Abstract
In insects, γ-aminobutyric acid (GABA) is the major inhibitory neurotransmitter, and GABA-gated ion channels are the target of different classes of insecticides, including fipronil. We report here the cloning of six subunits (four RDL, one LCCH3, and one GRD) that constitute the repertoire of the GABA-gated ion channel family of the Varroa mite (Varroa destructor), a honey bee ectoparasite. We also isolated a truncated GRD subunit with a premature stop codon. We found that when expressed in Xenopus laevis oocytes, three of the four RDL subunits (VdesRDL1, VdesRDL2, and VdesRDL3) formed functional, homomultimeric anionic receptors, whereas GRD and LCCH3 produced heteromultimeric cationic receptors. These receptors displayed specific sensitivities toward GABA and fipronil, and VdesRDL1 was the most resistant to the insecticide. We identified specific residues in the VdesRDL1 pore-lining region that explain its high resistance to fipronil. VdesRDL4 did not form a functional receptor when expressed alone, but it assembled with VdesRDL1 to form a heteromultimeric receptor with properties distinct from those of the VdesRDL1 homomultimeric receptor. Moreover, VdesRDL1 physically interacted with VdesRDL3, generating a heteromultimeric receptor combining properties of both subunits. On the other hand, we did not detect any functional interaction between VdesLCCH3 and the VdesRDL subunits, an observation that differed from what was previously reported for Drosophila melanogaster. In conclusion, this study provides insights relevant to improve our understanding of the precise role of GABAergic signaling in insects and new tools for the development of Varroa mite–specific insecticidal agents that do not harm honey bees.
Keywords: GABA receptor, cloning, electrophysiology, pharmacology, mutagenesis, protein assembly, Varroa mite, Xenopus leavis oocytes
Introduction
Varroa destructor is the most devastating pest for the Western honey bee Apis mellifera (1). The lifespan of infested honey bee colonies is significantly shorter unless they are treated with acaricides. Moreover, several honey bee viruses, including deformed wing virus or acute bee paralysis virus, are transmitted by Varroa mites (2). The combined effects of the parasite and the viral diseases play an important role in honey bee colony health, contributing to morphological deformities (small body, deformed wings) and immune system weakening. These infestations probably contribute to colony collapse disorder, a syndrome leading to the large-scale loss of managed bees and recorded in Europe and North America since 2006 (3). Colony collapse disorder is especially alarming because A. mellifera is commonly used for active crop pollination, and its role has been evaluated at over $200 billion worldwide. Synthetic agents, such as pyrethroids (fluvalinate) or organophosphates (coumaphos), are used to limit Varroa mite infestation. However, these treatments may have adverse effects on bee health (4). To minimize pesticide residues in bee colonies, beekeepers also use essential oil (thymol) and organic acids (formic acid, oxalic acid). Thymol, a constituent of thyme essential oil, is a positive allosteric modulator of GABA-gated receptors in humans and also in fruit flies (5). Moreover, insect GABA receptors are one of the major targets of insecticides including phenylpyrazoles (fipronil), cyclodienes (dieldrin), and metadiamides (broflanilide) (6).
GABA is the major inhibitory neurotransmitter in the vertebrate and invertebrate nervous systems. In insects, GABA is important for locomotion control, olfactory learning, and regulation of sleep and aggression (7–10). GABA receptors are members of the cysteine-loop ligand-gated ion channel superfamily that also includes nicotinic acetylcholine receptors, which permeate cations, and glutamate-gated channels, which permeate anions (11, 12). The receptors of this family are composed of five homologous subunits arranged around a central ion channel. Each subunit possesses an extracellular N-terminal domain that encompasses the ligand-binding domain (LBD),2 and four transmembrane segments (TM1–4), with a variable intracellular loop between TM3 and TM4. The TM2 segment lines the channel pore (13). In insects, three genes encoding GABA receptors have been identified: RDL (resistance to dieldrin), LCCH3 (ligand-gated chloride channel homolog 3), and GRD (GABA and glycine receptor-like subunit from Drosophila). Mutations in the TM2 of the RDL subunits have been detected in insecticide-resistant strains of many arthropod species (14). In heterologous expression systems, RDL subunits assemble into homopentameric, chloride-selective, and thymol-sensitive channels (5, 15–18). Less is known about LCCH3 and GRD. Only D. melanogaster subunits have been expressed in Xenopus laevis oocytes where they assemble into heteropentameric, cation-selective channels (19). Although a fragment encoding the TM2 of a RDL subunit was previously isolated in V. destructor (20), nothing is known about GABA-gated ion channels in this parasite, which is a major target of acaricides.
Here, we describe the molecular cloning of four RDL, one LCCH3, and two GRD subunits from V. destructor. We found that when expressed in X. laevis oocytes, the resulting GABA-gated channels displayed specific pharmacological and biophysical features. Moreover, RDL subunits could assemble in heteromultimeric receptors. The properties of the three homomultimeric RDL receptors expressed in X. laevis oocytes VdesRDL1, VdesRDL2, and VdesRDL3 were clearly distinct from those previously reported for the honey bee RDL ortholog (21). This suggests structural specificities that could be exploited in a pharmacological screen to identify insecticidal agents that will be more efficient on Varroa receptors than on their ortholog in honey bee and therefore could potentially be used to control Varroa mites without harming honey bees.
Results
Isolation of GABA-gated ion channel subunits from V. destructor
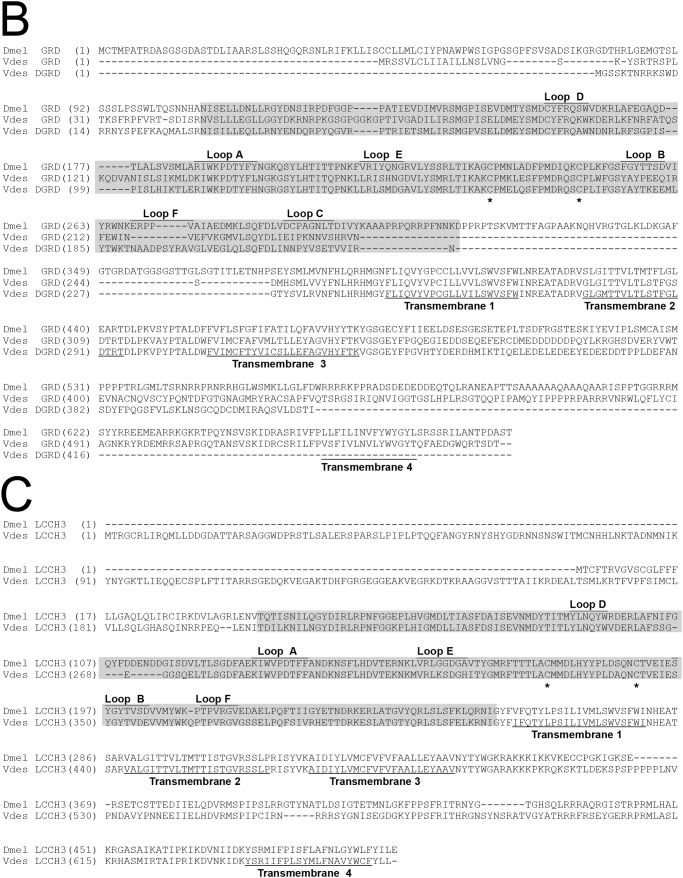
