritis using radiographic analysis. Ezh2 deficiency in murine chondrocytes reduced bone density at 4 weeks of age but caused no other gross developmental effects. Knockdown of Ezh2 in chondrocyte micromass cultures resulted in a global reduction in trimethylation of histone 3 lysine 27 (H3K27me3) and altered differentiation in vitro. RNA-Seq analysis revealed enrichment of an osteogenic gene expression profile in Ezh2-deficient chondrocytes. Joint development proceeded normally in the absence of Ezh2 in chondrocytes without inducing excessive hypertrophy or premature osteoarthritis in vivo. In summary, loss of Ezh2 reduced H3K27me3 levels, increased the expression of osteogenic genes in chondrocytes, and resulted in a transient post-natal bone phenotype. Remarkably, Ezh2 activity is dispensable for normal chondrocyte maturation and endochondral ossification in vivo, even though it appears to have a critical role during early stages of mesenchymal lineage commitment.
Keywords: chondrocyte, mesenchymal stem cells (MSCs), epigenetics, development, histone methylation, histone, conditional knockout mouse, skeletal development
Introduction
Epigenetic regulation of gene expression directs and maintains cellular phenotypes. Mechanisms of epigenetic regulation include post-translational modification of histones (e.g. methylation or acetylation), DNA methylation, and both small and long noncoding RNAs. Epigenetic signaling mechanisms are both dynamic and reversible, which makes them attractive targets for therapeutic drug discovery (1). Many different classes of epigenetic regulators, which include polycomb proteins and histone deacetylases, are required for normal skeletal development and differentiation of bone and cartilage (2–13). Therefore, studies of these proteins may lead to novel pharmacological strategies for the treatment of musculoskeletal disorders and injuries, including osteoporosis, osteoarthritis, scoliosis, and fractures, and would complement current cell-based strategies for musculoskeletal tissue regeneration (3, 14, 15).
Normal skeletal development requires the direct and coordinated differentiation of mesenchymal progenitor cells. In early skeletogenesis, mesenchymal progenitors may differentiate into chondrocytes and later form bone through endochondral ossification. Alternatively, they may differentiate into osteoblasts directly through the process of intramembranous bone formation. Mechanisms regulating differentiation include the expression of lineage-specific transcription factors (e.g. SOX9 and RUNX2) as well as autocrine or paracrine growth factor signaling (e.g. transforming growth factor β (TGF-β), bone morphogenic protein (BMP), wingless/integrated (WNT) (16). Recent studies have also identified epigenetic processes, including histone acetylation and methylation, that contribute to the regulation of differentiation and maturation of mesenchymal progenitors, chondrocytes, and osteoblasts (17–24).
The regulation of histone acetylation and methylation states has also been shown to control chondrogenesis (21, 25–27). Histone deacetylases, which remove acetyl groups from lysines, modulate proliferation, hypertrophy, and Wnt signaling pathways in chondrocytes (8, 21). Histone methylation at H3 lysine 9 (H3K9) regulates growth plate development and hypertrophic differentiation (28, 29), whereas inactivation of the histone 3 lysine 27 (H3K27) demethylase enzyme Jmjd3 (Kdm6b) inhibits endochondral bone formation (27).
Previous studies have identified the histone methyltransferase enzyme enhancer of zeste homolog 2 (EZH2) as an important regulator in bone development and osteoblast differentiation (2, 4, 30). EZH2 is the catalytic domain of the Polycomb repressive complex 2 (PRC2) and functions to trimethylate H3K27 (H3K27me3),3 resulting in chromatin compaction to repress gene expression during skeletal development (3, 31, 32). Global knockout of Ezh2 in mice is embryonic lethal and results in excessive accumulation of mesoderm cells in abnormal embryos (33). Conditional knockout studies of Ezh2 in the mesenchymal linage using the Prrx1-Cre driver revealed numerous skeletal abnormalities, including shortened limb segments, reduced vertebral height, and premature fusion of the cranial sutures (2, 4, 30). Similarly, conditional inactivation of Ezh2 in neural crest–derived cartilage modulated Hox gene expression and resulted in craniofacial defects in mice (10). These studies suggest that Ezh2 inactivation affects both endochondral and intramembranous bone formation. In this study, we conditionally inactivated Ezh2 in chondrocytes using the Col2a1-driven Cre recombinase to further define the role of Ezh2 during endochondral ossification.
Results
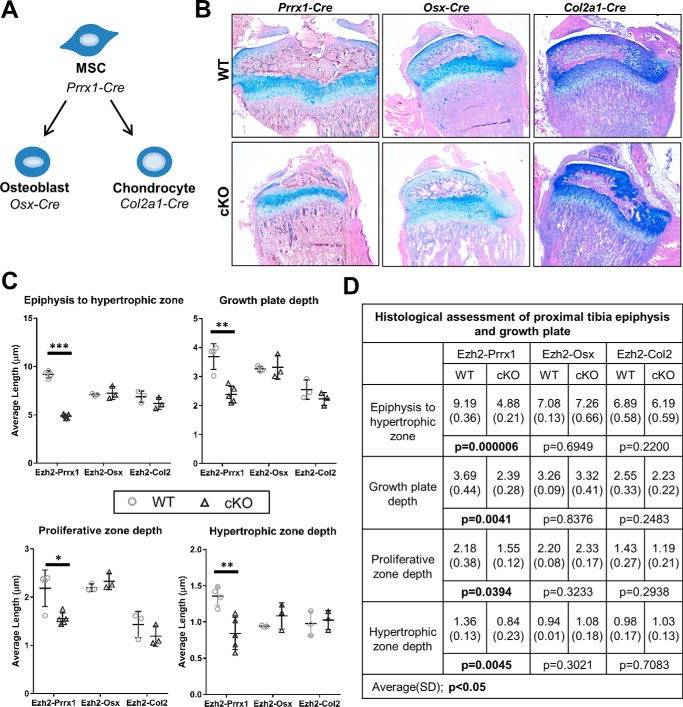
Ezh2 inactivation does not lead to skeletal defects in lineage-committed cells
Our previous work demonstrated that Ezh2 inactivation in mesenchymal progenitors (Prrx1-Cre) leads to skeletal malformations, in part because of reduced growth plate height and epiphyseal area (2). To evaluate whether Ezh2 deficiency also impacts skeletal development in linage-committed cells, we utilized the cartilage Col2a1-Cre driver and bone Osx1-Cre driver (Fig. 1A). Histological analysis of the proximal tibia of early adolescent (3–4 weeks of age) animals revealed that Ezh2-cKOOSX and Ezh2-cKOCol2 mice did not exhibit changes in cartilage morphology or growth plate depth compared with the respective WT-Cre+ animals (Fig. 1, B–D).
Figure 1.
