Abstract
The inflammasome is a critical molecular complex that activates interleukin-1 driven inflammation in response to pathogen- and danger-associated signals. Germline mutations in the inflammasome sensor NLRP1 cause Mendelian systemic autoimmunity and skin cancer susceptibility, but its endogenous regulation remains less understood. Here we use a proteomics screen to uncover dipeptidyl dipeptidase DPP9 as a novel interacting partner with human NLRP1 and a related inflammasome regulator, CARD8. DPP9 functions as an endogenous inhibitor of NLRP1 inflammasome in diverse primary cell types from human and mice. DPP8/9 inhibition via small molecule drugs and CRISPR/Cas9-mediated genetic deletion specifically activate the human NLRP1 inflammasome, leading to ASC speck formation, pyroptotic cell death, and secretion of cleaved interleukin-1β. Mechanistically, DPP9 interacts with a unique autoproteolytic domain (Function to Find Domain (FIIND)) found in NLRP1 and CARD8. This scaffolding function of DPP9 and its catalytic activity act synergistically to maintain NLRP1 in its inactive state and repress downstream inflammasome activation. We further identified a single patient-derived germline missense mutation in the NLRP1 FIIND domain that abrogates DPP9 binding, leading to inflammasome hyperactivation seen in the Mendelian autoinflammatory disease Autoinflammation with Arthritis and Dyskeratosis. These results unite recent findings on the regulation of murine Nlrp1b by Dpp8/9 and uncover a new regulatory mechanism for the NLRP1 inflammasome in primary human cells. Our results further suggest that DPP9 could be a multifunctional inflammasome regulator involved in human autoinflammatory diseases.
Keywords: inflammasome, inflammation, Nod-like receptor (NLR), cellular immune response, cell death, CARD8, DPP9, NLRP1
Introduction
The innate immune system employs a large array of pattern-recognition receptors to detect pathogen- or danger-associated molecules (1). A subset of immune sensor proteins belong to the NLR (Nod-like receptor) protein family and function as sensors and activators for the inflammasome complex, a conserved macromolecular platform that governs inflammation driven by the interleukin-1 family of cytokines. The canonical inflammasome complex minimally consists of an NLR sensor, the adaptor protein ASC (also known as PYCARD), and the effector inflammatory caspase, caspase-1 (2–4). Upon ligand binding by NLR sensors, the inflammasome complex initiates a distinct form of inflammatory cell death known as “pyroptosis,” which in concert with other innate immune pathways plays an important role in immune defense (5, 6).
In addition to recognizing infection- or danger-related molecules in a discriminatory manner, NLR proteins must avoid aberrant activation and tissue-damaging “sterile inflammation.” Recent work has revealed that human cells employ a network of post-transcriptional and post-translational regulatory “checkpoints” to guard against aberrant inflammasome activation, such as post-translational modification of the NLR proteins and obligate regulatory factors (7–13). The in vivo importance of the negative regulation of NLRs is illustrated by a number of Mendelian diseases caused by gain-of-function mutations in NLR sensors or loss-of-function mutations in their endogenous inhibitors. This group of diseases, or “inflammaosopathies” are characterized by periodic fever and sterile inflammatory response (14, 15) caused by aberrant inflammasome activation in multiple organs. In addition, dysregulation of NLR-driven inflammasome response has also been implicated in common, non-Mendelian diseases such as cancer, autoimmune, and neurodegenerative diseases (4, 16). Hence, there is an important need to fully understand how various NLR proteins are kept in the inactive state in the absence of pathogen- or danger-derived ligands (17).
We and others have recently characterized a unique member of the NLR family, NLRP1. Patients who have germline mutations in NLRP1 all experience early-onset epithelial hyperkeratosis/dyskeratosis, particularly on palmoplantar skin, whereas classical signs of periodic fever that define other inflammasome diseases are variable (17, 18). On the molecular level, human NLRP1 harbors an atypical pyrin domain (PYD)3 that is required for NLRP1 autoinhibition and is not present in rodent homologs (17, 19). In human cells, NLRP1 assembles the inflammasome adaptor protein ASC via its CARD in a noncanonical pathway that requires autoproteolysis within a domain of unknown function termed FIIND (17, 19, 20). Recent work has identified specific pathogen-derived proteases, such as the anthrax lethal toxin, that activate rodent Nlrp1b (21–23). However, no specific agonists or dedicated regulatory co-factors have been reported for human NLRP1. While this manuscript was in preparation, it was reported that chemical inhibitors of dipeptidyl peptidases, Dpp8 and 9 activate murine Nlrp1b inflammasome (24). These inhibitors have also previously been shown to cause Gasdermin D (GSDMD)- and caspase-1–dependent pyroptosis in human macrophage-like cells, albeit in an unusual mechanism that occurs independently of the inflammasome sensor protein ASC and without IL-1β cleavage (25, 26). Hence the exact mechanisms by which DPP8/9 regulate NLRP1, especially in human cells, remain to be clarified.
Here we report that DPP9 is an interacting partner of human NLRP1 and a related, human-specific inflammasome regulator, CARD8. Inhibition of DPP8/9 via chemical inhibitors and genetic deletion act as potent triggers for NLRP1-dependent inflammatory death, which proceeds via NLRP1 oligomerization, ASC speck assembly, and IL-1β cleavage in a range of human primary cell types. Mechanistically, the suppression of NLRP1 by DPP9 requires both its catalytic activity and its binding to NLRP1. We discovered that FIIND, an autoproteolytic domain shared between NLRP1 and CARD8 whose function was hitherto unknown, is a necessary and sufficient DPP9-binding domain. Disruption of NLRP1–DPP9 interaction by a patient-derived point mutation in the NLRP1 FIIND domain leads to spontaneous NLRP1 inflammasome activation without impacting NLRP1 autoproteolysis. This likely explains the persistent sterile inflammation seen in in the autoinflammatory/autoimmune syndrome autoinflammation with arthritis and dyskeratosis (AIADK; OMIM no. 617388) (18). In combination with recently published results on murine Dpp8/9 and Nlrp1b, our findings highlight an unprecedented, conserved peptidase-based regulatory checkpoint for an inflammasome sensor and suggest that DPP9 is a multifunctional inflammasome regulator that guards against human autoinflammatory diseases.
Results
Identification of DPP9 as a novel binding partner of full-length, autoinhibited human NLRP1
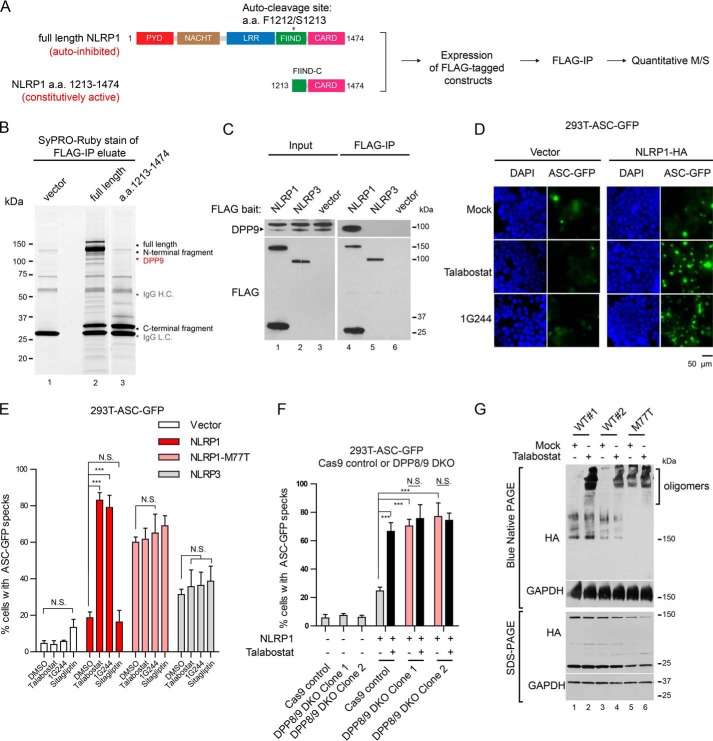
To search for novel proteins involved in NLRP1 regulation, we took advantage of the observation that full-length NLRP1 is minimally active when expressed in 293T cells, whereas the NLRP1 autoproteolytic fragment (a.a. 1213–1474) is constitutively active (17, 19). We thus hypothesized that 293T cells express unknown inhibitory factors that interact with the regulatory domains of NLRP1 (PYD, NACHT, LRR, and FIIND) to maintain NLRP1 inhibition. To identify such factors, we used immunoprecipitation (IP) followed by MS to compare the interacting partners of FLAG-tagged full-length NLRP1 and those of the constitutively active fragment (a.a. 1213–1474) (Fig. 1A). Direct protein staining of the IP eluates following SDS-PAGE revealed a prominent band at ∼100 kDa that co-purified only with full-length NLRP1 (Fig. 1B). Quantitative MS identified this candidate protein as the long isoform of dipeptidyl peptidase, DPP9 (Uniprot accession no. Q86TI2-2; Excel File S1). It was among the most enriched proteins that associated with full-length NLRP1 (Fig. S1A), but not with vector transfected cells, cells expressing NLRP1 a.a. 1213–1474, or an additional negative control, a.a. 1213–1373 (fold change > 16; Fig. S1, B and C, and Excel File S1). Human DPP9 is a dipeptidyl dipeptidase of the DPP-IV family with broad functions in immune regulation, growth factor signaling, adipocyte differentiation, and cellular metabolism (27–29). It shares a similar domain structure with other family members consisting of an N-terminal β-barrel (DPP-IV N) and a C-terminal S9 hydrolase domain (Fig. S1D). Of all the DPP-IV family members, only DPP9 was detected as a specific interacting protein with NLRP1 (Excel File S1). We further established that both DPP9 splice isoforms, DPP9L and DPP9S co-immunoprecipitate with full-length NLRP1 (Fig. S1E, lanes 6 and 7), but not DPP9L lacking the hydrolase domain (DPP9 Δhydrolase) (Fig. S1E, lane 6 versus lane 8). In addition, a related inflammasome sensor NLRP3 did not interact with endogenous DPP9 (Fig. 1C, lane 4 versus lane 5). These results suggest that DPP9 is a specific NLRP1-interacting partner and a candidate regulatory factor that helps maintain full-length NLRP1 inhibition.