We obtained the cDNAs of the V. destructor RDL (VdesRDL), GRD (VdesGRD), and LCCH3 (VdesLCCH3) subunits by rapid amplification of cDNA ends and PCR using gene specific oligonucleotides. For VdesRDL2, however, we could initially isolate only an incomplete ORF (GenBankTM accession number KY748051). We then took advantage of the large number of new sequences obtained from the transcriptomic analysis in Varroa jacobsoni (assembly vjacob_1.0 GCF_002532875) and V. destructor (Assembly Vdes3.0 GCF_002443255) deposited in the GenBankTM during this work to obtain a full-length ORF also for VdesRDL2. The cDNAs for the RDL subunits included ORFs of 1758, 1986, 1776, and 2193 bp encoding proteins of 585, 661, 591, and 730 amino acids for VdesRDL1 (KY748050), VdesRDL2 (MH423074), VdesRDL3 (KY748052), and VdesRDL4 (KY748053), respectively (Fig. 1). The predicted molecular masses were approximately 65 kDa for VdesRDL1 and VdesRDL3 and 72 and 80 kDa for VdesRDL2 and VdesRDL4, respectively. VdesRDL1 predicted sequences found in GenBankTM (XP_02265189 to XP_022651192) were 10 residues longer than the deduced amino acid sequence of our clone, but we found this small insertion in the TM3–TM4 loop in another clone not studied here. The predicted sequences for VdesRDL2 found in GenBankTM differed at the N terminus and the TM3–TM4 loop (XP_022658105 to XP_022658113). The deduced amino acid sequence of VdesRDL2 was similar to the sequence XP_022658108 at the TM3–TM4 loop and to the sequence XP_022658109 at the N terminus. The deduced amino acid sequence of VdesRDL3 was similar to the sequences XP_022644956 to XP_02264958. In the three other predicted sequences found in GenBankTM for VdesRDL3, the TM4 (XP_022644960 and XP_02264961), and a conserved segment into the LBD and the TM1 (XP_02264959) were missing. Like for VdesRDL2, the amino acid sequences predicted for VdesRDL4 differed mainly at the TM3–TM4 loop and at the N terminus (XP_022665348 to XP_022665350 and XP_022665352 to XP_022665356). Although most of the deduced amino acid sequence of VdesRDL4 was similar to XP_022665348, the N terminus was absent in any of the predicted sequences available in GenBankTM. The four VdesRDL subunits shared ∼50% identity. As expected, the LBD and the TM segments were well conserved between the VdesRDL subunits and the D. melanogaster RDL (DmelRDL) subunit (Fig. 1). Notably, all the residues involved in GABA binding in the pentameric receptor (Tyr109 and Arg111 in loop D, Ser176 at the end of loop E, Glu204 and Phe206 in loop B, and Thr251 and Tyr254 in loop C, according to DmelRDL sequence numbering) (22, 23) were conserved in the four VdesRDL subunits. However, VdesRDL1 sequence differed from the other sequences at residues critical for GABA receptor inhibition by the insecticides dieldrin and fipronil (14): a serine instead of alanine at 2′, a methionine instead of threonine at 6′ in TM2 (when the charged residue at the cytoplasmic end is numbered 0′), and an alanine instead of a threonine at the cytoplasmic end of the TM3 (Fig. 1). Two of these differences (Met at 6′ and Ala306) were also found in VdesRDL4. Therefore, specific pharmacology could be developed for multimeric receptors that include one of these subunits. The cDNAs for the VdesGRD (KY748054) and VdesLCCH3 (KY748055) subunits included ORFs of 1662 and 1980 bp encoding proteins of 553 and 659 amino acids, respectively (Fig. 1). The predicted molecular masses were 63 kDa for VdesGRD and 75 kDa for VdesLCCH3. We also cloned a truncated GRD variant that we named VdesΔGRD, but we did not investigate its features. It is noteworthy that three predicted sequences found in GenBankTM (XP_022664708 to XP_022664710) are similarly truncated. The deduced amino acid sequence of VdesGRD differed from that of the two predicted proteins found in GenBankTM (XP_022653054 and XP_022653055) that lack a fragment of the LBD. VdesLCCH3 deduced amino acid sequence was similar to that of XP_022672719. Two other predicted proteins differed from VdesLCCH3 at the TM3–TM4 loop and by an insertion of 17 residues between Loop E and Loop B (XP_022672700 and XP_022672710). VdesGRD and VdesLCCH3 sequences shared 33 and 50% identity with the D. melanogaster subunits, respectively. The residues identified in the RDL sequences as involved in GABA binding were not well conserved in VdesGRD or VdesLCCH3. However, because of the absence of structural data and mutagenic studies, GABA-interacting residues have been not precisely identify in these subunits (22).
Figure 1.
Alignment of the V. destructor and D. melanogaster RDL (A), GRD (B), and LCCH3 (C) sequences. The ligand-binding domain is shaded. The position of the two typical cysteine residues of the ligand-gated ion channel family is indicated by an asterisk (*). The amino acids involved in GABA binding in the RDL subunits are indicated by a pound sign (#). The amino acids involved in the binding/resistance to insecticides in the RDL subunits are boxed. The GenBankTM accession numbers for the D. melanogaster RDL, GRD, and LCCH3 subunits are NP523991, NP524131, and NP996469, respectively.
Functional expression of V. destructor GABA-gated channels
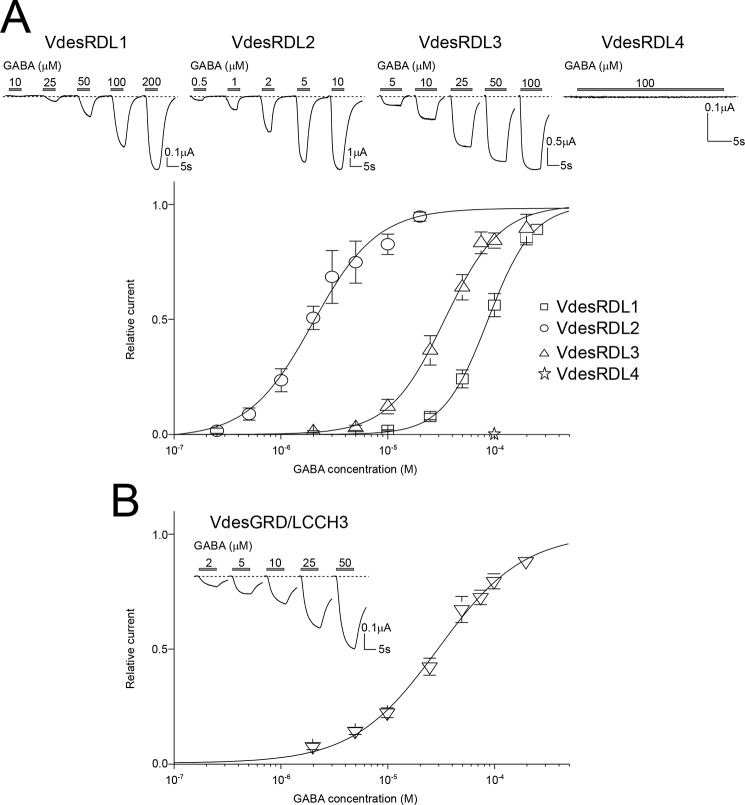
To evaluate the properties of the cloned V. destructor GABA-gated channels, we performed two-electrode voltage-clamp recordings in X. laevis oocytes after injection of the cRNAs encoding each individual VdesRDL subunit or of a mixture of cRNAs encoding the VdesGRD and VdesLCCH3 subunits because these subunits do not form functional receptors when expressed separately. When maintained at −60 mV, perfusion of GABA induced an inward current only in oocytes that expressed VdesRDL1, VdesRDL2, VdesRDL3, or VdesGRD/LCCH3, but not in oocytes that expressed VdesRDL4 (Fig. 2). This indicates that VdesRDL1, 2, and 3 subunits can form functional homomultimeric receptors. We then obtained concentration-response curves by challenging oocytes with GABA concentrations ranging from 0.1 to 250 μm. Each VdesRDL subunit displayed a different GABA EC50 with the smallest value for VdesRDL2 and the highest for VdesRDL1 (Table 1). In similar experiments, the GABA EC50 value of the A. mellifera RDL subunit (25.2 ± 3.9 μm, n = 5) was similar to value previously reported (21). The GABA EC50 value obtained with VdesGRD/LCCH3 was close to the one obtained with VdesRDL3 (Table 1). It is well established that homomultimeric RDL receptors permeate chloride ions (17, 24), but until now, only one study showed that the heteromultimeric GRD/LCCH3 receptor forms GABA-gated cation channels (19). In our recording conditions, the reversal potential (Erev) obtained with the VdesRDL subunits (−12.9 ± 1.3 mV, n = 11, −12.5 ± 2.1 mV, n = 7, and −14.5 ± 1.1 mV, n = 9, for VdesRDL1, VdesRDL2, and VdesRDL3, respectively) was similar to that obtained with the heteromultimeric VdesGRD/LCCH3 receptor (−10.3 ± 0.9 mV, n = 15). Erev increased by ∼50 mV for the VdesRDL subunits (+50 ± 2 mV, n = 5, +42 ± 1 mV, n = 4, and +51 ± 4 mV, n = 7, for VdesRDL1, VdesRDL2, and VdesRDL3, respectively) when we decreased the extracellular chloride concentration from 100 to 0 mm, in accordance with some Cl− permeability. Conversely, upon the same change, Erev was not affected for VdesGRD/LCCH3 (−1 ± 1 mV, n = 5; Fig. 3). Reciprocally when we decreased the extracellular sodium concentration from 100 to 0 mm, Erev measured for VdesRDL subunits was not strongly affected (−1 ± 2 mV, n = 4, +8 ± 1 mV, n = 4, and +3 ± 2 mV, n = 4, for VdesRDL1, VdesRDL2, and VdesRDL3, respectively), whereas that of VdesGRD/LCCH3 was reduced by −20 mV (−20 ± 2 mV, n = 6; Fig. 3). These results indicate that in our recording conditions, VdesGDR/LCCH3 permeates sodium ions (without being highly selective for this species) as previously shown for the D. melanogaster subunits (19).