Ezh2 inactivation in mesenchymal progenitor, osteoblast, and chondrocyte lineages. A, overview of mesenchymal lineage and mouse Cre drivers. Lineage-specific conditional inactivation of Ezh2 using the mesenchymal progenitor driver Prrx1-Cre, the osteoprogenitor driver Osx1-Cre, and the chondroprogenitor driver Col2a1-Cre was performed. B, Alcian blue/eosin analysis of proximal tibiae from WT and cKO mice at adolescence. C, average depth, growth plate depth, and hypertrophic zone depths within the growth plate were determined using ImageJ software. D, comparison of epiphysis and growth plate depth in WT and cKO animals. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
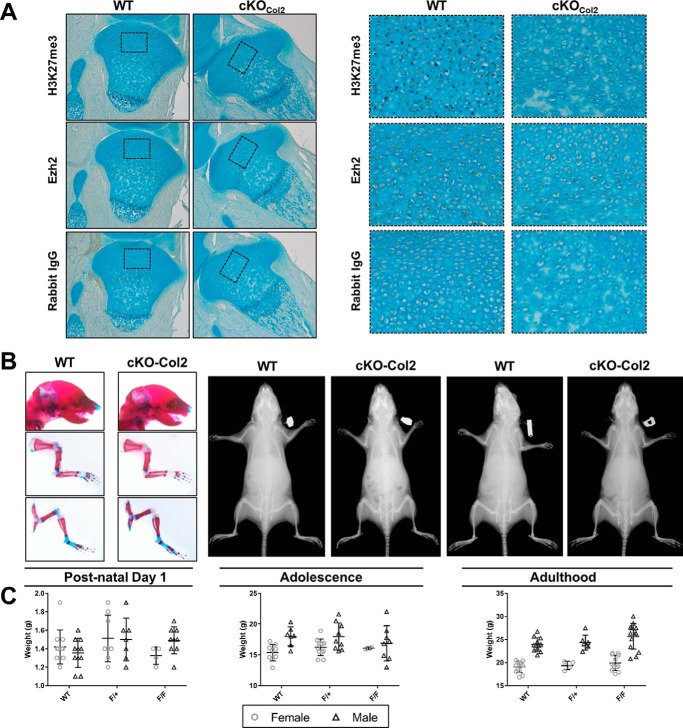
To validate Ezh2 inactivation, H3K27me3 and Ezh2 were measured by IHC in the proximal tibia from post-natal day 1 mice. Ezh2 and H3K27me3 were observed in the resting zone and hypertrophic chondrocytes of Ezh2-WTCol2 mice; however, Ezh2-cKOCol2 animals showed reduced nuclear staining of both H3K27me3 and Ezh2 Fig. 2A).
Figure 2.
Cartilage-specific deletion of Ezh2 does not affect skeletal developmental. A, immunohistochemical analysis of H3K27me3 and Ezh2 abundance in proximal tibiae from post-natal day 1 WT and cKOCol2 mice. B, Alcian blue/alizarin red whole-mount staining did not reveal growth abnormalities in post-natal day 1 mice (left panels), and radiographic analysis was similar between WT and cKOCol2 mice at 4 weeks (center panels) and 8 weeks of age (right panels). C, body weight was used as an indicator for altered growth. On post-natal day 1, adolescence, and adulthood, the body weights were similar between genotypes.
The development of mice with cartilage-specific inactivation of Ezh2 was also measured on postnatal day 1 and at 4 weeks and 8 weeks of age. The overall growth, stature, limb length, and weight of cKOCol2 mice were similar to control Ezh2-WTCol2 mice, and no sex-specific differences in these parameters were observed (Fig. 2, B and C). Together, these results indicate that Ezh2 is not essential for skeletal development in precommitted chondrocytes and osteoblasts compared with uncommitted mesenchymal progenitors.
Bone quality is reduced in Ezh2 cKOCol2 adolescent mice but normalized by adulthood
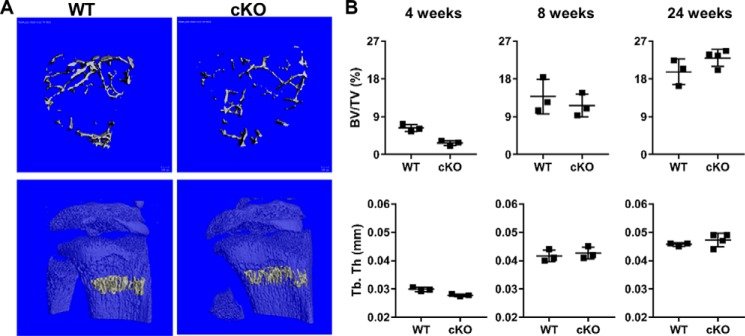
During the process of endochondral ossification, cartilage forms the framework for ossification by osteoblasts. To evaluate whether Ezh2 inactivation in chondrocytes affects matrix production and mineralization, we evaluated bone parameters in the proximal tibial metaphysis at 4 weeks and 8 weeks as well as L5 vertebrae at 24 weeks of age. At 4 weeks of age, cKOCol2 mice exhibited a 50% reduction in bone volume/tissue volume (BV/TV) compared with control animals, as well as reduced trabecular number (Tb.N) and trabecular thickness (Tb.Th) and increased trabecular spacing (Tb. Sp) (Fig. 3, A and B, and Table S1). Interestingly, cancellous bone parameters were not different between WT and cKOCol2 mice at 8 and 24 weeks of age. These data indicate that chondrocyte-specific deletion of Ezh2 reduces bone density in young mice, but this phenotype resolves during adulthood.
Figure 3.
Ezh2-deficient adolescent mice exhibit reduced bone density that is restored during adulthood. A, representative micro-CT 3D reconstruction of the proximal tibial metaphysis of WT and cKOCol2 mice. B, BV/TV and Tb.Th measured in the proximal tibial metaphysis were lower in Ezh2-deficient mice at 4 weeks of age (n = 3/group), although, upon skeletal maturity (8 weeks), no differences were observed (n = 3/group). Further, these parameters were also unchanged at 24 weeks of age when measured in the lumbar vertebrae (WT, n = 3; cKO, n = 4). *, p < 0.05.
Transcriptome profiling of immature mouse chondrocytes shows enhanced osteogenic potential of Ezh2 cKOCol2 animals
To characterize the chondrocyte-specific Ezh2 inactivation, we differentiated immature mouse chondrocytes (IMCs) from WT and cKOCol2 ex vivo for up to 14 days in micromass cultures (Fig. 4A). On day 3, the recombination event results in a reduction of Ezh2 exon counts, but a more robust loss in exon counts is observed across exons that encode the Ezh2 SET domain Fig. S1). Similar trends were also observed on days 7 and 14 (data not shown). The expression of Ezh2 protein as well as H3K27me3 was reduced in cKO IMC cultures on days 3 and 7 (Fig. 4B), further demonstrating inactivation of Ezh2 in chondrocytes. A minor increase in Ezh2 and H3K27me3 levels was observed in hypertrophic chondrocytes (day 14) in cKO IMC cultures that may be attributable to preferential proliferative expansion of cells with nonrecombined Ezh2 alleles. Histological analysis revealed that both cultures formed a hypertrophic chondrocyte-like phenotype, but staining for proteoglycans present in the extracellular matrix was reduced in cKO micromasses compared with those from WT mice Fig. 4C). Thus, loss of Ezh2 results in changes in the properties of the chondrocyte-related extracellular matrix ex vivo.