Figure 1.
Identification of dipeptidyl peptidase DPP9 as a specific interacting partner of human NLRP1 and inhibitor of the NLRP1 inflammasome. A, proteomics-based strategy to identify NLRP1-interacting proteins. 293T cells were transfected with full-length human NLRP1 and a.a. 1213–1474 constructs, expanded, and harvested 4 days post-transfection. Approximately 108 cells were used per immunoprecipitation. B, SyPRO-Ruby staining of 10% of FLAG immunoprecipitation eluates. C, 500,000 293T cells were transfected with FLAG-tagged NLRP1, NLRP3 or an empty vector. The cells were harvested 2 days after transfection, and 0.5 mg of whole cell lysate was used for FLAG IP. 20 μg of input was loaded with 50% of IP eluate for SDS-PAGE. D, 293T-ASC-GFP cells were transfected with empty vector or NLRP1-HA plasmid and treated with 3 μm talabostat or 10 μm 1G244 for 24 h. The cells were fixed with 4% formaldehyde and stained with DAPI. GFP fluorescence was visualized at 20× magnification on a wide-field fluorescent microscope. Scale bar, 50 μm. E, 293T-ASC-GFP cells were transfected with empty vector, WT NLRP1, NLRP1-M77T, or NLRP3 plasmids as in D. The cells were treated with 3 μm talabostat,10 μm 1G244, or 50 μm sitagliptin for 24 h. The percentage of cells with ASC-GFP specks were counted in three differential regions on mounted slides. Statistical significance was calculated with one-way ANOVA with a post hoc test to compare all treatment groups. N.S., not significant. Bar graphs represent data from one of two independent experiments. F, 293T-ASC-GFP control or DPP8/9 DKO cells were transfected with WT NLRP1 plasmid or an empty vector for 16 h and treated with 3 μm talabostat or DMSO for 24 h. The percentage of cells with ASC-GFP specks were counted in three different fields on the mounted slides after fixation. Statistical significance was calculated with one-way ANOVA with Bonferroni's multiple comparison test to compare all treatment groups. N.S., not significant. Bar graphs represent data from three independent experiments. G, 293T cells (without ASC-GFP) were transfected with WT NLRP1 or NLRP1-M77T expressing plasmids at a ratio of 2 μg/5 × 105 cells and treated with DMSO or talabostat (3 μm) for 2 days. Blue-Native PAGE was carried out using 20 μg of total cell lysate in 1% digitonin. Molecular mass of GAPDH tetramer is ∼150 kDa. NLRP1 oligomers appear at ∼1 MDa.
DPP9 inhibition triggers human NLRP1 oligomerization, ASC speck formation, and inflammasome activation
The chemical biology of DPP-IV family of peptidases has been extensively investigated (30), and a number of small molecule inhibitors for DPP8/9 have been developed (31). Pan-DPP-IV inhibitor talabostat (Val-boro-Pro) and DPP8/9-selective inhibitor 1G244 have recently been suggested to activate an atypical form of pyroptotic cell death that does not involve ASC or IL-1β cleavage in human macrophage-like cells, although the mechanism remains unclear (25, 26). While this manuscript was in preparation, a number of reports demonstrated that Dpp8/9 inhibitors activates murine Nlrp1b inflammasome (24, 47) and a noncanonical CARD8 inflammasome (32). Whether DPP9 regulates human NLRP1 remains to be determined, especially considering the divergent domain structure of human and rodent NLRP1 homologs. To test this, we reconstituted NLRP1 in a 293T reporter cell line that stably expressed GFP-tagged inflammasome adaptor, ASC (293T-ASC-GFP). When NLRP1-expressing 293T-ASC-GFP cells were treated with talabostat (3 μm) or DPP8/9 inhibitor 1G244 (10 μm), more than 70% of the cells formed ASC-GFP specks that represented activated, assembled inflammasome complexes (Fig. 1, D and E). This effect was not observed in the absence of NLRP1 (Fig. 1E, white bars) or in ASC-GFP cells reconstituted with NLRP3 (Fig. 1E, gray bars). We further confirmed that talabostat induced ASC polymerization using a DSS cross-linking assay (Fig. S1F, lane 5 versus lane 4), whereas a specific DPP4 inhibitor, sitagliptin, failed to nucleate ASC-GFP specks in NLRP1 inflammasome reconstituted cells (Fig. 1E) or induce ASC-GFP polymerization (Fig. S1F, lane 6 versus lane 3). Notably, talabostat or 1G244 did not enhance constitutive inflammasome activation by a known gain-of-function NLRP1 PYD mutation (p.M77T) found in patients with multiple self-healing palmoplantar carcinoma (MSPC) (Fig. 1E, pink bars, and Fig. S1F, lanes 7–9). Taken together, these results demonstrate that chemical inhibitors of DPP8/9 specifically activate the reconstituted human NLRP1 inflammasome and induce ASC speck formation, likely independently of the NLRP1 PYD. These results also suggest that the reported, pro-pyroptotic effect of talabostat in human cells likely occurs upstream of NLRP1 rather than downstream of ASC polymerization (25).
To rule out potential off-target effects of the chemical inhibitors, we generated DPP8 and DPP9 double-knockout (DPP8/9 DKO) 293T-ASC-GFP cells (Fig. S1G) because of their known functional redundancy (25). In contrast to control 293T-ASC-GFP cells, NLRP1 expression in DPP8/9 DKO cells led to constitutive ASC speck formation, which could not be further enhanced by talabostat treatment (Fig. 1F, pink bars). These results confirm that the effects of DPP8/9 inhibitors such as talabostat are on-target because they can be phenocopied by the genetic ablation of DPP9 and its homolog DPP8.
To determine the mechanism by which DPP9 inhibition activates NLRP1, we examined NLRP1 monomer-to-oligomer transition. Previously we established that all patient germline gain-of-function mutations in NLRP1 cause constitutive NLRP1 oligomer formation (17). When NLRP1-expressing 293T cells, which do not express endogenous ASC, were treated with talabostat, a substantial amount of NLRP1 shifted to ∼1 MDa high-molecular-mass species as detected by Blue-Native PAGE, whereas the total level of NLRP1 remained unchanged (Fig. 1G, lane 2 versus lane 1 and lane 4 versus lane 3). Because this occurred in the absence of ASC, these results suggest that endogenous DPP9 directly inhibits NLRP1 oligomerization.
NLRP1 autocleavage within the FIIND domain generates an active C-terminal fragment, a.a. 1213–1474 (Fig. 1A). We noted that this creates a potential DPP9 processing site with a proline residue at the P2 position. Hence it is possible that autocleaved NLRP1 fragment becomes a direct substrate of DPP9. In the MS analysis of full-length NLRP1 immunopurified from 293T cells, we readily detected tryptic peptides spanning and beginning at the autocleavage junction. However, we did not observe peptides that were consistent with DPP9 processing (i.e. starting at a.a. L1215) (Fig. S1H). Hence NLRP1 might not be a direct enzymatic substrate of DPP9, although we cannot rule out a noncanonical cleavage site at present. We also confirmed that talabostat did not directly engage NLRP1 itself using a cellular thermoshift assay (Fig. S1I).
DPP9 inhibition leads to NLRP1-dependent ASC oligomerization, mature IL-1β secretion, and lytic cell death in primary human cells
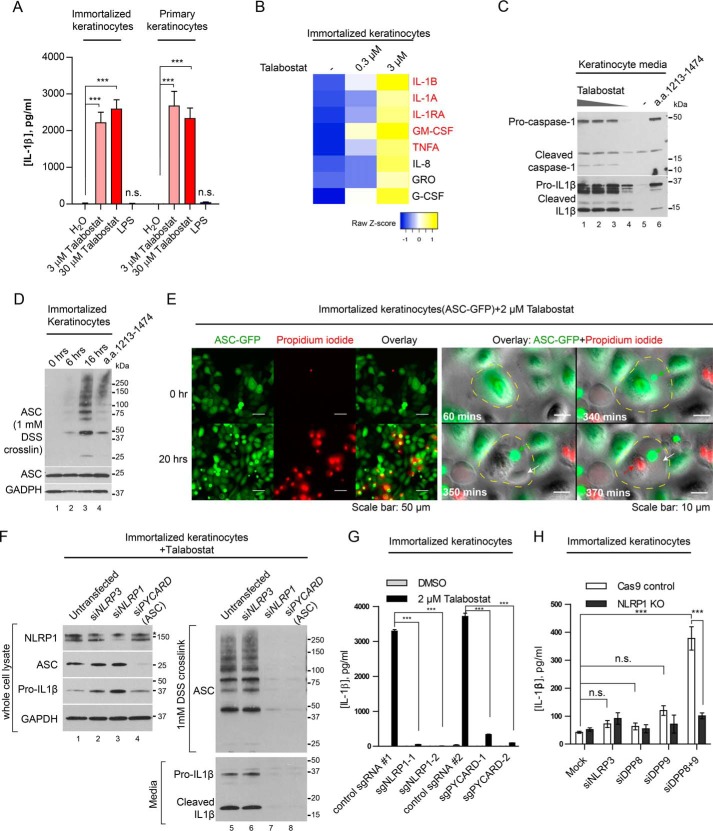
We next investigated the effect of DPP9 on NLRP1 in two primary human cell types that are of direct relevance to NLRP1-associated autoinflammatory diseases: skin keratinocytes and peripheral blood mononuclear cells (PBMCs). We and others have recently established that NLRP1 rather than NLRP3 is the most prominent if not the only inflammasome sensor expressed in resting human primary or immortalized, nontransformed keratinocytes (33) (17). This provides a unique system to directly and specifically investigate the regulation of NLRP1 without impinging upon other NLR sensors such as NLRP3. Both immortalized and primary keratinocytes secreted mature IL-1β into the culture medium across a range of talabostat concentrations (Fig. 2A and Fig. S2A). Talabostat also elicited dose-dependent lytic cell death in immortalized keratinocytes as measured by LDH release (Fig. S2C). Notably, the secretion of IL-1β was more pronounced than the increase in LDH release at lower talabostat doses (0.2–1 μm; Fig. S2, A and B). This could be result of the different sensitivity of the two assays or the fact that IL-1β secretion can precede cell lysis as shown for other cell types (34). When Luminex was used to characterize a larger panel of chemokine/cytokines caused by DPP9 inhibition, we found that IL-1β and IL-1α are the most significantly enriched of all chemokines/cytokines tested (Fig. 2B). Importantly, the chemokine/cytokine profile caused by talabostat treatment closely mimics that of primary keratinocytes derived from MSPC and FKLC patients with germline gain-of-function mutations in NLRP1 (Fig. 2B, red, and Fig. S2C) (17), demonstrating that DPP8/9 inhibition by talabostat has a strikingly similar effect as constitutive NLRP1 activation. These data support the notion that DPP8/9 inhibition is a highly specific trigger for an IL-1β–driven inflammatory response in human keratinocytes.