Figure 2.
GABA-evoked currents for VdesRDLs and VdesGRD/LCCH3 expressed in X. laevis oocytes. A, top panel, representative current traces obtained with increasing GABA concentrations in X. laevis oocytes after the injection of the cRNAs encoding VdesRDL1, VdesRDL2, VdesRDL3, or VdesRDL4. The duration of GABA perfusion was adjusted for each oocyte to reach the maximal current amplitude. Note the absence of current in oocytes injected with the cRNA encoding VdesRDL4. Bottom panel, GABA concentration-response curves obtained in X. laevis oocytes that express VdesRDL1, VdesRDL2, or VdesRDL3. B, same as in A but in X. laevis oocytes that express VdesGRD/LCCH3. The data are the means ± S.E. of n = 9–23 oocytes.
Table 1.
Parameters of the concentration-response curves obtained for GABA and fipronil in X. laevis oocytes
ND, not determined. The data are the means ± S.E. of n oocytes.
| RDL1 | RDL1 mut | RDL2 | RDL3 | GRD/LCCH3 | GRD/LCCH3 mut | RDL1/RDL4 | RDL1 mut/RDL4 | RDL1/RDL4 mut | RDL1 mut/RDL4 mut | RDL1/RDL3 | RDL3/RDL4 | RDL1/LCCH3 (1:5) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GABA | |||||||||||||
| EC50 μm | 92 ± 10 | ND | 2.4 ± 0.3 | 37 ± 4 | 35 ± 4 | ND | 102 ± 5 | ND | ND | ND | 107 ± 5 | 39 ± 10 | 131 ± 9 |
| nHill (n) | 2.3 ± 0.1 (11) | 2.5 ± 0.1 (23) | 2.5 ± 0.3 (19) | 1.5 ± 0.3 (9) | 2.3 ± 0.1 (13) | 2.2 ± 0.1 (6) | 2.7 ± 0.2 (14) | 3.0 ± 0.2 (3) | |||||
| Fipronil | |||||||||||||
| IC50 μm (n) | 55 ± 11 (12) | 7.2 ± 1.7a (21) | 3.7 ± 1.3 (8) | 1.1 ± 0.3 (12) | 0.9 ± 0.1 (10) | 2.6 ± 0.4b (17) | 15 ± 2 (15)a | 10.9 ± 3.7c (5) | 57 ± 13 (15) | 6.9 ± 1.2a (22) | 6.0 ± 1.2a (14) | 1.2 ± 0.2 (12) | 44 ± 6 (10) |
a Significantly different from VdesRDL1 (p < 0.01).
b Significantly different from VdesGRD/LCCH3 (p < 0.05).
c Significantly different from VdesRDL1 (p < 0.05).
Figure 3.
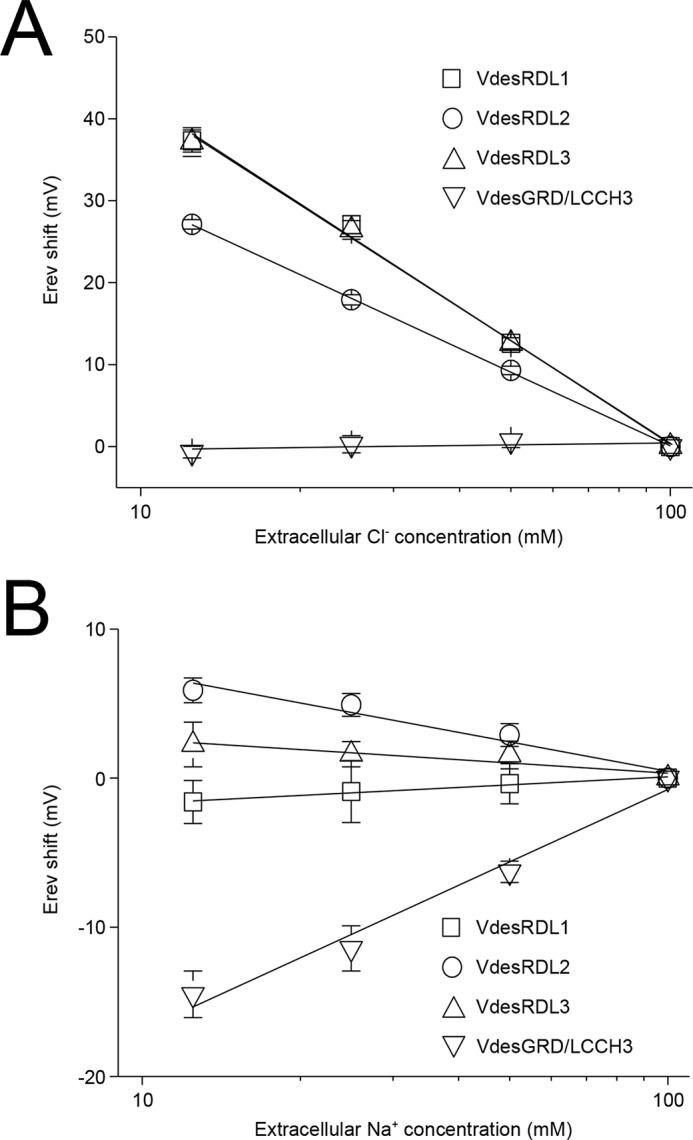
Semi-logarithmic plots showing the effects of the variation of extracellular chloride (A) or sodium (B) concentration on the reversal potential of the current evoked by GABA (EC30–50) determined in X. laevis oocytes that express VdesRDL1, VdesRDL2, VdesRDL3, or VdesGRD/LCCH3. The data are the means ± S.E. of n = 4–7 oocytes.
Inhibition of V. destructor GABA-gated channels by fipronil
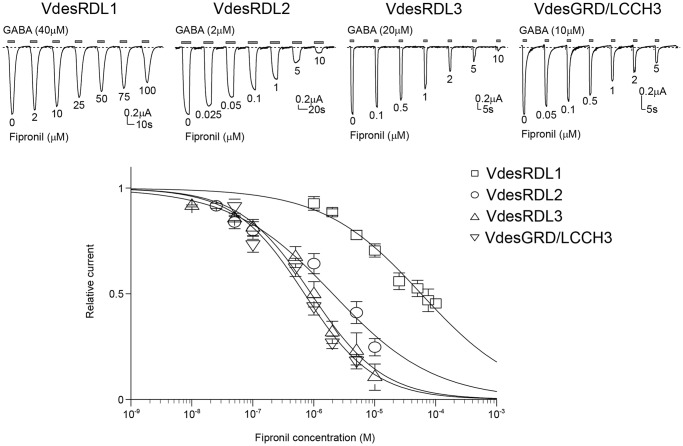
The presence of a 2′S and 6′M in the TM2 of the VdesRDL1 amino acid sequence suggested that this subunit may display a different sensitivity toward insecticides than the other VdesRDL subunits. Fipronil potently inhibited the GABA-evoked currents recorded in oocytes that expressed VdesRDL2, VdesRDL3, or VdesGDR/LCCH3 but not VdesRDL1 (Fig. 4). The VdesRDL1 IC50 value was ∼50 times higher than that of the other RDL subunits (Table 1). In our recording conditions, fipronil IC50 for A. mellifera RDL (0.066 ± 0.014 μm, n = 4) was similar to the value previously reported (21). To assess whether the specific differences in the TM2 sequence were responsible for VdesRDL1 resistance to fipronil, we determined the fipronil IC50 for VdesRDL1 that harbored the mutations S2′A and M6′T (VdesRDL1mut). The significantly smaller IC50 for VdesRDL1mut compared with VdesRDL1 (Table 1) demonstrated the importance of the 2′ and 6′ residues for RDL sensitivity toward fipronil. Our sequence analysis also indicated the presence of a glycine and an alanine at 2′ in VdesGRD and VdesLCCH3, respectively. Although the mutation A2′G in RDL subunit has been frequently identified in strains resistant to fipronil (14), our results indicate that its presence in VdesGRD does not abolish VdesGRD/LCCH3 sensitivity to fipronil.