Figure 4.
Chondrogenic genes exhibit normal expression upon loss of Ezh2 and H3K27me3. A, IMCs were isolated from WT (Ezh2f/f; Col2a1-Cre−/−) and cKOCol2 mice and plated in micromass culture for 14 days (D) in the presence of chondrogenic mixture (Chondro mix). B, abundance of Ezh2 and H3K27me3 was reduced in cKOCol2 micromasses compared with the WT. The numbers on the right indicate locations of molecular weight markers (kilodalton). C, histological staining of WT (Ezh2f/f; Col2a1-Cre−/−) and cKOCol2 micromasses show a reduced Alcian blue extracellular matrix in cKOCol2 chondrocytes. D, RNA-Seq analysis was performed on WT (Ezh2f/f; Col2a1-Cre−/−) and cKOCol2 IMCs on day 3, day 7, and day 14 of culture. Gene expression data were filtered to exclude genes that were not expressed in any of the samples (0 RPKM, 19,641 of 23,359 genes were visualized). Hierarchical clustering of the filtered RNA-Seq identified three clusters that grouped together WT and cKOCol2 samples taken at similar time points, indicating that the majority of expressed genes were similar between samples. E, chondrogenic genes associated with immature and proliferating chondrocytes (Col2a1, Acan, and Sox9) are highly expressed on day 3, whereas the hypertrophic markers Col10a1 and Vegfa are up-regulated on day 14. F, validation of RNA-Seq data shows good concordance with quantitative PCR.
We also performed whole-transcriptome analysis of IMC cultures from WT or cKO mice using RNA-Seq to determine the effect of Ezh2 inactivation in chondrocytes. Unsupervised hierarchical clustering analysis of genes expressed in any of the samples (19,641 of 23,359 genes) revealed distinct clustering of samples according to time point, where day 14 samples form a distinct outgroup (Fig. 4D). In particular, IMC cultures from both genotypes differentiated toward the hypertrophic chondrocyte phenotype as indicated by down-regulation of the extracellular matrix proteins Col2a1 and Acan and the transcription factor Sox9 (Fig. 4E). Conversely, the hypertrophy-associated extracellular matrix proteins Col10a1 and Vegfa were up-regulated on day 14 (Fig. 4E). Gene expression showed strong concordance between RNA-Seq and real-time quantitative PCR (Fig. 4, E and F).
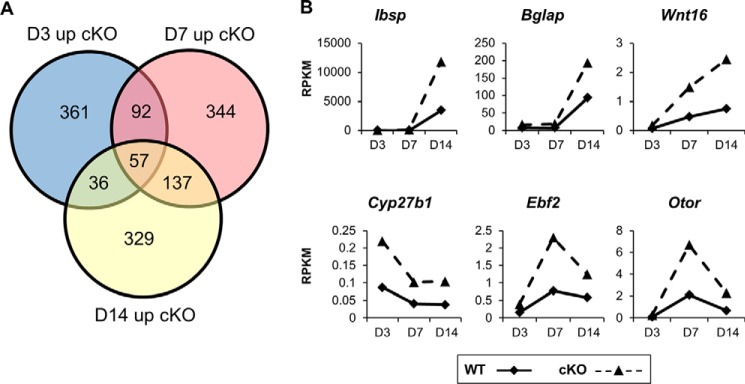
To further understand the transcriptional effects of Ezh2 inactivation, we performed differential gene expression analysis (>0.1 RPKM in cKO groups) between WT and cKO IMC cultures at each time point and observed 57 genes that were commonly up-regulated >1.4-fold in cKO compared with WT IMC cultures (Fig. 5A). These genes included osteogenic extracellular matrix proteins such as Ibsp, Bglap, and Wnt16 as well as Cyp27b1, Ebf2, and Otor (Fig. 5B).
Figure 5.
Differential gene expression analysis shows up-regulation of osteogenic genes in Ezh2-deficient immature chondrocyte cultures. A, differential gene expression analysis of RNA-Seq from WT (Ezh2wt/wt; Col2a1-Cre+/−) and cKOCol2 immature chondrocytes revealed 57 genes that were commonly up-regulated more than 1.4-fold in Ezh2-deficient cultures on days 3, 7, and 14. The analysis was limited to genes with a minimal expression of 0.1 RPKM in the cKO group at each time point. B, the osteogenic extracellular matrix genes Ibsp, Bglap, and Wnt16 were up-regulated in cKOCol2 chondrocytes as well as some nonosteogenic genes such as Cyp27b1, Ebf2, and Otor.
Functional gene annotation clustering using DAVID 6.7 (>0.1 RPKM expression of WT or cKO at each time point) shows that, in day 3 IMC cultures, Ezh2 inactivation led to the enrichment of functional categories, including “signal,” “extracellular matrix,” and “growth factor” (Table S2). Similar functional categories were also observed on day 7 of culture; however the “bone development” gene cluster was also notably enriched in cKO IMCs. Furthermore, Ezh2 inactivation led to the enrichment of a number of osteogenic functional categories on day 14, including “regulation of ossification,” “calcium signaling pathway,” “skeletal system development,” and “bone development” Table S2).
Functional annotation analysis suggests that Ezh2 inactivation enhances ossification; therefore, we re-evaluated the RNA-Seq results using a curated list of 93 genes that are relevant to skeletal development to determine whether osteogenesis was enhanced in cKO IMCs. Hierarchical clustering of this curated skeletal gene list revealed that day 3 IMC cultures expressed a distinct chondrogenic gene cluster that was down-regulated over time (Fig. 6A). Conversely, an osteogenic gene cluster including Sp7(Osx), Runx2, Ibsp, and Bglap was up-regulated in day 14 samples and had the greatest relative expression in cKO IMC cultures (Fig. 6A). Several osteogenic genes were expressed at 2-fold higher levels in cKO compared with WT IMC cultures on day 14 (Fig. 6B). As an important regulator of osteogenic differentiation, we evaluated whether the transcription factor Sp7(Osx) was also up-regulated at the protein level. Sp7(Osx) was present at higher levels in cKO IMC cultures compared with the WT on day 7 and furthermore at day 14 (Fig. 6C). Together, these data demonstrate that Col2a1-specific deletion of Ezh2 enhances the expression of osteogenic genes within the cartilage tissue.
Figure 6.