Figure 2.
Human keratinocytes undergo NLRP1-dependent ASC speck formation, lytic cell death and mature IL-1β secretion upon DPP8/9 inhibition or depletion. A, immortalized human keratinocytes (N/TERT-1) or primary foreskin keratinocytes were treated with 3 μm talabostat or 1 μg/ml LPS for 24 h. Culture media were harvested, clarified by centrifugation, and analyzed by IL-1β ELISA. Statistical significance was calculated using one-way ANOVA with Bonferroni's multiple comparison test between all treatment groups. Bar graph represents biological triplicates. B, immortalized human keratinocytes were treated with talabostat for 16 h. Cell culture media were collected and analyzed by Luminex assay. Heat map represents all cytokines or chemokines that showed >2-fold increase in the 3 μm talabostat treated group versus control-treated cells. Cytokines/chemokines previously shown to be up-regulated in MSPC patient-derived keratinocytes are shown in red (17). C, immortalized human keratinocytes (N/TERT-1) were treated 30, 3, and 0.3 μm of talabostat or DMSO for 24 h. In parallel, cells were transfected with an empty vector of NLRP1 a.a. 1213–1474, and media were harvested 24 h post-transfection. The cell culture media were concentrated 10 times using Amicon columns with a molecular mass cutoff of 3 kDa before SDS-PAGE. D, immortalized human keratinocytes were treated with 3 μm talabostat at the indicated times. In parallel, the cells were transfected with NLRP1 a.a. 1213–1474 and harvested 24 h later. Cell pellets were lysed in radioimmune precipitation assay buffer and insoluble materials were cross-linked with 1 mm DSS in PBS to visualize ASC polymers by SDS-PAGE. E, immortalized human keratinocytes (N/TERT-1) stably expressing ASC-GFP were treated with 2 μm talabostat or DMSO in the presence of propidium iodide. The cells were imaged using a wide-field fluorescent microscope equipped with a temperature- and CO2-controlled chamber at 20× magnification. Images were acquired every 10 min for 20 h. Left panel, cells before and ∼20 h after talabostat addition. Right panel, a close-up, time-course view of a cell undergoing pyroptotic cell death following talabostat treatment. Yellow dotted line delineates cell periphery. Green arrow, ASC speck; white arrow, membrane rupture; red arrow, PI influx. F, immortalized human keratinocytes (N/TERT-1) were treated with siRNAs against NLRP3, NLRP1, and ASC/PYCARD for 48 h before 3 μm talabostat treatment for 24 h. The cell pellets were lysed in radioimmune precipitation assay buffer, and the insoluble materials were cross-linked with 1 mm DSS in PBS. Cell culture media were concentrated 10 times with Amicon columns with a molecular mass cutoff of 3 kDa before SDS-PAGE. The asterisk indicates a nonspecific band with the NLRP1 antibody. G, immortalized keratinocytes stably expressing Cas9 or Cas9 with two independent guide RNAs against NLRP1 and ASC were treated with 3 μm talabostat for 24 h. IL-1β in the culture media was analyzed by ELISA. Statistical significance was calculated using one-way ANOVA with Bonferroni's multiple comparison test between all treatment groups. Bar graph represents four independent experiments. H, control or NLRP1 KO keratinocytes were treated with siRNAs against NLRP3, DPP8, DPP9, and DPP8 and DPP9 together for 72 h. IL-1β in the culture media was analyzed by ELISA. Statistical significance was calculated using one-way ANOVA with Bonferroni's multiple comparison test between all treatment groups. Bar graph represents two biologically independent experiments.
Previously it was reported that the pro-pyroptotic effects of DPP8/9 inhibitors in human cell lines did not involve ASC or IL-1β cleavage (25, 26). These observations are somewhat inconsistent with the involvement of human NLRP1, which requires ASC to activate the inflammasome response (17). To clarify the mechanism of DPP9 inhibition in human keratinocytes, we examined in greater detail the molecular hallmarks of “canonical” inflammasome activation, i.e. lytic cell death, IL-1β cleavage, and ASC polymerization. In immortalized keratinocytes, talabostat caused dose-dependent cleavage of pro-caspase-1 and pro–IL-1β into their respective mature forms (Fig. 2C, lanes 1–4 versus lane 5) and the polymerization of endogenous ASC in a time-dependent manner (Fig. 2D, lanes 2–4 versus lane 1). We next carried out live cell imaging to track the dynamics of two key events in pyroptosis, i.e. ASC speck formation and lytic cell death using keratinocytes expressing ASC-GFP. Over a course of ∼20 h, ∼30% of talabostat (2 μm) treated keratinocytes formed ASC specks and became permeable to propidium iodide (Fig. 2E, left panel, 20 h; Fig. S2D; and Videos S1 and S2), with some pyroptotic cell deaths apparent with the first 3 h (Video S2). When examined at the single cell level, the formation of ASC specks nearly always preceded the “ballooning” of the plasma membrane and the increase in PI fluorescence (Fig. 2E, right panel; Fig. S2D; and Video S3). Together, these results establish that DPP9 inhibition in human cells causes a conventional inflammasome response involving ASC speck formation, lytic cell death, and IL-1β cleavage and secretion.
To further prove that DPP8/9 inhibitors cause pyroptosis specifically upstream of NLRP1, we treated immortalized keratinocytes with NLRP1 and PYCARD (encoding ASC) siRNAs for 3 days before DPP9 inhibition for 24 h (Fig. 2F). siRNAs against NLRP3, which is not expressed in keratinocytes (RNAseq FKPM < 1) (17), were included as a negative control. In contrast to untransfected and siNLRP3-treated keratinocytes, NLRP1-depleted keratinocytes failed to secrete mature IL-1β into the culture media (Fig. 2F, lane 7 versus lanes 5 and 6, and Fig. S2, E and G) or undergo ASC polymerization (Fig. 2F, lane 7 versus lanes 5 and 6). NLRP1 depletion also significantly reduced cell death (Fig. S2F) after talabostat treatment. Similar results were obtained using 1G244, a more specific but less potent DPP8/9 inhibitor (Fig. S2E). To avoid nonspecific effects of siRNAs, we used CRISPR/Cas9 to knock out NLRP1 and PYCARD (encoding ASC) in immortalized keratinocytes. As shown with two guide RNAs, NLRP1 and ASC deletion abrogated IL-1β secretion following talabostat treatment (Fig. 2G). Despite repeated attempts, we failed in generating viable DPP8/9 knockout keratinocytes. This is consistent with their roles in inhibiting pyroptotic cell death, although we cannot rule out other essential functions of DPP8/9 in keratinocytes at the moment. To demonstrate that DPP8/9 are genetically required to inhibit NLRP1, we treated both control and NLRP1 KO keratinocytes with siRNAs against DPP8 and DPP9. Simultaneous depletion of both DPP8 and DPP9 caused a significant increase in IL-1β secretion in control keratinocytes but not in NLRP1 KO cells (Fig. 2H), in agreement with the effect of chemical inhibition of DPP8/9 with talabostat or 1G244 (Fig. 2G). These results provide orthogonal, genetic evidence that DPP9 inhibition specifically activates the NLRP1 inflammasome and IL-1β–driven inflammation in human keratinocytes cells in a manner that is contingent upon ASC and caspase-1.