Figure 4.
Effects of fipronil on GABA-evoked currents. Top panel, representative current traces evoked by GABA (EC30–50) in the presence of increasing concentrations of fipronil in X. laevis oocytes that express VdesRDL1, VdesRDL2, VdesRDL3, or VdesGRD/LCCH3. The duration of GABA perfusion was adjusted for each oocyte to reach the maximal current amplitude. Bottom panel, fipronil inhibition curves for the same receptors. The data are the means ± S.E. of n = 8–12 oocytes.
VdesRDL4 can form heteromultimeric receptors with VdesRDL1 in X. laevis oocytes
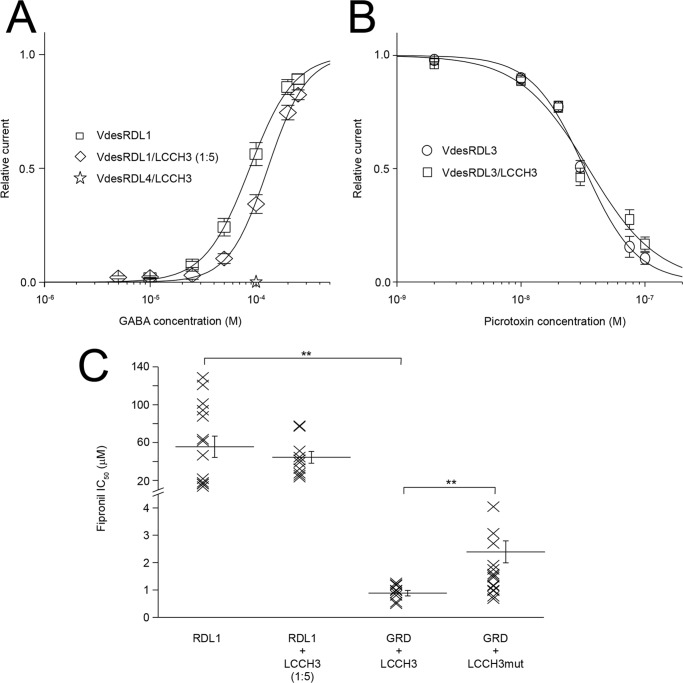
Western blotting analysis of lysates from X. laevis oocytes showed that oocytes injected with cRNA encoding VdesRDL4 tagged with GFP expressed a protein at the expected molecular mass (Fig. 5A). Because VdesRDL4 could not form functional homopentameric receptors, we tried to co-express this subunit with VdesRDL3 or VdesRDL1 (Fig. 5 and Table 1). We did not observe any significant differences between the GABA EC50 and fipronil IC50 values obtained with VdesRDL3/RDL4 or with VdesRDL3 alone (Table 1). However, these results do not exclude that these two subunits might form heteromultimeric receptors in X. laevis oocytes. Indeed, the sensitivity toward GABA or fipronil of the heteromultimeric receptor might be similar to that of VdesRDL3 alone. This possibility was suggested by the finding that co-expression of VdesRDL4 and VdesRDL1 increased the sensitivity to fipronil compared with VdesRDL1 expressed alone (Fig. 5C and Table 1). Conversely, the GABA EC50 value obtained with VdesRDL1/RDL4 was not significantly different from that of VdesRDL1 alone. To determine whether the higher sensitivity toward fipronil of the heteromultimer VdesRDL1/RDL4 was due to the presence of VdesRDL4, we introduced the mutation A2′S in this subunit (VdesRDL4mut). The fipronil IC50 value of VdesRDL1/RDL4mut was not different from that of VdesRDL1 alone (Fig. 5C and Table 1). As expected, the fipronil IC50 value obtained when the VdesRDL1mut was co-expressed with VdesRDL4 was comparable with that of VdesRDL1mut alone (Fig. 5C and Table 1). This demonstrates that VdesRDL1 can assemble in functional receptors with VdesRDL4 and that these heteromultimeric receptors display different features compared with the VdesRDL1 homopentameric receptors.
Figure 5.
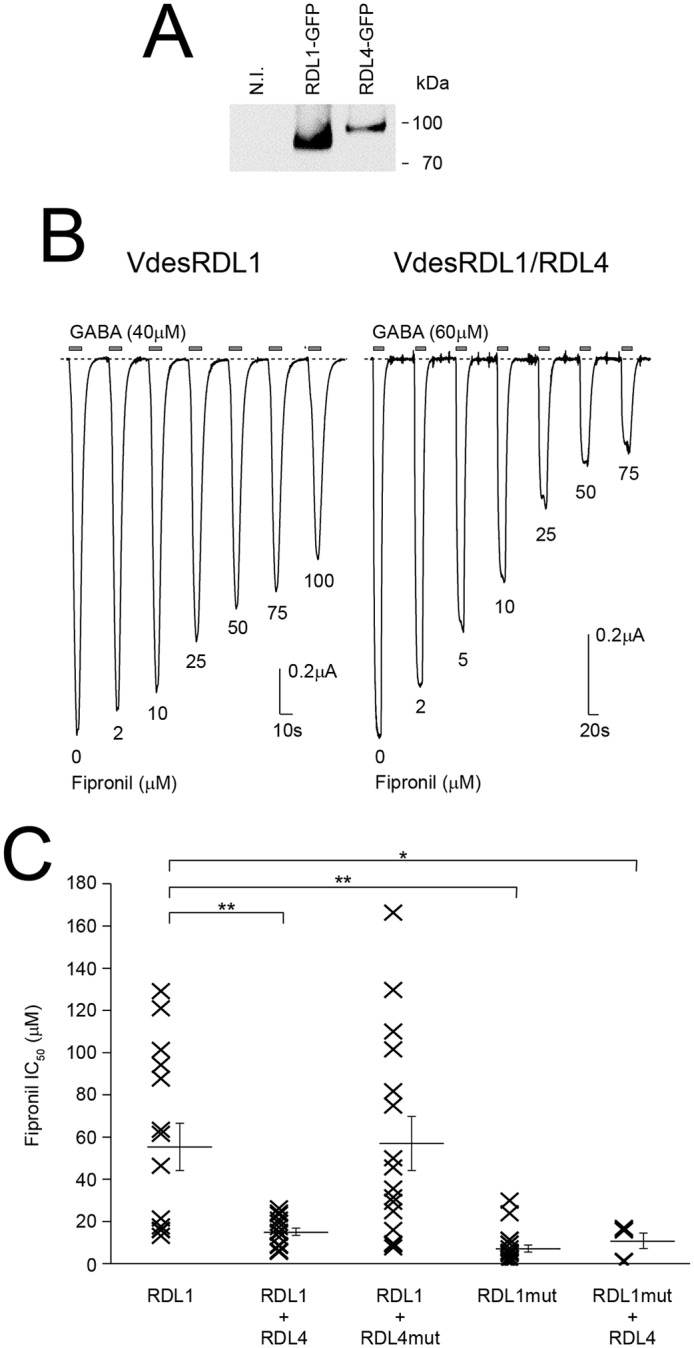
VdesRDL1 and VdesRDL4 assemble in functional heteromultimers. A, Western blotting analysis of lysates from X. laevis oocytes that express GFP-tagged VdesRDL1 or VdesRDL4 showing the expression of the two proteins. The expected molecular masses are 93 kDa for VdesRDL1 and 108 kDa for VdesRDL4. N.I., noninjected oocytes. B, representative current traces evoked by GABA in the presence of increasing concentrations of fipronil in X. laevis oocytes that express VdesRDL1 alone or with VdesRDL4. C, scatter plot showing the fipronil IC50 values obtained in X. laevis oocytes that express different combinations of RDL subunits. Bars show means ± S.E. of 5–21 oocytes. *, p < 0.05; **, p < 0.01.