Ezh2-deficient chondrocytes exhibit enhanced expression of an osteogenic-program in vitro, but genetic inactivation does not result in osteoarthritis in vivo. A, expression of chondrogenic, osteogenic, cell–cell contact/extracellular matrix, and other genes by RNA-Seq. Ezh2-deficient IMCs exhibit up-regulation of osteogenic genes on day 14 compared with WT (Ezh2wt/wt; Col2a1-Cre+/−) IMC cultures. B, osteoblast-associated genes, including Sp7(Osx) and Bmp2, the osteocyte-associated extracellular matrix gene Mepe, as well as the WNT pathway inhibitors Sost and Dkk1 are up-regulated in cKOCol2 IMC cultures. C, protein levels of the transcription factor Sp7(Osx) were also enhanced in Ezh2-deficient IMC cultures. The numbers on the right indicate the location of molecular weight markers (kilodalton). D, WT and cKOCol2 mice were aged to 24 weeks, and micro-CT analysis was performed on the right knee joint. No changes in the subchondral bone were observed (n = 4/group).
Ezh2 inactivation does not promote ossification of cartilage in vivo
Because Ezh2 inactivation lead to enhanced expression of osteogenic factors in vitro, we screened 6-month-old WT and cKO mice for early bony changes that are seen in early osteoarthritis. Radiographic analysis of WT and cKO mice did not show evidence of abnormalities consistent with early osteoarthritic change (Fig. 2). In addition, micro-CT analysis of the knee joint of WT and cKO mice did not reveal the presence of osteophytes or irregularities within the subchondral bone that would be expected with bony osteoarthritic change (Fig. 6D). These results suggest that Ezh2 loss in Col2a1-expressing cells does not promote cartilage mineralization and formation of hypertrophic chondrocytes.
Discussion
The results of this investigation, in which Ezh2 was knocked down using the Col2a1 driver, demonstrate that Ezh2 deficiency leads to a global reduction in H3K27me3 in chondrocytes. This decrease in H3K27me3 does not appear to impact normal skeletal development or impair the protective function of mature articular cartilage compared with Ezh2 inactivation in mesenchymal progenitor cells. Notably, transcriptome analysis of in vitro IMC cultures derived from WT and cKO mice shows that chondrocytes with Ezh2 knockdown display enhanced osteogenesis. This program is characterized by increased expression of bone-related genes, including the transcription factors Sp7(Osx), extracellular matrix genes, including Ibsp and Bglap, and signaling factors such as Bmp2 and Wnt16. This study suggests that H3K27me3 mediated by Ezh2 has only minimal impact and appears to be dispensable for chondrogenic differentiation. Our study also provides evidence for Ezh2 as an important factor in regulating osteogenic differentiation of chondrocytes.
Ezh2 is critical for early embryonic development (32, 33). However, mice are able to survive to adulthood when Ezh2 is inactivated in mesenchymal progenitors using the Prrx1-Cre driver (2, 4, 30). Although viable, these mice exhibit significant skeletal developmental abnormalities, including short stature and limb segments, premature closure of the cranial sutures, and clinodactyly, findings reminiscent of Weaver syndrome in humans (34). Interestingly, loss of Ezh2 in osteoblasts results in normal-looking animals that exhibit a low bone mass phenotype (35). Our findings that loss of Ezh2 activity in the chondrocyte linage does not lead to alterations in skeletal development indicates that previous observations of skeletal abnormalities are the result of Ezh2 effects on mesenchymal progenitor cells and skeletal patterning (2, 4, 30).
While this study was in progress, Lui et al. (36) published a study of the phenotype of mice with dual inactivation of Ezh1 and Ezh2 in chondrocytes. The chondrocyte-specific loss of Ezh2 alone was not specifically reported (36), but the dual knockout mice exhibit significant growth retardation. However, the data presented by Lui et al. (36) indicates that Ezh1 and Ezh2 may potentially compensate for each other during skeletal development. The skeletal role of Eed, a member of the PRC2 complex that is required for Ezh1 or Ezh2 activity, was also recently evaluated (31). Inactivation of this scaffolding protein produced severe skeletal deformities, including kyphosis, short stature, and an increase in hypertrophic chondrocyte differentiation. These changes were similar but distinctly different from the changes observed in the dual Ezh1/Ezh2 knockout mice (36). In the context of our work, these studies indicate that Ezh1 and Ezh2 have redundant functionality within chondrocytes and may compensate, at least partially for the loss of each other.
Ezh1 and Ezh2 have been shown previously to regulate H3K27 methylation and chromatin compaction by different mechanisms (37). Specifically, the histone methyltransferase activity of Ezh1 was found to be 20-fold weaker compared with Ezh2. However, Ezh1 was more potent than Ezh2 in compacting chromatin independent of histone methylation. Despite mechanistic differences in chromatin regulation, Ezh1 was shown to regulate a subset of Ezh2 target genes, reinforcing the idea that these proteins may potentially compensate for each other. In addition to mechanism-related differences, Ezh1 and Ezh2 may be expressed at different stages of mesenchymal differentiation (2, 19). Previous studies suggest that Ezh2 is expressed in proliferating uncommitted mesenchymal stem cells, whereas Ezh1 may be more important for post-proliferative lineage-committed cells. Although these proteins are partially redundant, it appears that Ezh1 and Ezh2 differ in spatio-temporal expression and may have different mechanistic activities during mesenchymal differentiation and skeletal development.
In this study, in vivo immunohistochemical staining resulted in a modest reduction of Ezh2 protein but a more robust depletion of its enzymatic target H3K27me3. It is possible that chondrocytes that express one or both copies of the Ezh2 WT allele have a selective growth advantage. The discrepancy between modest Ezh2 loss and more robust H3K27me3 loss suggests that the residual Ezh2 protein may lack enzymatic activity. Indeed, we have shown recently that recombinase activity with this Ezh2 conditional model results in the production of the truncated mRNA and protein (35). In this study, we also provide evidence indicating that the truncated Ezh2 mRNA is detectable after recombination. Thus, although Ezh2 protein is detected by immunohistochemical staining in vivo, it is anticipated that a portion of this protein lacks enzymatic activity. Thus, the cumulative effects of Ezh2 protein loss and the presence of the enzymatically inactive Ezh2 may account for a significant loss of H3K27me3 within chondrocytes.
In our study, modification of Ezh2's enzymatic function through genetic inactivation resulted in reduced H3K27me3 in chondrocytes, but the expression of chondrogenic genes was unchanged. A previous study utilizing the Ezh2 inhibitor UNC1999 to treat chondrocytes in vitro observed a slight reduction in hypertrophic cell size and decreased expression of the hypertrophic markers Col10a1 and Ihh (36). We also observed reduced Col10a1 and Ihh (data not shown) in IMC cultures, although the impact of these changes in vivo appears to be minimal. Conversely, global knockout of the H3K27me3 demethylase Jmjd3(Kdm6b) severely impaired the proliferation and differentiation of chondrocytes, resulting in reduced endochondral ossification (27). Collectively, these data suggest that Ezh2 is dispensable for chondrocyte differentiation, including early hypertrophic differentiation during endochondral ossification.