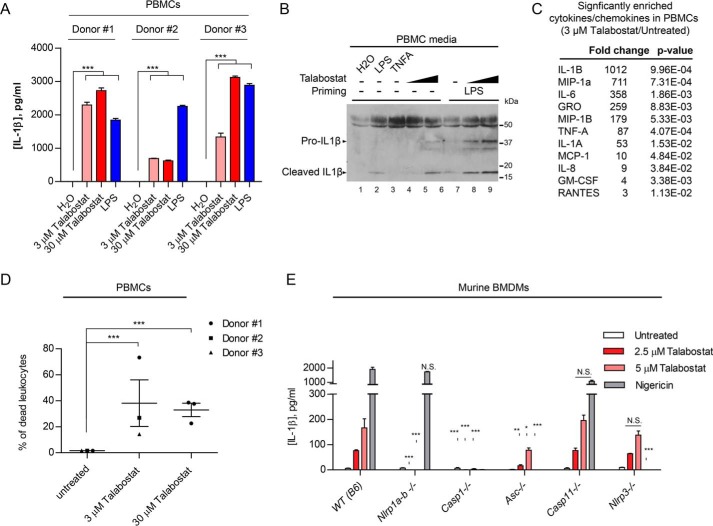
Because germline gain-of-function NLRP1 mutations have been shown to cause systemic autoinflammation and autoimmunity in addition to skin dyskeratosis in the inherited disease AIADK (OMIM no. 617388) (18) and murine NLRP1a activation causes pyroptosis in hematopoietic stem cells (35), we tested whether DPP8/9 inhibition could induce inflammasome activation and pyroptosis in human PBMCs. In three unrelated healthy donors, talabostat treatment alone caused IL-1β secretion to a similar extent to or at higher than levels than LPS (Fig. 3A). Importantly, similar to keratinocytes, talabostat induced the cleavage of pro-IL-1β into its mature form (Fig. 3B, lanes 4 and 5 versus lanes 1 and 3), and this effect could be boosted by prior LPS priming (Fig. 3B, lanes 8 and 9 versus lane 7). Luminex analysis of PBMC-conditioned media revealed that IL-1β was the most enriched cytokine after talabostat treatment (Fig. 3C), similar to keratinocytes (Fig. 2B). PBMCs underwent significant cell deaths as measured by PI influx, particularly in the leukocyte compartment, after talabostat treatment (Fig. 3D). Taken together, these results demonstrate that DPP8/9 inhibition by talabostat activates the inflammasome and pyroptosis in PBMCs, similar to its effects on primary keratinocytes (Fig. 2). Together with recent reports showing that Dpp8/9 inhibition activates murine Nlrp1b, our present results demonstrate that DPP9 is a conserved regulator of NLRP1 inflammasome in mammals, despite divergent ligand specificities across species. Using a panel of inflammasome knockout murine bone marrow-derived macrophages, we independently confirmed that talabostat induces IL-1β secretion in mice and established that the effect of talabostat depends on Nlrp1a-b, Asc, and caspase-1, but not Nlrp3 or caspase-11 (Fig. 2E). Therefore, the downstream mechanisms of inflammasome activation following DPP9 inhibition are also likely conserved between humans and mice.
Figure 3.
DPP8/9 inhibition causes inflammasome activation in human PBMCs. A, PBMCs from three independent healthy donors were isolated using Ficoll gradients and treated with talabostat or LPS (1 μg/ml) for 16 h. Conditioned media were analyzed by IL-1β ELISA. Statistical significance was calculated using one-way ANOVA with Bonferroni's multiple comparison test between all treatment groups for each donor. Bar graph represents technical triplicates for each comparison group. B, PBMCs (donor 1) was left untreated or primed with 1 μg/ml LPS for 1 h before talabostat treatment. 20 μl of unconcentrated medium was used for SDS-PAGE. The band at ∼50 kDa likely represents albumin present in the FBS growth medium. Tumor necrosis factor α (100 ng/ml) was included as a negative control. C, Luminex analysis of chemokines and cytokines induced in talabostat-treated PBMCs from all donors. p values were calculated using Student's t test after log transformation from all three donors. D, PBMCs from three healthy donors were primed and treated as in A. Cell death was measured by LIVE/DEAD fixable Green Dead cell stain followed by FACS. The p values were calculated by one-way ANOVA after log transformation. E, murine BMDMs from the WT (B6), Nlrp1a/b−/−, Casp1−/−, Asc/Pycard−/−, Casp11−/−, and Nlrp3−/− mice were primed with LPS (200 ng/ml) and treated with the 2.5 μm, 5 μm talabostat or 5 ng/ml nigericin 24 h. Cell culture medium was analyzed by murine IL-1β ELISA. Bar graphs represent data from biological triplicates. Statistical significance was calculated with one-way ANOVA comparing the knockout groups to WT control.
Human DPP9 represses NLRP1 via both its catalytic function and NLRP1 binding
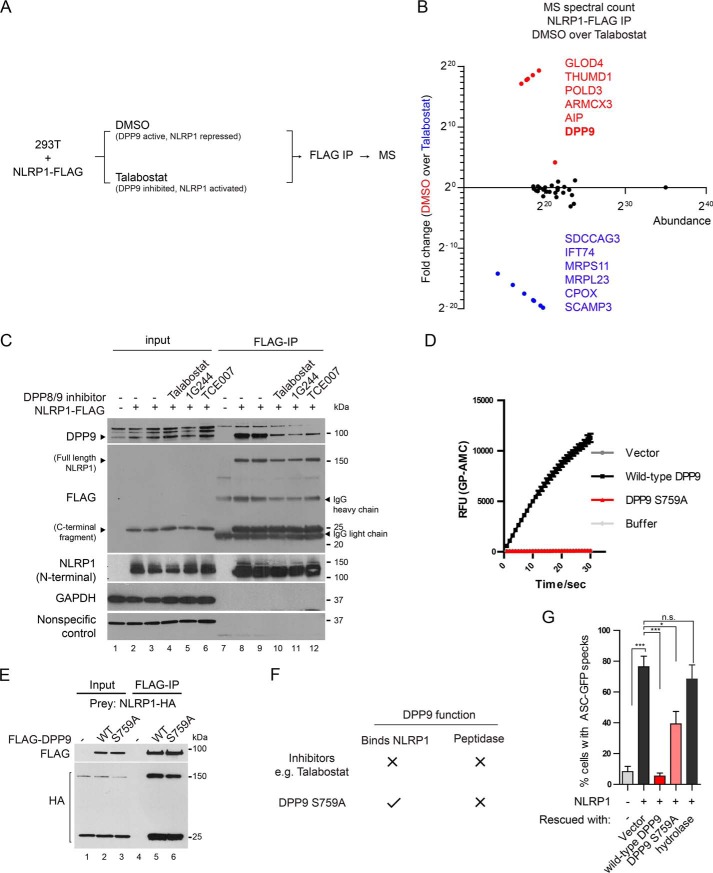
Our biochemical findings demonstrate that the inhibitory function of DPP9 on NLRP1 may not involve the direct cleavage of NLRP1 itself (Fig. S1, H and I). We therefore postulated that DPP9 inhibition might activate human NLRP1 indirectly by modulating its interacting partners. To test this, we immunoprecipitated FLAG-tagged full-length NLRP1 from DMSO- or talabostat-treated human cells and used MS to identify selectively enriched NLRP1-interacting proteins (Fig. 4A and Fig. S3A, lane 2 versus lane 3). We discovered multiple proteins that selectively associate NLRP1 in untreated cells or following talabostat treatment (Fig. 4B). Although the exact functions of these proteins with regard to NLRP1 remain to be fully characterized, we were surprised to identify DPP9 itself as one of the most abundant, differentially enriched NLRP1-binding protein in untreated cells. Talabostat treatment caused a ∼17-fold reduction in the level of DPP9 associated with NLRP1 (Fig. 4B, Fig. S3A, and Excel File S2). This effect was not restricted to talabostat, because other DPP8/9 inhibitors including 1G244 and TCE007, which inhibit DPP8/9 in distinct modes, also caused the dissociation DPP9 from NLRP1 (Fig. 4C, lanes 10–12 versus lanes 8 and 9). These results suggest that the inhibition of DPP9 catalytic activity is not the only effect of the chemical DPP8/9 inhibitors, because they also significantly affect the binding between DPP9 and NLRP1.
Figure 4.
DPP9 enzymatic activity and NLRP1 binding both contribute to NLRP1 inhibition by DPP9. A, experimental strategy to identify talabostat-dependent NLRP1-interacting proteins. 293T cells were transfected with NLRP1-FLAG and treated with DMSO or 3 μm talabostat for 24 h. Approximately 108 cells were used for anti-FLAG immunoprecipitation and MS. B, MS spectral counts of NLRP1-interacting proteins in control versus talabostat-treated cells. Proteins that are most enriched in untreated cells are in red. Proteins most enriched in talabostat-treated cells are in blue. C, 293T cells were transfected with NLRP1-FLAG and treated with 3 μm talabostat, 10 μm 1G244, or 100 μm TCE007 for 24 h. 0.5 mg of whole cell lysate was used for anti-FLAG IP. D, 293T cells were transfected with an empty vector, WT DPP9, or DPP9-S759A and lysed in PBS with 1% Tween 20 48 h afterward. 0.3 μg of total lysate was incubated with a fluorescent test substrate, Gly-Pro-AMC. AMC fluorescence was measured for 30 mins at 25 °C in a 50-μl reaction every minute on a spectrometer. E, 293T cells were transfected with NLRP1-HA, FLAG-tagged WT DPP9, or DPP9 S759A for 48 h. 0.5 mg of whole cell lysate was used for anti-FLAG IP. F, chemical inhibitors and S759A mutation have different effects on the catalytic and NLRP1-binding function of DPP9. G, 293T-ASC-GFP DPP8/9 DKO cells were transfected with an empty vector control or WT NLRP1 together with WT DPP9, DPP9 S759A, and DPP9 Δhydrolase. The percentage of cells with ASC-GFP specks were counted in three different fields on the mounted slides after fixation. Statistical significance was calculated with one-way ANOVA with Bonferroni's multiple comparison test to compare with all treatment groups. N.S., not significant. Bar graphs represent data from biological triplicates.