VdesRDL1 assembles with VdesRDL3 to form heteromultimeric receptors in X. laevis oocytes
To test whether VdesRDL1 and VdesRDL3 could assemble in heteromultimeric receptors, we co-injected equimolar concentration of RNAs encoding VdesRDL1 and VdesRDL3 in X. laevis oocytes. Indeed the expression of both receptors was similar with current amplitude values ranging from −635 to −8235 nA (mean −5087 ± 679 nA, n = 13) for VdesRDL1 and from −1026 to −5802 nA (mean −2882 ± 335 nA, n = 21) for VdesRDL3, 1–3 days after injection. The GABA EC50 value obtained from oocytes that expressed VdesRDL1/RDL3 (89.2–121.2 μm) was not significantly different from the value obtained for VdesRDL1 alone (Fig. 6A and Table 1). Conversely, the fipronil IC50 value for VdesRDL1/RLD3 (1.4–14.2 μm) was close to the value obtained for VdesRDL3 alone (Fig. 6B and Table 1). These results are compatible with the expression of a homogenous population of heteromultimeric receptors rather than two populations of homomultimers. They also suggests that in X. laevis oocytes, VdesRDL1 and VdesRDL3 preferentially assemble to form heteromultimers, with properties different from those of the single receptors. Co-immunoprecipitation experiments using VdesRDL1 and VdesRDL3 tagged with GFP and mCherry, respectively, confirmed the direct interaction between VdesRDL1 and VdesRDL3 (Fig. 6C). This strongly supports that when the two RDL subunits are expressed together, they preferentially form heteromultimeric receptors.
Figure 6.
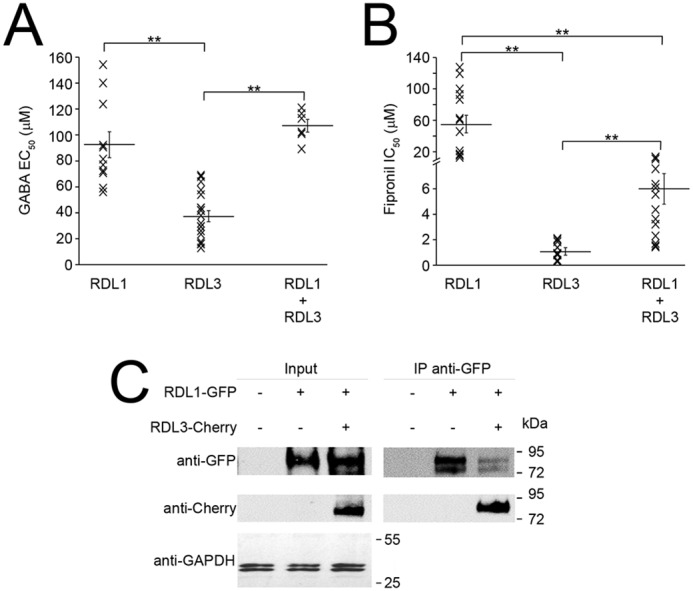
VdesRDL1 and VdesRDL3 assemble in heteromultimers rather than homomultimers when expressed together in X. laevis oocytes. A, scatter plot showing the GABA EC50 values obtained in X. laevis oocytes that express the indicated RDL subunits. B, scatter plot showing the fipronil IC50 values. Bars show means ± S.E. of n = 6–14 oocytes. **, p < 0.01. C, co-immunoprecipitation experiment using HEK293 cells transfected with VdesRDL1-GFP alone or with VdesRDL3-Cherry showing the physical interaction between these subunits. The expected molecular mass is 93 kDa for both proteins.
VdesLCCH3 does not assemble in functional heteromultimers with VdesRDL subunits
Despite the fact that VdesRDL and VdesGRD/LCCH3 permeate different ions (anions/Cl− ions and cations/Na+ ions, respectively), a functional interaction between RDL and LCCH3 has been suggested in flies (25) and honey bees (8). Because the VdesGRD/LCCH3 heteromultimer was highly sensitive to fipronil inhibition (Table 1), VdesRDL1/LCCH3 heteromultimers should be fipronil sensitive on the basis of our results with VdesRDL1/RDL4. First, introduction of the mutation A2′S in VdesLCCH3 (VdesLCCH3mut) confirmed that this subunit was, at least in part, responsible for the VdesGRD/LCCH3 sensitivity to fipronil. Indeed, the fipronil IC50 value of the VdesGRD/LCCH3mut heteromultimer was significantly higher than that of VdesGRD/LCCH3 (Fig. 7C and Table 1). Then we co-injected into X. laevis oocytes the cRNA encoding VdesRDL1 with a 5-fold excess of the cRNA encoding VdesLCCH3 to promote the formation of heteromultimeric receptors. However, we did not observe any significant difference between the GABA EC50 and fipronil IC50 values for VdesRDL1/LCCH3 and for VdesRDL1 alone (Fig. 7 and Table 1). On the basis of the results obtained after co-expression of VdesRDL1 with VdesRDL4 or VdesRDL3, this finding seems to exclude the formation of a functional heteromultimeric receptor between VdesRDL1 and VdesLCCH3. We next assessed whether VdesRDL4, which cannot form functional homomultimeric receptors, could assemble with VdesLCCH3 to form functional heteromultimeric receptors. We could not record any GABA-evoked current in oocytes co-injected with these subunits, suggesting that VdesRDL4 and VdesLCCH3 did not assemble or that the heteromultimeric receptors were not functional (Fig. 7A). In flies, the co-expression of LCCH3 decreased the picrotoxin sensitivity of RDL (25). Because VdesRDL1 (like VdesRDL4) harbors a methionine at 6′ in TM2 that confers picrotoxin resistance (20), we tested whether VdLCCH3 could form functional heteromultimers with VdesRDL3 by evaluating the picrotoxin sensitivity (Fig. 7B). The picrotoxin IC50 value determined in X. laevis oocytes that expressed VdesRDL3/LCCH3 (33 ± 2 nm, n = 7) was not significantly different from that of VdesRDL3 alone (37 ± 3 nm, n = 7). These results seem to exclude a functional interaction between VdesLCCH3 and any of the VdesRDL subunits.
Figure 7.
VdesLCCH3 does not assemble in functional heteromultimers with VdesRDL1, VdesRDL3, or VdesRDL4 in X. laevis oocytes. A, GABA concentration-response curves obtained in X. laevis oocytes that express the indicated combinations of GABA receptor subunits. VdesLCCH3 cRNA was injected in 5-fold molar excess compared with VdesRDL1 cRNA. Note that the co-expression of VdesLCCH3 and VdesRDL4 does not lead to functional expression of heteromultimers. B, picrotoxin concentration-response curves obtained in X. laevis oocytes that express VdesRDL3 alone or with VdesLCCH3. C, scatter plot showing fipronil IC50 values in X. laevis oocytes that express the indicated subunit combinations. Bars show means ± S.E. of n = 7–17 oocytes. **, p < 0.01.
Discussion
In insects, there are four genes encoding potential GABA-gated channels: RDL, LCCH3, GRD, and CG8916. Since their identification in D. melanogaster (15), many RDL orthologs have been isolated from various arthropod species. However, only few of them, for example in the tobacco budworm Heliothis virescens (Lepidoptera) (16), the American dog tick Dermacentor variabilis (Acari) (17), the planthopper Sogatella furcifera (Hemiptera) (18), or the honey bee A. mellifera (Hymenoptera) (21), have been functionally characterized in heterologous expression systems. Moreover, functional expression of the GRD/LCCH3 heteromultimer has only been done with the D. melanogaster subunits (19), although these subunits have been isolated in other arthropod species, such as the silkworm Bombyx mori (Lepidoptera) (26) or the planthopper Laodelphax striatellus (Hemiptera) (27). On the other hand, functional expression of the CG8916 subunit alone, as homomultimer, or with LCCH3 has never been obtained (19).