Chondrocytes deficient in Ezh2 exhibit enhanced expression of osteogenic genes in vitro, including Bmp2, Ibsp, Sp7(Osx), and others. Our results support previous observations in which siRNA or pharmacological inactivation of Ezh2 accelerates osteogenic differentiation of human mesenchymal stromal cells and mouse preosteoblast cells (2, 3, 22, 31, 38, 39). Furthermore, a pharmacological inhibitor of Ezh2 in vivo was also found to prevent bone loss in estrogen deficiency models (3, 39, 40). Although Fang et al. (40) have proposed that loss of Ezh2 activity leads to inhibition of osteoclastogenesis, our results also support a direct bone anabolic mechanism for Ezh2 inhibition (3). Thus, Ezh2 inhibitors may be effective therapeutic agents for osteoporosis treatment via dual mechanisms: enhancement of bone anabolic activity and inhibition of bone resorption.
Even though enhanced osteogenic potential of chondrocytes was observed in vitro in this study, this biological effect did not result in early-onset osteoarthritis in Ezh2 KO mice. Consistent with these findings, Chen et al. (41) observed that EZH2 expression is increased in human osteoarthritis and up-regulated in response to inflammation (interleukin 1β). In addition, they also evaluated the Ezh2 inhibitor EPZ005687 as a therapeutic agent for treating cartilage degeneration and observed delayed osteoarthritis development in vivo. These results are encouraging and warrant further investigation of the specific role of Ezh2 in the development of osteoarthritis (41).
In summary, Ezh2 regulates H3K27me3 in chondrocytes, and its cartilage-specific inactivation does not lead to skeletal defects observed in early mesenchymal progenitors. Furthermore, Ezh2 deficiency does not modulate the expression of chondrogenic genes in vitro. However, in both the chondrogenic and osteogenic lineages, we found that Ezh2 inactivation promotes an osteogenic gene expression program. Thus, Ezh2 may normally function to suppress the bone phenotype in distinct mesenchymal lineages.
Experimental procedures
Ezh2-deficient mice
The Mayo Clinic Institutional Animal Care and Use Committee preapproved all animal experimentation, and studies were conducted according to guidelines provided by the National Institutes of Health and the Institute of Laboratory Animal Resources, National Research Council. Mice harboring two copies of the Ezh2 allele with loxP sites flanking the SET domain (Ezh2fl/fl) were obtained from the Mutant Mouse Regional Resource Centre (B6;129P2-Ezh2tm1Tara/Mmnc, University of North Carolina, Chapel Hill, NC). Homozygous animals (Ezh2fl/fl) were crossed with Col2a1-Cre (21, 42), Prrx1-Cre (2, 43), or Osx1-Cre (44) mice to inactivate Ezh2 function in, respectively, developing chondrocytes, mesenchymal progenitor cells, or osteoprogenitor cells. Animals were housed in an accredited facility under a 12-h light/dark cycle and provided water and food (PicoLab Rodent Diet 20, LabDiet) ad libitum. These crosses generated WT (WT, Ezh2wt/wt: Col2a1-Cre+; WT, Ezh2wt/wt: Prrx1-Cre+; WT, Ezh2wt/wt: Osx1-Cre+), Ezh2fl/fl: Col2a1-Cre−/−, and conditional knockout (cKO-Col2, Ezh2fl/fl: Col2a1-Cre+; cKO-Prrx1, Ezh2fl/fl: Prrx1-Cre+; cKO-Osx, Ezh2fl/fl; Osx1-Cre+) animals. The following primers were used for genotyping: Ezh2, TGTCATGTCTGGGTCTAATGCTAC (forward) and GGAACCTCGCTATGTGTAACCA (reverse); Col2a1-Cre, CCAATTTACTGACCGTACACCAA (forward) and CCTGATCCTGGCAATTTCGGCTA (reverse). Prrx1-Cre and Osx1-Cre mice were genotyped as described previously (2, 21).
Isolation and culture of immature mouse chondrocytes
Primary IMCs were isolated from post-natal day 5 mice as described previously by Gosset et al. (45). Freshly isolated IMCs were resuspended at a concentration of 2 × 107 cells/ml in growth medium (Dulbecco's modified Eagle's medium with 2% fetal bovine serum and 1% anti-mycotic/antibiotic) and plated in micromass culture by pipetting 10 μl of cell suspension onto a low-adhesion tissue culture dish with up to 3 micromasses/dish. Micromasses formed by incubation for 1 h at 37 °C and 5% CO2, after which growth medium was carefully added and left undisturbed for 3 days. Following micromass formation, the growth medium was replaced with differentiation medium (growth medium with 1× insulin–transferrin–selenium solution (Life Technologies), 0.05 mg/ml ascorbic acid, and 10 mm β-glycerol phosphate) on day 3 of culture, and the differentiation medium was replaced every 3–4 days.
Radiographic and micro-CT analysis
Radiographic analysis was performed using a Faxitron X-ray imaging cabinet (Faxitron Bioptics, Tucson, AZ). Quantitative analysis of the spine was performed using a vivaCT 40 scanner (SCANCO Medical AG) with the following parameters: E = 55 kVp, I = 145 μA, integration time = 300 ms (where E = energy, kVp = kilovoltage peak, and I = intensity). A 10.5-μm voxel size using a threshold of 220 was applied to all scans at high resolution. Two-dimensional data from scanned slices were used for 3D analysis of morphometric parameters of trabecular bone mass and microarchitecture, including BV/TV, Tb.N, Tb.Th, Tb.Sp, and the structure model index (SMI), an indicator of plate-like versus rod-like trabecular architecture.
Morphological and histology analysis
Whole-mount staining of skeletal preparations was performed as described previously (2). For histological analyses, mice were sacrificed either on post-natal day 1, 3, 4, or 8 or at 24 weeks of age, and the right hind limb was fixed in 10% neutral buffered formalin, decalcified with 15% EDTA, and embedded in paraffin. Micromass cultures were also fixed with 10% neutral buffered formalin before dehydration and paraffin embedding. Serial sections were taken at 5-μm thickness and stained with hematoxylin and eosin, Alcian blue/eosin, or Safranin O/fast green/hematoxylin.
Immunohistochemical staining
The right hind limbs or tibiae from mice on post-natal day 1, 4, or 8 or from 24-week-old mice were prepared as described above. IHC was performed on serial sections of 5-μm thickness using the Mouse- and Rabbit-Specific HRP (ABC) Detection IHC Kit (Abcam) and the DAB Enhanced Liquid Substrate System (Sigma-Aldrich) according to the instruction manuals. The primary antibodies used were Ezh2 (1:50, 07-689, Millipore), H3K27me3 (1:150, 17-622, Millipore), and rabbit isotype control (rabbit IgG, 1:50, AB-150-C, R&D Systems). Slides were counterstained with Alcian blue.