To examine how DPP9 catalytic activity and NLRP1 binding each contributes to NLRP1 repression, we studied the catalytically inactive DPP9 point mutant S759A. In contrast to chemical inhibitors such as talabostat, the S759A mutation does not compromise the ability of DPP9 to bind NLRP1 (Fig. 4, E, lane 5 versus lane 6, and F), but it completely eliminated the enzymatic activity of DPP9 toward a model substrate (Fig. 4D). Thus DPP9 S759A genetically dissociates the peptidase activity of DPP9 from its NLRP1 binding affinity (Fig. 4F). We next measured the ability of WT and DPP9 S759A mutant to repress NLRP1 when re-expressed in DPP8/9 DKO 293T-ASC-GFP reporter cells. Although WT DPP9 fully repressed NLRP1-dependent ASC speck formation in DPP8/9 DKO cells (Fig. 4G, red bar, and Fig. S3B), DPP9 S759A also led to significant but partial repression (Fig. 4G, pink bar, and Fig. S3B). In contrast, the DPP9 Δhydrolase mutant, which lacks the catalytic domain and does not bind NLRP1 (Figs. S1E, lane 8 versus lanes 6 and 7, and S3B), failed to repress NLRP1-dependent ASC speck formation. Taken together, these experiments reveal that the catalytic function of DPP9 and its physical interaction with NLRP1 both contribute to the inhibition of NLRP1 inflammasome.
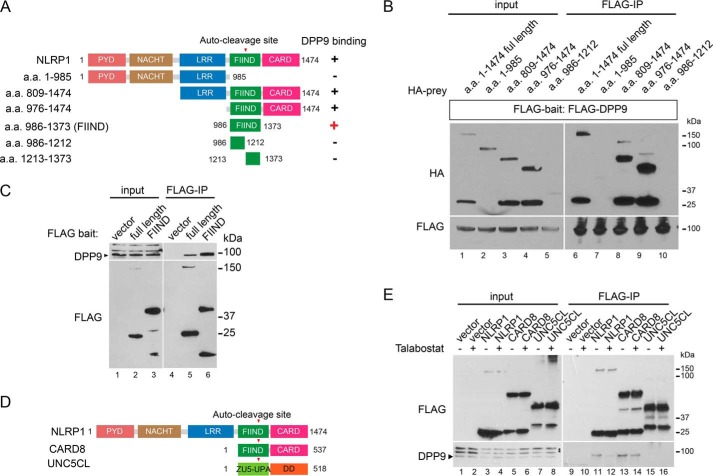
NLRP1 FIIND is a necessary and sufficient DPP9-binding domain
To understand NLRP1–DPP9 interaction in greater detail, a series of NLRP1 truncation mutants were used to identify the DPP9-binding domain in NLRP1 (summarized in Fig. 5A). This analysis revealed that NLRP1 FIIND, an autoproteolytic domain whose function had remained elusive, is the minimal domain that is both necessary (Fig. 5, A and B) and sufficient (Fig. 5C, lane 6 versus lane 5) for DPP9 binding. In addition, this interaction requires the full FIIND, because neither the N-terminal nor the C-terminal post-proteolytic fragment alone was sufficient (Fig. 5, A and B, lanes 7 and 10 versus lane 6, and Fig. S1C, lane 5). NLRP1 FIIND is conserved between human and rodent NLRP1 and also in a putative zebrafish homolog (36). Although its autoproteolytic function has been shown to be required for NLRP1 activation (17, 19), its overall role in NLRP1 regulation remains uncertain. Our results thus ascribe a new function to the FIIND as a necessary and sufficient DPP9-binding module. In support with this notion, another inflammasome regulator, CARD8, which also harbors an autoproteolytic FIIND domain (Fig. 5D), also binds DPP9 in an talabostat-dependent manner, but a more distantly related autoproteolytic protein UNC5CL does not (Fig. 5, D and E, lanes 13 and 14 versus lanes 15 and 16). In fact, DPP9 was identified the most abundant CARD8-interacting partner through an independent, unbiased IP-MS proteomic experiment using FLAG–CARD8 as the bait (Fig. S4, A and B). Notably, while this manuscript was in revision, an independent study reported the role of DPP8/9 on CARD8-dependent pyroptosis in leukemia cells (32). Our findings suggest that DPP9–CARD8 FIIND association likely contributes to this effect.
Figure 5.
NLRP1 FIIND is a necessary and sufficient DPP9-binding domain. A, summary of mapping experiments to identify the DPP9-binding domain in NLRP1. B, 293T cells were transfected with FLAG-DPP9 and HA-tagged NLRP1 domain truncation mutants and harvested 48 h after transfection. 0.5 mg of whole cell lysate was used for anti-FLAG IP. Note that the post-cleavage N-terminal portion of the FIIND domain (a.a. 986–1212 has reduced abundance but was not enriched by FLAG-DPP9 IP. C, 293T cells were transfected as in B with full-length NLRP1 or NLRP1 FIIND (a.a. 986–1373). 0.5 mg of total cell lysate was used for anti-FLAG IP. D, domain structures of human NLRP1, CARD8, and UNC5CL. E, 293T cells were transfected with an empty vector, FLAG-tagged NLRP1, CARD8, and UNC5CL. Transfected cells were treated with 3 μm talabostat for 24 h. 0.5 mg of whole cell lysate was used for anti-FLAG IP.
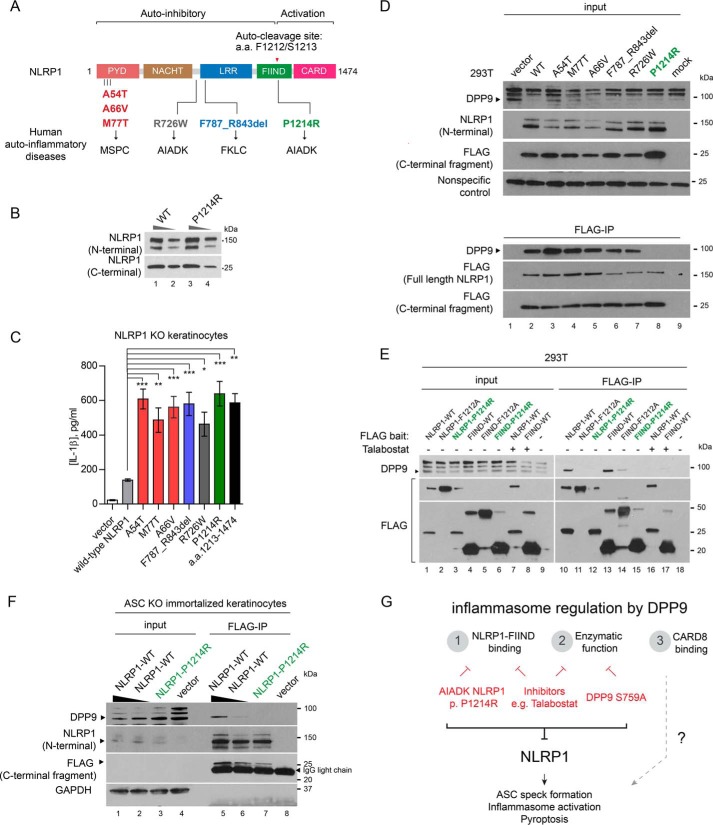
Disruption of DPP9-NLRP1 FIIND association by a patient-derived mutation in a human autoinflammatory disease
Recently our group and others have discovered that distinct germline mutations in NLRP1 cause three human Mendelian diseases, MSPC, familial keratosis lichenoid chronica, and NLRP1-associated AIADK (Fig. 6A). In an effort to comprehensively compare the effects of these mutations, we constituted all NLRP1 mutants in NLRP1 KO keratinocytes. Similar to the constitutively active NLRP1 fragment a.a. 1213–1474, all patient-derived mutations led to ligand-independent IL-1β secretion (Fig. 6B). These results biochemically confirm that MSPC, familial keratosis lichenoid chronica, and AIADK are indeed allelic diseases caused by constitutive NLRP1 inflammasome activation, despite distinct NLRP1 domains in which these mutations are found. At the mechanistic level, the gain-of-function effects of most of these mutations could be explained by the inactivation one of the NLRP1 autoinhibitory domains (Fig. 6A); however, one mutation P1214R found in an AIADK patient (OMIM no. 617388) stands out. P1214R is found in the NLRP1 FIIND domain (Fig. 6A) whose autoproteolytic function is required for NLRP1 activation. Its pathogenic mechanism therefore remains unclear. Our discovery of NLRP1 FIIND as a DPP9-binding domain prompted us to examine whether this mutation impacts NLRP1–DPP9 association. Despite the fact that Pro1214 is adjacent to the autocleavage (Phe1212–Ser1213) site, the P1214R mutation did not cause an appreciable difference in the degree of autocleavage relative to WT NLRP1 when expressed in 293T cells (Fig. 6C). However, it is the only patient-derived mutation that disrupted DPP9 binding, both in the context of full-length NLRP1 (Fig. 6D, lane 8 versus lanes 2–7) and the isolated FIIND domain (Fig. 6E, green, lane 12 versus lane 10 and lane 15 versus lane 13). Its gain-of-function effects in keratinocytes still requires ASC, because ectopic expression of NLRP1 P1214R failed to elicit IL-1β secretion in ASC KO cells (Fig. S4C, green). We took advantage of this observation and confirmed that the P1214R abrogated the binding between NLRP1 and endogenous DPP9 in ASC KO keratinocytes (Fig. 6F, lane 7 versus lanes 5 and 6), because mutant NLRP1 expression caused massive pyroptosis in WT keratinocytes,
Figure 6.