In this study, we report the complete cDNA sequences of four RLD, one LCCH3, and one GRD subunits from V. destructor. This is the largest family of GABA-gated ion channel subunits cloned to date in insects. We also isolated a cDNA encoding an additional GRD subunit in which a large part of the TM3–TM4 loop and the TM4 segment are lacking. We identified a splice variant only for VdesRDL1, but future experiments will undoubtedly isolate other variants for the GABA receptors in V. destructor, like in other arthropod species. In many Lepidoptera, several genes encode RDL subunits. This is the case for the diamondback moth Plutella xylostella (28) and the rice stem borer Chilo suppressalis (29), in which two RDL subunits have been isolated, or the silkworm B. mori, in which three RDL subunits have been cloned (26). In Arachnoidea also several genes encode RDL subunits, for instance in V. destructor (four, this study) and in the two-spotted spider mite Tetranychus urticae (30), in which three RDL subunits have been isolated. This characteristic should be confirmed thanks to the availability of the genomic sequence of other Arachnoidea (31). Conversely, there is only a single gene encoding RDL subunit in Diptera (e.g. fruit fly) (12), in Hymenoptera (e.g. honey bee) (11), and in Hemiptera, (e.g. the brown planthopper Nilaparvata lugens) (32).
It has been hypothesized that the existence of multiple RDL subunits could result from gene duplication to enhance the tolerance to naturally occurring (e.g. picrotoxin- and picrotoxin-like molecules of plant origin) and synthetic insecticides (e.g. dieldrin and fipronil) (26, 33). Differences in the amino acid sequence at 2′ in TM2 support this hypothesis. Indeed, the three RDL subunits isolated in B. mori have alanine, serine, or glutamine residues at this position (26), and the two RDL subunits of C. suppressalis harbor an alanine or serine residue (29). In V. destructor, the VdesRDL1 subunit has a serine residue, and the other three RDL subunits have alanines. Mutations at this specific position in the inner mouth of the channel pore have been observed in many insecticidal-resistant arthropod strains (14, 34). In a homology model, fipronil docks best in the channel pore, interacting with 2′A, 3′L, and 6′T (33). C. suppressalis RDL1 and RDL2 subunits have different residues at 2′ (alanine and serine) in an otherwise conserved TM2, but they display a similar sensitivity toward fipronil (29). We did not evaluate the role of the specific residues found in VdesRDL1 in fipronil sensitivity, but it seems that the combination of 2′S, 6′M, and Ala355, at the end of TM3, is involved in the sensitivity to this insecticide (34).
Multiple RDL subunits with specific pharmacological or biophysical properties could also be advantageous for GABA signaling. The two RDL subunits isolated from C. suppressalis have a specific affinity for GABA (the GABA EC50 is 38.3 μm for CsRDL1 and 21.6 μm for CsRDL2) (29). The situation is even more complex with the Varroa mite RDL subunits because the GABA sensitivity of VdesRDL2 was 50 times higher than that of VdesRDL1, and the GABA affinity of VdesRDL3 was intermediate. We also found that in X. laevis oocytes, the VdesRDL subunits can assemble in heteromultimeric receptors with specific properties. Therefore, in insects, multiple RLD subunits might provide a large toolbox to finely tune GABA signaling. Studies on the physiological roles of GABA receptors in insects are scarce, and it is difficult to attribute these roles to a specific GABA-gated receptor (35). This could be improved by the analysis of the expression pattern of the different GABA receptor subunits in the different tissues and during development. Such analysis has been partially done in Leodelphax striatellus (36) and in C. suppressalis where CsRDL1 and CsRDL2 show similar expression pattern but different expression levels (29). This suggests that in C. suppressalis, the two RDL subunits could assemble in heteromultimers. Additional experiments are needed to confirm the existence of such heteromultimeric receptors in vivo.
In mammals, GABA can also play the role of an excitatory neurotransmitter, and this effect results from the inversion of the chloride gradient that causes cellular depolarization (37). Excitatory GABA receptors have been described in Caenorhabditis elegans in which they play important roles in muscular contraction (38). Conversely, GABA-mediated excitatory neurotransmission has never been observed in insects (39, 40), although it has been clearly demonstrated that the D. melanogaster GRD and LCCH3 subunits assemble in cationic ligand-gated channels in X. laevis oocytes (19). Similarly, the VdesGRD/LCCH3 heteromultimer shows a cationic selectivity suggesting that this feature could be shared by other arthropod species in which these subunits are found. However, the existence of such heteromultimeric receptors in vivo remains an open question. Indeed, overlapping expression of these two subunits in the same cells has never been detected (8, 27). It has been suggested that the LCCH3 subunit may assemble with RDL in A. mellifera antennal lobe (8), but in D. melanogaster LCCH3 and RDL exhibit a different pattern of expression (41). Although co-expression of LCCH3 with RDL in Sf21 cells generates an unusual receptor (25), we did not find any evidence for such an association in X. laevis oocytes for the V. destructor LCCH3 and RDL subunits. More experiments are needed to confirm the existence of cationic ligand-gated ion channel in vivo and to determine how subunits with opposite selectivity might assemble to form functional receptors.
In conclusion, we cloned eight GABA-gated ion channel subunits from the honey bee mite V. destructor and demonstrated the expression of three homomultimeric RDL receptors and one heteromultimeric GRD/LCCH3 receptor in X. laevis oocytes. These receptors have distinct biophysical and pharmacological features suggesting that they could specifically regulate GABA signaling in vivo. We also found that in X. laevis oocytes, the VdesRDL subunits assemble in heteromultimeric receptors with specific features. The existence of these heteromultimers in vivo, and their consequence for pest control should be investigated.
Experimental procedures
Molecular biology
Total RNA was isolated from whole V. destructor specimens or from honey bee brain, and first strand cDNAs were obtained as previously described (42). The 5′ and 3′ end regions of the different cDNAs were obtained by rapid amplification of cDNA ends–PCR using the GeneRacer kit (Thermo Fisher) and primers designed based on the sequences deposited in BeeBase, with the exception of VdesRDL2 for which we designed specific primers to obtain the 5′-end based on the predicted sequence obtained from transcriptomic data deposited in GenBankTM. The cDNA of amRDL (KJ485710) was cloned in the pBS-SK vector (Agilent). For the V. destructor subunits, full-length cDNAs covering the entire ORF were cloned in the pcDNA3.1(+) vector, with the alfalfa mosaic virus sequence immediately before the start codon and the 3′-UTR sequence of the X. laevis β-globin gene immediately after the stop codon. The cDNAs encoding VdesRDL1 and VdesRDL4 were cloned in-frame with the GFP in pEGFP-N1 vector (Clontech) to obtain RDL1-GFP and RDL4-GFP, and the cDNA of VdesRDL3 was cloned in-frame with mCherry in pmCherry-N1 (Clontech) to obtain RDL3-Cherry. The mutants VdesRDL1mut (S306A and M310T), VdesRDL4mut (A257S), and VdesLCCH3mut (A444S) were obtained with the GeneEditorTM in vitro site-directed mutagenesis system (Promega), following the manufacturer's instructions. Mutations and cDNA integrity were checked by sequencing the entire ORF. For X. laevis oocyte injection, cRNAs were obtained from linearized plasmids using the mMessage mMachine transcription kit (Thermo Fisher) following the manufacturer's instructions. The cRNA concentration was adjusted to 1 μg/μl. For co-injection of subunits, we prepared a mixture of cRNA at a weight/weight ratio of 1:1, unless otherwise indicated.
X. laevis oocyte preparation and injection
Preparation and injection of X. laevis oocytes were done as previously described (43). The oocytes were injected with ∼30 ng of cRNA. Injected oocytes were maintained at 19 °C in NDS (96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm Hepes, 2.5 mm sodium pyruvate, 0.05 mm gentamycin, pH 7.2, with NaOH) renewed daily.
Electrophysiology and data analysis
Expressed currents were recorded at room temperature using the two-electrode voltage clamp method 1–3 days after injection. Electrodes were pulled from borosilicate glass and filled with 3 m KCl. Oocytes were clamped at −60 mV, and GABA-gated currents were recorded with a Geneclamp 500 amplifier (Molecular Devices) and digitized with a Digidata 1200 converter (Molecular Devices) using Clampex software (Molecular Devices). The external solution, NDherg, (96 mm NaCl, 3 mm KCl, 0.5 mm CaCl2, 1 mm MgCl2, 5 mm Hepes, pH 7.4, with NaOH) was continuously perfused in the recording chamber at the rate of 1 ml/min. GABA (stock solution 500 mm in H2O), fipronil (stock solution 200 mm in DMSO), and picrotoxin (stock solution 10 mm in DMSO) were diluted in NDherg solution.