RNA isolation, reverse transcription, and real-time quantitative PCR
IMC cultures were harvested on days 3, 7, and 14 for RNA analysis. Briefly, micromasses were washed with PBS to remove medium and mechanically disrupted with a scalpel, and then 700 μl of QIAzol was added to lyse cells. Total RNA was extracted using the miRNeasy MicroKit (Qiagen), and 1 μg of RNA was used as template for reverse transcription reactions. The Superscript III First Strand Synthesis System (Life Technologies) was used to convert RNA to complementary DNA. Real-time PCR reactions were composed of 10 ng of complementary DNA, gene-specific primers (Table S3), and 1× QuantiTect SYBR® Green PCR Master Mix (Qiagen) with a final volume of 10 μl and detected using the CFX384 real time system machine (Bio-Rad). Relative transcript abundance of genes of interest was normalized to AKT1 expression using the 2−ΔΔCt method.
Western blotting
Total protein was harvested from IMC cultures on days 3, 7, and 14 of culture. Micromasses were washed with PBS, mechanically disrupted with a scalpel in the presence of radioimmunoprecipitation buffer (150 mm NaCl, 50 mm Tris (pH 7.4), 1% w/v sodium deoxycholate, 0.1% w/v SDS, and 1% v/v Triton X-100) supplemented with protease inhibitor mixture (Sigma) and phenylmethylsulfonyl fluoride (Sigma) and stored at −80 °C until quantification. Protein content was quantified using the DCTM protein assay (Bio-Rad) according to the manufacturer's protocol. Western blotting and membrane development were performed as described previously (2), and the following primary antibodies were used to detect proteins: Ezh2 (1:10,000, 5848, Cell Signaling Technology), H3K27me3 (1:5000, 17-622, Millipore), SP7 (1:1000, ab22552, Abcam), and tubulin (1:10,000, E7, Developmental Studies Hybridoma Bank (DSHB), University of Iowa).
RNA-Seq and bioinformatics analysis
Whole-transcriptome sequencing and bioinformatics analyses were performed as described previously (46). The TruSeq RNA sample Prep Kit v2 (Illumina) was used to prepare RNA samples, and then samples were analyzed using the Illumina HiSeq 2000 with the TruSeq SBS sequencing kit version 3 and HCS v2.0.12 data collection software. Following data collection, sequence data were processed using MAPRSeq (v.1.2.1) and the bioinformatics workflow (TopHat 2.0.6, HTSeq, and edgeR 2.6.2), where expression data were normalized using the reads per kilobase per million (RPKM) method.
Hierarchical clustering was performed using GENE-E (version 3.0.228, Broad Institute, Cambridge, MA), and DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov) was used to perform gene functional annotation analysis (47, 49).
Image quantitation
Images were digitally photographed using the AxioVert A1 (Zeiss) and Zen software packages (Zeiss). Growth plate depth was evaluated using National Institutes of Health ImageJ software (48).
Statistical analysis
Data obtained are presented as the mean ± S.D. Statistical analysis was performed using JMP® Pro v10.0.0 (SAS Institute), and p values were determined with the nonparametric Wilcoxon test. Significance is noted in the figures when applicable (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Author contributions
E. C., A. D., S. M. R., C. G.-G., and A. J. v. W. conceptualization; E. C., A. D., S. M. R., C. G.-G., C. R. P., E. W. B., M. E. M.-L., and H.-J. I. data curation; E. C., A. D., J. J. W., and A. N. L. formal analysis; E. C., M. K., A. J. K., J. J. W., A. N. L., and A. J. v. W. supervision; E. C., A. D., S. M. R., and A. J. v. W. funding acquisition; E. C. and A. D. investigation; E. C., A. D., S. M. R., C. G.-G., C. R. P., E. W. B., M. E. M.-L., H.-J. I., M. K., A. J. K., and J. J. W. methodology; E. C., A. D., S. M. R., C. G.-G., and A. J. v. W. writing-original draft; E. C. and A. J. v. W. project administration; E. C., A. D., S. M. R., C. G.-G., C. R. P., E. W. B., M. E. M.-L., H.-J. I., M. K., A. J. K., J. J. W., A. N. L., and A. J. v. W. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Oksana Pichurin, David Razidlo, Bridget Stensgard, and Bashar Hassan for technical support and the members of our laboratories for stimulating discussions. We also acknowledge support from the Bioinformatics Core, Medical Genome Facility, and Robert and Arlene Kogod Center on Aging at the Mayo Clinic.
This work was supported by NIAMS, National Institutes of Health Grants R01 AR049069 (to A. J. v. W.), R03 AR066342 (to A. N. L.), F32 AR066508 (to A. D.), K01 AR065397 (to E. W. B.), and R01 AR068103 (to J. J. W.) as well as by intramural grants from the Center for Regenerative Medicine at the Mayo Clinic and Mayo Graduate School (Clinical and Translational Sciences Track) and the Center for Clinical and Translational Science (UL1 TR000135). We also appreciate generous philanthropic support from William H. and Karen J. Eby and the charitable foundation in their name. A. J. K. is a paid consultant for Arthrex, Inc., and M. K. is a founder and chair holder of Hy2Care B.V. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1–S3.
The RNA-Seq data have been deposited in the publicly available Gene Expression Omnibus (GEO) repository with accession number GSE97118.
- H3K27me3
- histone H3 lysine 27 trimethylation
- IHC
- immunohistochemistry
- cKO
- conditional knockout
- BV
- bone volume
- TV
- tissue volume
- Tb.N
- trabecular number
- Tb.Th
- trabecular thickness
- Tb.Sp
- trabecular spacing
- IMC
- immature mouse chondrocyte
- RPKM
- reads per kilobase per million
- micro-CT
- micro-computed tomography.