Patient-derived NLRP1 FIIND mutation P1214R abrogates NLRP1–DPP9 binding and causes aberrant inflammasome activation. A, summary of NLRP1 domain structure and all known NLRP1 mutations in human autoinflammatory diseases. B, 293T cells were transfected with WT NLRP1 or AIADK mutant P1214R. 10 or 2 μg of total lysate was used for SDS-PAGE and Western blotting. C, NLRP1 KO immortalized keratinocytes were transfected with WT NLRP1 or NLRP1 disease-associated mutants. Conditioned media were analyzed by IL-1β ELISA 24 h post-transfection. Statistical significance was calculated with one-way ANOVA with Bonferroni's multiple comparison test to compare with all treatment groups. Bar graph represents data from one representative experiment from two biological replicates. D, 293T cells were transfected with WT NLRP1 or NLRP1 disease-associated mutants. Cell pellets were harvested 48 h post-transfection. 0.5 mg of total cell lysate was used for anti-FLAG IP. E, 293T cells were transfected with full-length NLRP1 or FIIND mutants and treated with DMSO or 3 μm talabostat. 0.5 mg of total cell lysate was used for anti-FLAG IP. Full-length NLRP1 P1214R and FIIND P1214R are highlighted in green. F, ASC KO keratinocytes were transfected with an empty vector, WT NLRP1, or NLRP1 P1214R. Two concentrations of the WT NLRP1 constructs were used, as P1214R demonstrated reduced accumulation in multiple experiments. 2 mg of total cell extract was used for anti-FLAG IP. G, proposed model for the role of DPP9 as a multifunctional inflammasome regulator in primary human cells.
Taken together, these results suggest that the gain-of-function effect of AIADK mutation P1214R could be explained by the derepression of NLRP1 inflammasome via loss of NLRP1–FIIND and DPP9 binding. In addition, because other patient-derived mutations do not impinge upon NLRP1–DPP9 association (Fig. 6D, lane 2 versus lanes 3–7), our findings suggest that the regulation of NLRP1 by DPP9 must function in parallel with additional regulatory mechanisms, which likely involve other NLRP1 domains such as PYD and LRR, to maintain NLRP1 inhibition in primary human cells. In agreement with this hypothesis, DPP9 overexpression in keratinocytes did not alleviate the aberrant inflammasome activation caused by two representative patient-derived non-FIIND NLRP1 mutations (Fig. S4, D and E).
Discussion
Although NLRP1 was one of the first inflammasome sensors to be identified, its function in innate immunity and human inflammatory conditions has not been characterized in detail, presumably because of the lack of knowledge of its endogenous regulatory mechanisms and ligand specificity. We and others have identified three human Mendelian diseases caused by NLRP1 mutations, which demonstrate remarkable differences from other inflammasome disorders (17, 18). This likely reflects the distinct regulatory mechanisms governing NLRP1 function in humans. Recently it was reported that chemical inhibitors of dipeptidases Dpp8 and 9 activate murine Nlrp1b inflammasome and a noncanonical inflammasome consisting a related sensor CARD8 (24, 25, 32). In this manuscript, we report the findings from an independent investigation on human NLRP1. Using biochemical reconstitution and functional assays in primary human cells, we found that DPP9 is a novel binding partner for human NLRP1 and a related inflammasome regulator CARD8. DPP9 inhibition causes NLRP1-dependent ASC speck formation and IL-1β cleavage and primary human keratinocytes and PBMCs. Mechanistically, DPP9 binds to the autoproteolytic NLRP1 FIIND domain, and the disruption of the NLRP1–DPP9 interaction by a patient-derived mutation in the FIIND domain causes spontaneous inflammasome activation, thereby offering a mechanistic explanation for the Mendelian autoinflammatory disease, AIADK.
Our findings extend previous observations using chemical inhibition of Dpp8 and 9 in rodent systems. Using genetic separation of function mutants, we found that both NLRP1 binding and DPP9 enzymatic function must act together to repress NLRP1 (Fig. 6G). This is consistent with one previous report demonstrating that DPP9 regulates SYK level and B-cell activation through binding to a nonsubstrate interacting partner, FLNA (28). However, it is currently unclear which DPP9 enzymatic substrate functions as the direct activator of NLRP1. Our data are consistent with the model proposed by Okondo et al. (24) in the murine system: it is likely that a yet-to-be-identified protein, which is continuously inactivated by DPP8/9, serves as an endogenous NLRP1 activator. A more detailed analysis of our proteomic data on the NLRP1 interactome (Fig. S1A and Excel File S1) would help elucidate this missing link in future investigations.
Our results have a number of implications for the regulation of human inflammasome in general. We propose that NLRP1 acts as a sensor for a homeostasis-altering molecular process (37) involving alterations of DPP9 function, which might be suppressed during infection by an unknown foreign pathogen effector. In this case, alterations of DPP9 function would alert the host to a loss of cellular homeostasis. As a result, DPP9 function must require constant monitoring by a dedicated sensor, NLRP1. In the absence of any trigger, enzymatically active DPP9 ensures that NLRP1 is kept in the inactive state in a manner that requires DPP9–FIIND domain interaction (Fig. 6G). Our work also identified a second FIIND-containing inflammasome regulator, CARD8 as a DPP9-binding partner. Although the role of CARD8 in human inflammasome regulation remains to be clarified, germline mutations in CARD8 have been shown to cause a Mendelian autoinflammatory syndrome via the NLRP3 inflammasome (38). Because CARD8 does not exist in rodents, it is very likely that DPP9 has broader functions in the human innate immune system than previous mouse experiments have demonstrated through its combined action on human NLRP1 and CARD8 (Fig. 6G). This is supported by a paper published recently (32) while this paper was under revision, demonstrating that DPP8/9 inhibitors elicit leukemia cell pyroptosis via CARD8.
Our findings might help explain the potent immune-stimulating effect of DPP8/9 inhibitor talabostat in vivo (39). Because talabostat is currently undergoing preclinical evaluation as a novel combination agent for cancer immunotherapy, our findings raise the interesting possibility that the activation of NLRP1 inflammasome is at least in part responsible for the anti-cancer properties of talabostat. Furthermore, the human-specific biology of DPP9 reported here, e.g. its effects on primary keratinocytes, should be carefully considered in moving current DPP8/9 inhibitors from mouse models into patient trials.
Experimental procedures
Cell culture
293Ts (laboratory stock, validated by ATCC cell authentication) was cultured in DMEM supplemented with 10% fetal bovine serum (FBS) without antibiotics. Immortalized keratinocytes (N/TERT-1) (a generous gift from Rheinwald JG under Material Transfer Agreement) (40) were cultured in keratinocyte serum-free medium with bovine pituary extract epidermal growth factor in the presence of 0.3 mm CaCl2. PBMCs were obtained from donor using Ficoll and cultured in RPMI medium with 10% FBS. Primary human keratinocytes were cultured using methods described by Rheinwald and Green (41). BMDM were prepared from the bone marrow of Nlrp1(abc)−/− (35), Casp1−/− (42), Asc−/− (43), Casp11−/− (44), and Nlrp3−/− (45) mice, cultured in DMEM supplemented with 10% FBS and 10% L929-cell conditioned medium for 6 days.
Immunoprecipitation and MS
Small-scale immunoprecipitations were carried out by incubating overnight 0.5 mg of total cell lysate prepared in 1× TBS–Nonidet P-40 buffer (20 mm Tris-HCl, 150 mm NaCl, 0.5% Nonidet P-40) and 10 μl of anti-FLAG-M2-agarose resin (Sigma–Aldrich) in ∼300–500 μl of total volume. Bound proteins were eluted in 1× Laemmli's buffer at 95 °C for 5 min.
For MS, ∼108 transfected 293T cells were lysed in 3 ml of 1× TBS–Nonidet P-40 buffer, and 100 μl of anti-FLAG-M2-agaorase beads were used for immunoprecipitation and directed subjected to trypsin digestion after washes in lysis buffer. An equal amount of peptides was taken for Tandem Mass Tag (TMT) isobaric tag (Thermo) labeling. Following labeling, the samples were combined, desalted, vacuum-dried, and subsequently resuspended in 10 mm ammonia and using step gradient fractionated on C18 high pH reverse phase material using self-packed column (C-18 ReproSil-Pur Basic, Dr. Maisch, 10 μm) with 12, 25, and 50% of acetonitrile in 10 mm ammonia formate. Fractions were washed with 70% acetonitrile with 0.1% formic acid, vacuum-dried, and subsequently analyzed using Easy nLC1000 (Thermo) chromatography system coupled with Orbitrap Fusion (Thermo). Each sample was separated in 120-min gradient (0.1% formic acid in water and 99.9% acetonitrile with 0.1% formic acid) using 50-cm × 75-m inner diameter Easy-Spray column (C-18, 2m particles; Thermo). Data-dependent mode was used with a 3-s cycle and Orbitrap analyzer (ion targets and resolution OT-MS 4xE5 ions, resolution 60K, OT-MS/MS 6E4 ions, resolution 15k). Peak lists were generated with Proteome Discoverer 2.2 software (Thermo) followed by searches with Mascot 2.6.1 (Matrix Science) against concatenated forward/decoy Human Uniprot database with following parameters: precursor mass tolerance (MS) 20 ppm, OT-MS/MS 0.05 Da, three miss cleavages; static modifications: carboamidomethyl (C), TMT6plex; variable modifications: oxidation (M), deamidated (NQ), Acetyl N-terminal protein. Forward/decoy searches were used for false discovery rate estimation (1%).
Plasmid transfection and lentiviral transduction
All expression plasmids for transient expression was constructed based on the pCS2+ backbone and cloned using InFusion HD (Clontech). All 293T transfection experiments were performed with Lipofectamine 2000 (ThermoFisher). Keratinocytes were transfected with FuGENE HD (Promega). Lentiviral constructs were based on pCDH-puro (System Biosciences) and packaged with the third generation packaging plasmids.
Chemical compounds
The small molecule inhibitors used are TC-E 5007 (Tocris), sitagliptin (MedChemExpress), talabostat (MedChemExpress), 1G244 (Santa Cruz Biotechnology), LPS (Ultrapure, Escherichia coli O111:B4, Sigma–Aldrich), and nigericin (Invivogen, catalog no. tlrl-nig).