GABA concentration response curves were generated by challenging oocytes with increasing concentrations of GABA. Peak current amplitudes were plotted against GABA concentrations. The curves were fitted with a logistic function using Origin 6.0 (Microcal Software) to obtain the extrapolated maximal response that was used to normalize GABA response curves. Fipronil and picrotoxin inhibition curves were generated by inhibiting the response to a GABA concentration between its EC30 and EC50 for the tested receptor. Inhibition curves were normalized to the response induced by GABA in the absence of inhibitor. The concentration of GABA required to obtain 50% of the maximum response (EC50), the concentration of inhibitor required to inhibit 50% of the GABA response (IC50), and the Hill coefficient (nHill) were determined with a logistic function using Origin 6.0. The data are presented as the means ± S.E. of n individual oocytes. The statistical significance of the difference between data were determined using the nonpaired Student's t test. The reversal potential of the different receptors was measured in a solution made of 100 mm NaCl, 2 mm MgCl2, 5 mm Hepes, pH 7.4, with NaOH with a 400-ms-long ramp of voltage from −80 to +80 mV. For ion exchange experiments, 50, 75, 87.5, and 100% NaCl was replaced by sodium acetate for chloride exchange and by TEACl for sodium exchange. The differences between the reversal potential measured in the different solutions and that measured in control solution (Erev shift) were plotted against the chloride or sodium concentration.
Western blotting and immunoprecipitation experiments
At day 3 after injection of RDL1-GFP or RDL4-GFP cRNAs, ∼40 X. laevis oocytes were homogenized in 10 mm Tris-HCl, pH 7.5, 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, pH 8, and Complete protease inhibitor (Roche). A volume of lysate corresponding to approximately three oocytes were separated on 10% SDS-PAGE. Immunoblotting was performed with anti-GFP (1:5000, Sigma) and horseradish peroxidase-conjugated goat anti-rabbit (1:5000, Sigma) antibodies. HEK293 cells were transfected with RDL1-GFP alone or together with RDL3-Cherry using Lipofectamine (Thermo Fisher). At day 4 after transfection, the cells were harvested in radioimmune precipitation assay lysis buffer (Boster) with 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, pH 8, and Complete protease inhibitor. Cell lysates were incubated with a prewashed slurry of 20 μl of anti-GFP-Trap® A beads (Chromotek) at 4 °C for 2 h. Immunoprecipitates were recovered in 0.2 m glycine, pH 2.5. Cell lysates and immunoprecipitates were analyzed separately using anti-GFP (1:5000), anti-mCherry (1:5000, Thermo Fisher), anti-GAPDH (1:5000, Sigma), and horseradish peroxidase-conjugated goat anti-rabbit (1:5000), or goat anti-mouse (1:5000, Sigma) antibodies. Detection was performed with the Western Lightning Plus (Perkin Elmer) reagents.
Author contributions
C. M., M. F., L. B., M. C., C. C., R. M., M. R., J.-B. T., M. V., and P. C. investigation; P. C. writing-original draft; T. C. conceptualization; T. C. supervision.
This work was supported by INSERM, CNRS, the Université de Montpellier, and Agence Nationale de la Recherche Grant ANR-13-BSV7-0010 (bee channel). The authors declare that they have no conflicts of interest with the contents of this article.
- LBD
- ligand-binding domain.
References
- 1. Le Conte Y., Ellis M., and Ritter W. (2010) Varroa mites and honey bee health: can Varroa explain part of the colony losses? Apidologie 41, 353–363 10.1051/apido/2010017 [DOI] [Google Scholar]
- 2. Francis R. M., Nielsen S. L., and Kryger P. (2013) Varroa–virus interaction in collapsing honey bee colonies. PLoS One 8, e57540 10.1371/journal.pone.0057540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fairbrother A., Purdy J., Anderson T., and Fell R. (2014) Risks of neonicotinoid insecticides to honeybees. Environ. Toxicol. Chem. 33, 719–731 10.1002/etc.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hawthorne D. J., and Dively G. P. (2011) Killing them with kindness?: in-hive medications may inhibit xenobiotic efflux transporters and endanger honey bees. PLoS One 6, e26796 10.1371/journal.pone.0026796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Priestley C. M., Williamson E. M., Wafford K. A., and Sattelle D. B. (2003) Thymol, a constituent of thyme essential oil, is a positive allosteric modulator of human GABA A receptors and a homo-oligomeric GABA receptor from Drosophila melanogaster. Br. J. Pharmacol. 140, 1363–1372 10.1038/sj.bjp.0705542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casida J. E., and Durkin K. A. (2015) Novel GABA receptor pesticide targets. Pestic. Biochem. Physiol. 121, 22–30 10.1016/j.pestbp.2014.11.006 [DOI] [PubMed] [Google Scholar]
- 7. Choudhary A. F., Laycock I., and Wright G. A. (2012) γ-Aminobutyric acid receptor A-mediated inhibition in the honeybee's antennal lobe is necessary for the formation of configural olfactory percepts. Eur. J. Neurosci. 35, 1718–1724 10.1111/j.1460-9568.2012.08090.x [DOI] [PubMed] [Google Scholar]
- 8. Dupuis J. P., Bazelot M., Barbara G. S., Paute S., Gauthier M., and Raymond-Delpech V. (2010) Homomeric RDL and heteromeric RDL/LCCH3 GABA receptors in the honeybee antennal lobes: two candidates for inhibitory transmission in olfactory processing. J. Neurophysiol. 103, 458–468 10.1152/jn.00798.2009 [DOI] [PubMed] [Google Scholar]
- 9. Chung B. Y., Kilman V. L., Keath J. R., Pitman J. L., and Allada R. (2009) The GABAA receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr. Biol. 19, 386–390 10.1016/j.cub.2009.01.040,10.1016/j.sbi.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yuan Q., Song Y., Yang C.-H., Jan L. Y., and Jan Y. N. (2014) Female contact modulates male aggression via a sexually dimorphic GABAergic circuit in Drosophila. Nat. Neurosci. 17, 81–88 10.1038/ncb3082,10.1038/nn.3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jones A. K., and Sattelle D. B. (2006) The cys-loop ligand-gated ion channel superfamily of the honeybee, Apis mellifera. Invert. Neurosci. 6, 123–132 10.1007/s10158-006-0026-y [DOI] [PubMed] [Google Scholar]
- 12. Knipple D. C., and Soderlund D. M. (2010) The ligand-gated chloride channel gene family of Drosophila melanogaster. Pestic. Biochem. Physiol. 97, 140–148 10.1016/j.pestbp.2009.09.002 [DOI] [Google Scholar]
- 13. Nys M., Kesters D., and Ulens C. (2013) Structural insights into Cys-loop receptor function and ligand recognition. Biochem. Pharmacol. 86, 1042–1053 10.1016/j.bcp.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 14. Feyereisen R., Dermauw W., and Van Leeuwen T. (2015) Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic. Biochem. Physiol. 121, 61–77 10.1016/j.pestbp.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 15. Ffrench-Constant R. H., Rocheleau T. A., Steichen J. C., and Chalmers A. E. (1993) A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature 363, 449–451 10.1038/363449a0 [DOI] [PubMed] [Google Scholar]
- 16. Wolff M. A., and Wingate V. P. (1998) Characterization and comparative pharmacological studies of a functional γ-aminobutyric acid (GABA) receptor cloned from the tobacco budworm, Heliothis virescens. Invert. Neurosci. 3, 305–315 10.1007/BF02577690 [DOI] [PubMed] [Google Scholar]
- 17. Zheng Y., Priest B., Cully D. F., and Ludmerer S. W. (2003) RdlDv, a novel GABA-gated chloride channel gene from the American dog tick Dermacentor variabilis. Insect Biochem. Mol. Biol. 33, 595–599 10.1016/S0965-1748(03)00038-9 [DOI] [PubMed] [Google Scholar]