References
- 1. Arrowsmith C. H., Bountra C., Fish P. V., Lee K., and Schapira M. (2012) Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 10.1038/nrd3674 [DOI] [PubMed] [Google Scholar]
- 2. Dudakovic A., Camilleri E. T., Xu F., Riester S. M., McGee-Lawrence M. E., Bradley E. W., Paradise C. R., Lewallen E. A., Thaler R., Deyle D. R., Larson A. N., Lewallen D. G., Dietz A. B., Stein G. S., Montecino M. A., et al. (2015) Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. J. Biol. Chem. 290, 27604–27617 10.1074/jbc.M115.672345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Camilleri E. T., Gustafson M. P., Dudakovic A., Riester S. M., Garces C. G., Paradise C. R., Takai H., Karperien M., Cool S., Sampen H. J., Larson A. N., Qu W., Smith J., Dietz A. B., and van Wijnen A. J. (2016) Identification and validation of multiple cell surface markers of clinical-grade adipose-derived mesenchymal stromal cells as novel release criteria for good manufacturing practice-compliant production. Stem Cell Res. Ther. 7, 107 10.1186/s13287-016-0370-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hemming S., Cakouros D., Codrington J., Vandyke K., Arthur A., Zannettino A., and Gronthos S. (2017) EZH2 deletion in early mesenchyme compromises postnatal bone microarchitecture and structural integrity and accelerates remodeling. FASEB J. 31, 1011–1027 10.1096/fj.201600748R [DOI] [PubMed] [Google Scholar]
- 5. Carpio L. R., Bradley E. W., McGee-Lawrence M. E., Weivoda M. M., Poston D. D., Dudakovic A., Xu M., Tchkonia T., Kirkland J. L., van Wijnen A. J., Oursler M. J., and Westendorf J. J. (2016) Histone deacetylase 3 supports endochondral bone formation by controlling cytokine signaling and matrix remodeling. Sci. Signal. 9, ra79 10.1126/scisignal.aaf3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldring M. B., and Marcu K. B. (2012) Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol. Med. 18, 109–118 10.1016/j.molmed.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barter M. J., Bui C., and Young D. A. (2012) Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthritis Cartilage 20, 339–349 10.1016/j.joca.2011.12.012 [DOI] [PubMed] [Google Scholar]
- 8. Bradley E. W., Carpio L. R., van Wijnen A. J., McGee-Lawrence M. E., and Westendorf J. J. (2015) Histone deacetylases in bone development and skeletal disorders. Physiol. Rev. 95, 1359–1381 10.1152/physrev.00004.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gordon J. A. R., Stein J. L., Westendorf J. J., and van Wijnen A. J. (2015) Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease. Bone 81, 739–745 10.1016/j.bone.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schwarz D., Varum S., Zemke M., Schöler A., Baggiolini A., Draganova K., Koseki H., Schübeler D., and Sommer L. (2014) Ezh2 is required for neural crest-derived cartilage and bone formation. Development 141, 867–877 10.1242/dev.094342 [DOI] [PubMed] [Google Scholar]
- 11. Pike J. W., Meyer M. B., St John H. C., and Benkusky N. A. (2015) Epigenetic histone modifications and master regulators as determinants of context dependent nuclear receptor activity in bone cells. Bone 81, 757–764 10.1016/j.bone.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meyer M. B., Benkusky N. A., Sen B., Rubin J., and Pike J. W. (2016) Epigenetic plasticity drives adipogenic and osteogenic differentiation of marrow-derived mesenchymal stem cells. J. Biol. Chem. 291, 17829–17847 10.1074/jbc.M116.736538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu H., Gordon J. A., Whitfield T. W., Tai P. W., van Wijnen A. J., Stein J. L., Stein G. S., and Lian J. B. (2017) Chromatin dynamics regulate mesenchymal stem cell lineage specification and differentiation to osteogenesis. Biochim. Biophys. Acta 1860, 438–449 10.1016/j.bbagrm.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riester S. M., Denbeigh J. M., Lin Y., Jones D. L., de Mooij T., Lewallen E. A., Nie H., Paradise C. R., Radel D. J., Dudakovic A., Camilleri E. T., Larson D. R., Qu W., Krych A. J., Frick M. A., et al. (2017) Safety studies for use of adipose tissue-derived mesenchymal stromal/stem cells in a rabbit model for osteoarthritis to support a phase I clinical trial. Stem Cells Transl. Med. 6, 910–922 10.5966/sctm.2016-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Caplan A. I. (2016) MSCs: The sentinel and safe-guards of injury. J. Cell. Physiol. 231, 1413–1416 10.1002/jcp.25255 [DOI] [PubMed] [Google Scholar]
- 16. Deng Z. L., Sharff K. A., Tang N., Song W. X., Luo J., Luo X., Chen J., Bennett E., Reid R., Manning D., Xue A., Montag A. G., Luu H. H., Haydon R. C., and He T. C. (2008) Regulation of osteogenic differentiation during skeletal development. Front. Biosci. 13, 2001–2021 10.2741/2819 [DOI] [PubMed] [Google Scholar]
- 17. Dudakovic A., Camilleri E. T., Lewallen E. A., McGee-Lawrence M. E., Riester S. M., Kakar S., Montecino M., Stein G. S., Ryoo H. M., Dietz A. B., Westendorf J. J., and van Wijnen A. J. (2015) Histone deacetylase inhibition destabilizes the multi-potent state of uncommitted adipose-derived mesenchymal stromal cells. J. Cell. Physiol. 230, 52–62 10.1002/jcp.24680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dudakovic A., Evans J. M., Li Y., Middha S., McGee-Lawrence M. E., van Wijnen A. J., and Westendorf J. J. (2013) Histone deacetylase inhibition promotes osteoblast maturation by altering the histone H4 epigenome and reduces Akt phosphorylation. J. Biol. Chem. 288, 28783–28791 10.1074/jbc.M113.489732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dudakovic A., Gluscevic M., Paradise C. R., Dudakovic H., Khani F., Thaler R., Ahmed F. S., Li X., Dietz A. B., Stein G. S., Montecino M. A., Deyle D. R., Westendorf J. J., and van Wijnen A. J. (2017) Profiling of human epigenetic regulators using a semi-automated real-time qPCR platform validated by next generation sequencing. Gene 609, 28–37 10.1016/j.gene.2017.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khani F., Thaler R., Paradise C. R., Deyle D. R., Kruijthof-de Julio M., Galindo M., Gordon J. A., Stein G. S., Dudakovic A., and van Wijnen A. J. (2017) Histone H4 methyltransferase Suv420h2 maintains fidelity of osteoblast differentiation. J. Cell. Biochem. 118, 1262–1272 10.1002/jcb.25787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bradley E. W., Carpio L. R., Olson E. N., and Westendorf J. J. (2015) Histone deacetylase 7 (Hdac7) suppresses chondrocyte proliferation and β-catenin activity during endochondral ossification. J. Biol. Chem. 290, 118–126 10.1074/jbc.M114.596247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hemming S., Cakouros D., Isenmann S., Cooper L., Menicanin D., Zannettino A., and Gronthos S. (2014) EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 32, 802–815 10.1002/stem.1573 [DOI] [PubMed] [Google Scholar]