Blue-Native and SDS-PAGE
Blue-Native PAGE was carried out using the Native-PAGE system (ThermoFisher) with 10–20 μg of total cell lysate followed by dry transfer (TurboBlot, Bio-Rad) and Western blotting. SDS-PAGE was carried out using precast TGX 4–20% gels (Bio-Rad).
CRISPR/Cas9 gene editing
DPP8/9 deletion in 293T-ASC-GFP cells was carried out using a co-editing and positive selection protocol published by the Doyon lab (46). Parental cells were co-transfected with plasmids (Plasmid catalog no. 62988) encoding guide RNAs against DPP8, DPP9, and ATP1A (G2) in a ratio of 3:1. Clonal selection was carried out in the presence of 1 μm ouabain (Sigma–Aldrich). Lentiviral Cas9 and guide RNA plasmids (Addgene plasmid catalog nos. 52962 and 52963) were used to create stable deletions of NLRP1 and PYCARD in keratinocytes. The guide RNA target sequences are NLRP1 (CTATCAGCTGCTCTCGATAC, AGCCCGAGTGACATCGGTGG); ASC (CGCTAACGTGCTGCGCGACA, GCTAACGTGCTGCGCGACAT); DPP9 (ATCCATGGCTGGTCCTACGG, TGTGTCGTAGGCCATCCAGA); and DPP8 (GAGGAGATAGCAATTACCAG, TAGTTGTCCCAACATACGGA, GTGACAAGGCTGACTGACCG, ACCTGAAAAAGATCCAAAAG).
IL-1B ELISA and Luminex cytokine/chemokine array
Human IL-1B was measured with human IL-1B BD OptEIA ELISA kit. Mouse IL-1B ELISA was measured with R&D DY401 ELISA kit. Luminex cytokine/chemokine array was carried out using standard manufacturer-supplied protocol without modification.
Measurement of cell death in PBMCs
After harvesting supernatants from PBMCs, the remaining cell pellets were used for quantification of cell death. The cells were washed in PBS, resuspended, and incubated for 10 min in PBS containing 1:1000 LIVE/DEAD fixable green dead cell stain (Thermo Fisher). The cells were then washed in staining buffer containing PBS, 0.2% (v/v) FBS, and 2 mm EDTA. To identify immune cell lineages, a surface stain was performed for 20 min in Brilliant stain buffer (BD Biosciences) containing the following antibodies: CD4-BV786, CD8-Alexa Fluor 700, CD19-BUV496 (BD Biosciences), CD11c-BV421, CD123-BV650, CD56-BV711 (Biolegend), and CD14-Viogreen (Miltenyi Biotec). Samples were acquired using a FACSymphony flow cytometer (Becton-Dickinson) and analyzed with FlowJo V10 (FlowJo LLC).
Live-cell imaging
Live-cell imaging was performed on an Olympus IX-81 high content screening, inverted microscope using a 300W metal halide fluorescence lamp with enhanced EGFP and TRITC filters and an Olympus 20× objective with 0.17-nm glass correction. NTERT/1 cells engineered to express GFP-ASC were seeded into chamber slides. The next day cells were either treated with talabostat or left untreated. 1 μg/ml propidium iodide was added to all wells. Live imaging was then conducted in keratinocyte serum-free medium (ThermoFisher, 17005042) at 37 °C within XL3 chamber (Pecon) with 5% CO2 supplied for a total of ∼21 h, with images collected every 10 min. The images were visualized and analyzed using Fiji (Image J) and are presented as representative images of three experiments.
LDH assay
NTERT/1 cells were seeded at 0.3 × 106 cells/well in 12-well plates in KSFM 24 h prior to treatment. Supernatants were then harvested and analyzed for LDH activity using an LDH cytotoxicity assay kit (Pierce, ThermoScientific) according to the manufacturer's instructions. The percentage cytotoxicity of talabostat was then calculated based on the percentage difference compared with the LDH-positive control provided with the kit.
Author contributions
F. L. Z., K. R., and B. R. conceptualization; F. L. Z., A. D., J. C. R., J. E. C., and S. L. M. resources; F. L. Z., K. R., D. E. T. T., K.-Y. T., C. L., C. R. H., C.-H. Y., W. H. X., R. M. S., V. B. A., R. H., A. D., J. C. R., J. E. C., and S. L. M. data curation; F. L. Z., K. R., D. E. T. T., C. L., C. R. H., W. H. X., J. E. C., and S. L. M. formal analysis; F. L. Z., J. C. R., J. E. C., S. L. M., and B. R. supervision; F. L. Z. and B. R. funding acquisition; F. L. Z. validation; F. L. Z., K. R., C. R. H., C.-H. Y., W. H. X., R. M. S., V. B. A., R. H., A. D., J. C. R., J. E. C., S. L. M., and B. R. investigation; F. L. Z. methodology; F. L. Z. writing-original draft; F. L. Z. and B. R. project administration; F. L. Z. and B. R. writing-review and editing.
Supplementary Material
Acknowledgments
We are grateful to all members of the Reversade laboratory for support.
This work was supported by core funding from the Institute of Molecular and Cell Biology (IMCB) and Institute of Medical Biology (IMB) Strategic Positioning Fund (Biomedical Research Council (BMRC), A*STAR) (to B. R. and R. M. S.), Young Investigator Grant YIG 2015 (BMRC, A*STAR) (to B. R. and R. M. S.), National Medical Research Council MS-CETSA platform Grant MOHIAFCAT2/004/2015 (to B. R. and R. M. S.), National Medical Research Council Young Investigator Grant NMRC/OFYIRG/0046/2017 (to F. L. Z.), National Health and Medical Research Council Grants 1142354 and 1099262 (to S. L. M.), funds from the Sylvia and Charles Viertel Foundation (to S. L. M.), a Howard Hughes Medical Institute–Wellcome International Research Scholarship (to S. L. M.), and funds from GlaxoSmithKline (to S. L. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Videos S1–S3, Figs. S1–S4, and Excel Files S1 and S2.
- PYD
- pyrin domain
- IL
- interleukin
- AIADK
- autoinflammation with arthritis and dyskeratosis
- a.a.
- amino acid(s)
- IP
- immunoprecipitation
- MSPC
- multiple self-healing palmoplantar carcinoma
- DKO
- double-knockout
- PBMC
- peripheral blood mononuclear cells
- FBS
- fetal bovine serum
- TRITC
- tetramethylrhodamine isothiocyanate
- ANOVA
- analysis of variance
- OMIM
- Online Mendelian Inheritance in Man
- PI
- propidium iodide.
References
- 1. Janeway C. A. Jr., and Medzhitov R. (2002) Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 10.1146/annurev.immunol.20.083001.084359 [DOI] [PubMed] [Google Scholar]
- 2. Martinon F., Burns K., and Tschopp J. (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- 3. Tschopp J., Martinon F., and Burns K. (2003) NALPs: a novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 4, 95–104 10.1038/nrm1019 [DOI] [PubMed] [Google Scholar]
- 4. Davis B. K., Wen H., and Ting J. P. (2011) The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29, 707–735 10.1146/annurev-immunol-031210-101405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lamkanfi M., and Dixit V. M. (2011) Modulation of inflammasome pathways by bacterial and viral pathogens. J. Immunol. 187, 597–602 10.4049/jimmunol.1100229 [DOI] [PubMed] [Google Scholar]
- 6. Lamkanfi M., Kanneganti T. D., Franchi L., and Núñez G. (2007) Caspase-1 inflammasomes in infection and inflammation. J. Leukocyte Biol. 82, 220–225 10.1189/jlb.1206756 [DOI] [PubMed] [Google Scholar]
- 7. Xu H., Yang J., Gao W., Li L., Li P., Zhang L., Gong Y. N., Peng X., Xi J. J., Chen S., Wang F., and Shao F. (2014) Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 513, 237–241 10.1038/nature13449 [DOI] [PubMed] [Google Scholar]
- 8. Guo C., Xie S., Chi Z., Zhang J., Liu Y., Zhang L., Zheng M., Zhang X., Xia D., Ke Y., Lu L., and Wang D. (2016) Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity 45, 802–816 10.1016/j.immuni.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 9. Qu Y., Misaghi S., Izrael-Tomasevic A., Newton K., Gilmour L. L., Lamkanfi M., Louie S., Kayagaki N., Liu J., Kömüves L., Cupp J. E., Arnott D., Monack D., and Dixit V. M. (2012) Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490, 539–542 10.1038/nature11429 [DOI] [PubMed] [Google Scholar]
- 10. Shoham N. G., Centola M., Mansfield E., Hull K. M., Wood G., Wise C. A., and Kastner D. L. (2003) Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc. Natl. Acad. Sci. U.S.A. 100, 13501–13506 10.1073/pnas.2135380100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spalinger M. R., Kasper S., Gottier C., Lang S., Atrott K., Vavricka S. R., Scharl S., Raselli T., Frey-Wagner I., Gutte P. M., Grütter M. G., Beer H. D., Contassot E., Chan A. C., Dai X., et al. (2016) NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J. Clin. Invest. 126, 1783–1800 10.1172/JCI83669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stutz A., Kolbe C. C., Stahl R., Horvath G. L., Franklin B. S., van Ray O., Brinkschulte R., Geyer M., Meissner F., and Latz E. (2017) NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J. Exp. Med. 214, 1725–1736 10.1084/jem.20160933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim M. L., Chae J. J., Park Y. H., De Nardo D., Stirzaker R. A., Ko H. J., Tye H., Cengia L., DiRago L., Metcalf D., Roberts A. W., Kastner D. L., Lew A. M., Lyras D., et al. (2015) Aberrant actin depolymerization triggers the pyrin inflammasome and autoinflammatory disease that is dependent on IL-18, not IL-1β. J. Exp. Med. 212, 927–938 10.1084/jem.20142384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moghaddas F., and Masters S. L. (2015) Monogenic autoinflammatory diseases: cytokinopathies. Cytokine 74, 237–246 10.1016/j.cyto.2015.02.012 [DOI] [PubMed] [Google Scholar]