- 18. Nakao T., Hama M., Kawahara N., and Hirase K. (2012) Fipronil resistance in Sogatella furcifera: molecular cloning and functional expression of wild-type and mutant RDL GABA receptor subunits. J. Pestic. Sci. 37, 37–44 10.1584/jpestics.D11-018 [DOI] [Google Scholar]
- 19. Gisselmann G., Plonka J., Pusch H., and Hatt H. (2004) Drosophila melanogaster GRD and LCCH3 subunits form heteromultimeric GABA-gated cation channels. Br. J. Pharmacol. 142, 409–413 10.1038/sj.bjp.0705818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Price K. L., and Lummis S. C. (2014) An atypical residue in the pore of Varroa destructor GABA-activated RDL receptors affects picrotoxin block and thymol modulation. Insect Biochem. Mol. Biol. 55, 19–25 10.1016/j.ibmb.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taylor-Wells J., Hawkins J., Colombo C., Bermudez I., and Jones A. K. (2017) Cloning and functional expression of intracellular loop variants of the honey bee (Apis mellifera) RDL GABA receptor. Neurotoxicology 60, 207–213 10.1016/j.neuro.2016.06.007 [DOI] [PubMed] [Google Scholar]
- 22. Ashby J. A., McGonigle I. V., Price K. L., Cohen N., Comitani F., Dougherty D. A., Molteni C., and Lummis S. C. (2012) GABA binding to an insect GABA receptor: a molecular dynamics and mutagenesis study. Biophys. J. 103, 2071–2081 10.1016/j.bpj.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Comitani F., Limongelli V., and Molteni C. (2016) The free energy landscape of GABA binding to a pentameric ligand-gated ion channel and its disruption by mutations. J. Chem. Theory Comput. 12, 3398–3406 10.1021/acs.jctc.6b00303 [DOI] [PubMed] [Google Scholar]
- 24. Narusuye K., Nakao T., Abe R., Nagatomi Y., Hirase K., and Ozoe Y. (2007) Molecular cloning of a GABA receptor subunit from Laodelphax striatella (Fallén) and patch clamp analysis of the homo-oligomeric receptors expressed in a Drosophila cell line. Insect Mol. Biol. 16, 723–733 10.1111/j.1365-2583.2007.00766.x [DOI] [PubMed] [Google Scholar]
- 25. Zhang H. G., Lee H. J., Rocheleau T., Ffrench-Constant R. H., and Jackson M. B. (1995) Subunit composition determines picrotoxin and bicuculline sensitivity of Drosophila γ-aminobutyric acid receptors. Mol. Pharmacol. 48, 835–840 [PubMed] [Google Scholar]
- 26. Yu L.-L., Cui Y.-J., Lang G.-J., Zhang M.-Y., and Zhang C.-X. (2010) The ionotropic γ-aminobutyric acid receptor gene family of the silkworm, Bombyx mori. Genome 53, 688–697 10.1139/G10-056 [DOI] [PubMed] [Google Scholar]
- 27. Wei Q., Wu S.-F., and Gao C.-F. (2017) Molecular characterization and expression pattern of three GABA receptor-like subunits in the small brown planthopper Laodelphax striatellus (Hemiptera: Delphacidae). Pestic. Biochem. Physiol. 136, 34–40 10.1016/j.pestbp.2016.08.007 [DOI] [PubMed] [Google Scholar]
- 28. Yuan G., Gao W., Yang Y., and Wu Y. (2010) Molecular cloning, genomic structure, and genetic mapping of two Rdl-orthologous genes of GABA receptors in the diamondback moth, Plutella xylostella. Arch. Insect Biochem. Physiol. 74, 81–90 [DOI] [PubMed] [Google Scholar]
- 29. Sheng C.-W., Jia Z.-Q., Ozoe Y., Huang Q.-T., Han Z.-J., and Zhao C.-Q. (2018) Molecular cloning, spatiotemporal and functional expression of GABA receptor subunits RDL1 and RDL2 of the rice stem borer Chilo suppressalis. Insect Biochem. Mol. Biol. 94, 18–27 10.1016/j.ibmb.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 30. Dermauw W., Ilias A., Riga M., Tsagkarakou A., Grbić M., Tirry L., Van Leeuwen T., and Vontas J. (2012) The cys-loop ligand-gated ion channel gene family of Tetranychus urticae: implications for acaricide toxicology and a novel mutation associated with abamectin resistance. Insect Biochem. Mol. Biol. 42, 455–465 10.1016/j.ibmb.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 31. Dong X., Armstrong S. D., Xia D., Makepeace B. L., Darby A. C., and Kadowaki T. (2017) Draft genome of the honey bee ectoparasitic mite, Tropilaelaps mercedesae, is shaped by the parasitic life history. Gigascience 6, 1–17 , , , , , , , , , , , , , , , , , , , [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garrood W. T., Zimmer C. T., Gutbrod O., Lüke B., Williamson M. S., Bass C., Nauen R., and Emyr Davies T. G. (2017) Influence of the RDL A301S mutation in the brown planthopper Nilaparvata lugens on the activity of phenylpyrazole insecticides. Pestic. Biochem. Physiol. 142, 1–8 10.1016/j.pestbp.2017.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Remnant E. J., Morton C. J., Daborn P. J., Lumb C., Yang Y. T., Ng H. L., Parker M. W., and Batterham P. (2014) The role of Rdl in resistance to phenylpyrazoles in Drosophila melanogaster. Insect Biochem. Mol. Biol. 54, 11–21 10.1016/j.ibmb.2014.08.008 [DOI] [PubMed] [Google Scholar]
- 34. Nakao T. (2017) Mechanisms of resistance to insecticides targeting RDL GABA receptors in planthoppers. Neurotoxicology 60, 293–298 10.1016/j.neuro.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 35. Ozoe Y. (2013) γ-Aminobutyrate- and glutamate-gated chloride channels as targets of insecticides. Adv. Insect Phys. 44, 211–286 10.1016/B978-0-12-394389-7.00004-1 [DOI] [Google Scholar]
- 36. Wei Q., Wu S.-F., Niu C.-D., Yu H.-Y., Dong Y.-X., and Gao C.-F. (2015) Knockdown of the ionotropic γ-aminobutyric acid receptor (GABAR) rdl gene decreases fipronil susceptibility of the small brown planthopper, Laodelphax striatellus (Hemiptera: Delphacidae). Arch. Insect Biochem. Physiol. 88, 249–261 10.1002/arch.21232 [DOI] [PubMed] [Google Scholar]
- 37. Raimondo J. V., Richards B. A., and Woodin M. A. (2017) Neuronal chloride and excitability: the big impact of small changes. Curr. Opin. Neurobiol. 43, 35–42 10.1016/j.conb.2016.11.012 [DOI] [PubMed] [Google Scholar]
- 38. Nicholl G. C., Jawad A. K., Weymouth R., Zhang H., and Beg A. A. (2017) Pharmacological characterization of the excitatory “Cys-loop” GABA receptor family in Caenorhabditis elegans. Br. J. Pharmacol. 174, 781–795 10.1111/bph.13736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shimizu K., and Stopfer M. (2017) A population of projection neurons that inhibits the lateral horn but excites the antennal lobe through chemical synapses in Drosophila. Front. Neural Circuits 11, 30 10.3389/fncir.2017.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buckingham S. D., Biggin P. C., Sattelle B. M., Brown L. A., and Sattelle D. B. (2005) Insect GABA receptors: splicing, editing, and targeting by antiparasitics and insecticides. Mol. Pharmacol. 68, 942–951 10.1124/mol.105.015313 [DOI] [PubMed] [Google Scholar]
- 41. Aronstein K., Auld V., and Ffrench-Constant R. (1996) Distribution of two GABA receptor-like subunits in the Drosophila CNS. Invert. Neurosci. 2, 115–120 10.1007/BF02214114 [DOI] [PubMed] [Google Scholar]
- 42. Cens T., Rousset M., Collet C., Charreton M., Garnery L., Le Conte Y., Chahine M., Sandoz J.-C., and Charnet P. (2015) Molecular characterization and functional expression of the Apis mellifera voltage-dependent Ca2+ channels. Insect Biochem. Mol. Biol. 58, 12–27 10.1016/j.ibmb.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 43. Cens T., Mangoni M. E., Nargeot J., and Charnet P. (1996) Modulation of the alpha 1A Ca2+ channel by β subunits at physiological Ca2+ concentration. FEBS Lett. 391, 232–237 10.1016/0014-5793(96)00704-1 [DOI] [PubMed] [Google Scholar]