- 23. Spaapen F., van den Akker G. G., Caron M. M., Prickaerts P., Rofel C., Dahlmans V. E., Surtel D. A., Paulis Y., Schweizer F., Welting T. J., Eijssen L. M., and Voncken J. W. (2013) The immediate early gene product EGR1 and polycomb group proteins interact in epigenetic programming during chondrogenesis. PLoS ONE 8, e58083 10.1371/journal.pone.0058083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang H., Meng Y., Cui Q., Qin F., Yang H., Chen Y., Cheng Y., Shi J., and Guo Y. (2016) MiR-101 targets the EZH2/Wnt/β-catenin pathway to promote the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Sci. Rep. 6, 36988 10.1038/srep36988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bradley E. W., Carpio L. R., and Westendorf J. J. (2013) Histone deacetylase 3 suppression increases PH domain and leucine-rich repeat phosphatase (Phlpp)1 expression in chondrocytes to suppress Akt signaling and matrix secretion. J. Biol. Chem. 288, 9572–9582 10.1074/jbc.M112.423723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakatani T., Chen T., and Partridge N. C. (2016) MMP-13 is one of the critical mediators of the effect of HDAC4 deletion on the skeleton. Bone 90, 142–151 10.1016/j.bone.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang F., Xu L., Xu L., Xu Q., Li D., Yang Y., Karsenty G., and Chen C. D. (2015) JMJD3 promotes chondrocyte proliferation and hypertrophy during endochondral bone formation in mice. J. Mol. Cell. Biol. 7, 23–34 10.1093/jmcb/mjv003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ideno H., Shimada A., Imaizumi K., Kimura H., Abe M., Nakashima K., and Nifuji A. (2013) Predominant expression of H3K9 methyltransferases in prehypertrophic and hypertrophic chondrocytes during mouse growth plate cartilage development. Gene Expr. Patterns 13, 84–90 10.1016/j.gep.2013.01.002 [DOI] [PubMed] [Google Scholar]
- 29. Lawson K. A., Teteak C. J., Zou J., Hacquebord J., Ghatan A., Zielinska-Kwiatkowska A., Fernandes R. J., Chansky H. A., and Yang L. (2013) Mesenchyme-specific knockout of ESET histone methyltransferase causes ectopic hypertrophy and terminal differentiation of articular chondrocytes. J. Biol. Chem. 288, 32119–32125 10.1074/jbc.M113.473827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wyngaarden L. A., Delgado-Olguin P., Su I. H., Bruneau B. G., and Hopyan S. (2011) Ezh2 regulates anteroposterior axis specification and proximodistal axis elongation in the developing limb. Development 138, 3759–3767 10.1242/dev.063180 [DOI] [PubMed] [Google Scholar]
- 31. Mirzamohammadi F., Papaioannou G., Inloes J. B., Rankin E. B., Xie H., Schipani E., Orkin S. H., and Kobayashi T. (2016) Polycomb repressive complex 2 regulates skeletal growth by suppressing Wnt and TGF-β signalling. Nat. Commun. 7, 12047 10.1038/ncomms12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O'Connor M. B., Kingston R. E., and Simon J. A. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 10.1016/S0092-8674(02)00976-5 [DOI] [PubMed] [Google Scholar]
- 33. O'Carroll D., Erhardt S., Pagani M., Barton S. C., Surani M. A., and Jenuwein T. (2001) The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21, 4330–4336 10.1128/MCB.21.13.4330-4336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen A. S., Yap D. B., Lewis M. E., Chijiwa C., Ramos-Arroyo M. A., Tkachenko N., Milano V., Fradin M., McKinnon M. L., Townsend K. N., Xu J., Van Allen M. I., Ross C. J., Dobyns W. B., Weaver D. D., and Gibson W. T. (2016) Weaver syndrome-associated EZH2 protein variants show impaired histone methyltransferase function in vitro. Hum. Mutat. 37, 301–307 10.1002/humu.22946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dudakovic A., Camilleri E. T., Paradise C. R., Samsonraj R. M., Gluscevic M., Paggi C. A., Begun D. L., Khani F., Pichurin O., Ahmed F. S., Elsayed R., Elsalanty M., McGee-Lawrence M. E., Karperien M., Riester S. M., et al. (2018) Enhancer of zeste homolog 2 (Ezh2) controls bone formation and cell cycle progression during osteogenesis in mice. J. Biol. Chem. 293, 12894–12907 10.1074/jbc.RA118.002983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lui J. C., Garrison P., Nguyen Q., Ad M., Keembiyehetty C., Chen W., Jee Y. H., Landman E., Nilsson O., Barnes K. M., and Baron J. (2016) EZH1 and EZH2 promote skeletal growth by repressing inhibitors of chondrocyte proliferation and hypertrophy. Nat. Commun. 7, 13685 10.1038/ncomms13685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C. L., Dynlacht B. D., and Reinberg D. (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hemming S., Cakouros D., Vandyke K., Davis M. J., Zannettino A. C., and Gronthos S. (2016) Identification of novel EZH2 targets regulating osteogenic differentiation in mesenchymal stem cells. Stem Cells Dev. 25, 909–921 10.1089/scd.2015.0384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jing H., Liao L., An Y., Su X., Liu S., Shuai Y., Zhang X., and Jin Y. (2016) Suppression of EZH2 prevents the shift of osteoporotic MSC fate to adipocyte and enhances bone formation during osteoporosis. Mol. Ther. 24, 217–229 10.1038/mt.2015.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fang C., Qiao Y., Mun S. H., Lee M. J., Murata K., Bae S., Zhao B., Park-Min K. H., and Ivashkiv L. B. (2016) Cutting edge: EZH2 promotes osteoclastogenesis by epigenetic silencing of the negative regulator IRF8. J. Immunol. 196, 4452–4456 10.4049/jimmunol.1501466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen L., Wu Y., Wu Y., Wang Y., Sun L., and Li F. (2016) The inhibition of EZH2 ameliorates osteoarthritis development through the Wnt/β-catenin pathway. Sci. Rep. 6, 29176 10.1038/srep29176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ovchinnikov D. A., Deng J. M., Ogunrinu G., and Behringer R. R. (2000) Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 26, 145–146 10.1002/(SICI)1526-968X(200002)26:2%3C145::AID-GENE14%3E3.0.CO%3B2-C [DOI] [PubMed] [Google Scholar]
- 43. Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N., and Tabin C. J. (2002) Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77–80 10.1002/gene.10092 [DOI] [PubMed] [Google Scholar]
- 44. Rodda S. J., and McMahon A. P. (2006) Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231–3244 10.1242/dev.02480 [DOI] [PubMed] [Google Scholar]
- 45. Gosset M., Berenbaum F., Thirion S., and Jacques C. (2008) Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 3, 1253–1260 10.1038/nprot.2008.95 [DOI] [PubMed] [Google Scholar]
- 46. Dudakovic A., Camilleri E., Riester S. M., Lewallen E. A., Kvasha S., Chen X., Radel D. J., Anderson J. M., Nair A. A., Evans J. M., Krych A. J., Smith J., Deyle D. R., Stein J. L., Stein G. S., et al. (2014) High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J. Cell. Biochem. 115, 1816–1828 10.1002/jcb.24852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 48. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to Image J: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.