- 15. Kastner D. L., Aksentijevich I., and Goldbach-Mansky R. (2010) Autoinflammatory disease reloaded: a clinical perspective. Cell 140, 784–790 10.1016/j.cell.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Venegas C., Kumar S., Franklin B. S., Dierkes T., Brinkschulte R., Tejera D., Vieira-Saecker A., Schwartz S., Santarelli F., Kummer M. P., Griep A., Gelpi E., Beilharz M., Riedel D., Golenbock D. T., et al. (2017) Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer's disease. Nature 552, 355–361 10.1038/nature25158 [DOI] [PubMed] [Google Scholar]
- 17. Zhong F. L., Mamaï O., Sborgi L., Boussofara L., Hopkins R., Robinson K., Szeverényi I., Takeichi T., Balaji R., Lau A., Tye H., Roy K., Bonnard C., Ahl P. J., Jones L. A., et al. (2016) Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167, 187–202.e17 10.1016/j.cell.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 18. Grandemange S., Sanchez E., Louis-Plence P., Tran Mau-Them F., Bessis D., Coubes C., Frouin E., Seyger M., Girard M., Puechberty J., Costes V., Rodière M., Carbasse A., Jeziorski E., Portales P., et al. (2017) A new autoinflammatory and autoimmune syndrome associated with NLRP1 mutations: NAIAD (NLRP1-associated autoinflammation with arthritis and dyskeratosis). Ann. Rheum. Dis. 76, 1191–1198 10.1136/annrheumdis-2016-210021 [DOI] [PubMed] [Google Scholar]
- 19. Finger J. N., Lich J. D., Dare L. C., Cook M. N., Brown K. K., Duraiswami C., Bertin J. J., and Gough P. J. (2012) Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem. 287, 25030–25037 10.1074/jbc.M112.378323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D'Osualdo A., Weichenberger C. X., Wagner R. N., Godzik A., Wooley J., and Reed J. C. (2011) CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS One 6, e27396 10.1371/journal.pone.0027396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chavarría-Smith J., and Vance R. E. (2013) Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathogens 9, e1003452 10.1371/journal.ppat.1003452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cirelli K. M., Gorfu G., Hassan M. A., Printz M., Crown D., Leppla S. H., Grigg M. E., Saeij J. P., and Moayeri M. (2014) Inflammasome sensor NLRP1 controls rat macrophage susceptibility to Toxoplasma gondii. PLoS Pathogens 10, e1003927 10.1371/journal.ppat.1003927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ewald S. E., Chavarria-Smith J., and Boothroyd J. C. (2014) NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect. Immun. 82, 460–468 10.1128/IAI.01170-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okondo M. C., Rao S. D., Taabazuing C. Y., Chui A. J., Poplawski S. E., Johnson D. C., and Bachovchin D. A. (2018) Inhibition of Dpp8/9 activates the Nlrp1b inflammasome. Cell Chem. Biol. 25, 262–267.e5 10.1016/j.chembiol.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okondo M. C., Johnson D. C., Sridharan R., Go E. B., Chui A. J., Wang M. S., Poplawski S. E., Wu W., Liu Y., Lai J. H., Sanford D. G., Arciprete M. O., Golub T. R., Bachovchin W. W., and Bachovchin D. A. (2017) DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat. Chem. Biol. 13, 46–53 10.1038/nchembio.2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taabazuing C. Y., Okondo M. C., and Bachovchin D. A. (2017) Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem. Biol. 24, 507–514.e4 10.1016/j.chembiol.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gall M. G., Chen Y., Vieira de Ribeiro A. J., Zhang H., Bailey C. G., Spielman D. S., Yu D. M., and Gorrell M. D. (2013) Targeted inactivation of dipeptidyl peptidase 9 enzymatic activity causes mouse neonate lethality. PLoS One 8, e78378 10.1371/journal.pone.0078378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Justa-Schuch D., Silva-Garcia M., Pilla E., Engelke M., Kilisch M., Lenz C., Möller U., Nakamura F., Urlaub H., and Geiss-Friedlander R. (2016) DPP9 is a novel component of the N-end rule pathway targeting the tyrosine kinase Syk. Elife 5, e16370 10.7554/eLife.16370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim M., Minoux M., Piaia A., Kueng B., Gapp B., Weber D., Haller C., Barbieri S., Namoto K., Lorenz T., Wirsching J., Bassilana F., Dietrich W., Rijli F. M., and Ksiazek I. (2017) DPP9 enzyme activity controls survival of mouse migratory tongue muscle progenitors and its absence leads to neonatal lethality due to suckling defect. Dev. Biol. 431, 297–308 10.1016/j.ydbio.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 30. Pratley R. E., and Salsali A. (2007) Inhibition of DPP-4: a new therapeutic approach for the treatment of type 2 diabetes. Curr. Med. Res. Opin 23, 919–931 10.1185/030079906X162746 [DOI] [PubMed] [Google Scholar]
- 31. Yazbeck R., Howarth G. S., and Abbott C. A. (2009) Dipeptidyl peptidase inhibitors, an emerging drug class for inflammatory disease? Trends Pharmacol. Sci. 30, 600–607 10.1016/j.tips.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 32. Johnson D. C., Taabazuing C. Y., Okondo M. C., Chui A. J., Rao S. D., Brown F. C., Reed C., Peguero E., de Stanchina E., Kentsis A., and Bachovchin D. A. (2018) DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat. Med. 24, 1151–1156 10.1038/s41591-018-0082-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sand J., Haertel E., Biedermann T., Contassot E., Reichmann E., French L. E., Werner S., and Beer H. D. (2018) Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death Dis. 9, 24 10.1038/s41419-017-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evavold C. L., Ruan J., Tan Y., Xia S., Wu H., and Kagan J. C. (2018) The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity 48, 35–44.e6 10.1016/j.immuni.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Masters S. L., Gerlic M., Metcalf D., Preston S., Pellegrini M., O'Donnell J. A., McArthur K., Baldwin T. M., Chevrier S., Nowell C. J., Cengia L. H., Henley K. J., Collinge J. E., Kastner D. L., Feigenbaum L., et al. (2012) NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity 37, 1009–1023 10.1016/j.immuni.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang M. X., Chen W. Q., and Nie P. (2010) Structure and expression pattern of teleost caspase recruitment domain (CARD) containing proteins that are potentially involved in NF-κB signalling. Dev. Comp. Immunol. 34, 1–13 10.1016/j.dci.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 37. Liston A., and Masters S. L. (2017) Homeostasis-altering molecular processes as mechanisms of inflammasome activation. Nat. Rev. Immunol. 17, 208–214 10.1038/nri.2016.151 [DOI] [PubMed] [Google Scholar]
- 38. Mao L., Kitani A., Similuk M., Oler A. J., Albenberg L., Kelsen J., Aktay A., Quezado M., Yao M., Montgomery-Recht K., Fuss I. J., and Strober W. (2018) Loss-of-function CARD8 mutation causes NLRP3 inflammasome activation and Crohn's disease. J. Clin. Invest. 128, 1793–1806 10.1172/JCI98642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams S., Miller G. T., Jesson M. I., Watanabe T., Jones B., and Wallner B. P. (2004) PT-100, a small molecule dipeptidyl peptidase inhibitor, has potent antitumor effects and augments antibody-mediated cytotoxicity via a novel immune mechanism. Cancer Res. 64, 5471–5480 10.1158/0008-5472.CAN-04-0447 [DOI] [PubMed] [Google Scholar]
- 40. Dickson M. A., Hahn W. C., Ino Y., Ronfard V., Wu J. Y., Weinberg R. A., Louis D. N., Li F. P., and Rheinwald J. G. (2000) Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20, 1436–1447 10.1128/MCB.20.4.1436-1447.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rheinwald J. G., and Green H. (1975) Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6, 331–343 10.1016/S0092-8674(75)80001-8 [DOI] [PubMed] [Google Scholar]
- 42. Schott W. H., Haskell B. D., Tse H. M., Milton M. J., Piganelli J. D., Choisy-Rossi C. M., Reifsnyder P. C., Chervonsky A. V., and Leiter E. H. (2004) Caspase-1 is not required for type 1 diabetes in the NOD mouse. Diabetes 53, 99–104 10.2337/diabetes.53.1.99 [DOI] [PubMed] [Google Scholar]
- 43. Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., Roose-Girma M., Erickson S., and Dixit V. M. (2004) Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- 44. Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., Zhang J., Lee W. P., Roose-Girma M., and Dixit V. M. (2011) Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 10.1038/nature10558 [DOI] [PubMed] [Google Scholar]
- 45. Martinon F., Pétrilli V., Mayor A., Tardivel A., and Tschopp J. (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 10.1038/nature04516 [DOI] [PubMed] [Google Scholar]
- 46. Agudelo D., Duringer A., Bozoyan L., Huard C. C., Carter S., Loehr J., Synodinou D., Drouin M., Salsman J., Dellaire G., Laganière J., and Doyon Y. (2017) Marker-free coselection for CRISPR-driven genome editing in human cells. Nat. Methods 14, 615–620 10.1038/nmeth.4265 [DOI] [PubMed] [Google Scholar]
- 47. Bachovchin D., Chui A. J., Okondo M., Rao S., Gai K., Griswold A., and Vittimberga B. (2018) N-terminal degradation activates the Nlrp1b inflammasome. bioRxiv 10.1101/317826 10.1101/317826 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.