Abstract
A cascade of events leads to the development of microbial biofilm communities that are thought to be responsible for over 80% of infections in humans. However, not all surface-growing bacteria reside in a stationary biofilm state. Here, we have employed confocal Raman microscopy to analyze and compare variations in the alkyl quinolone (AQ) family of molecules during the transition between surface-attached motile-swarming and stationary biofilm communities. The AQs have been established previously as important to Pseudomonas aeruginosa biofilms, interspecies competition, and virulence. The AQ Pseudomonas quinolone signal (PQS) is also a known quorum-sensing signal. We detail spatial identification of AQ, PQS, and 2-alkyl-4-hydroxyquinoline N-oxide (AQNO) metabolites in both swarm and biofilm communities. We find that AQNO metabolites are abundant signatures in active swarming communities.
Keywords: Pseudomonas aeruginosa, swarming, biofilms, PQS, AQNO, confocal Raman microscopy
Introduction
Development of antibiotic tolerant biofilms is aided by the motility appendage actions of many motile pathogenic bacteria. For the opportunistic pathogen, Pseudomonas aeruginosa, both the polar flagellum and type IV pili contribute to biofilm development in multiple ways. In addition to participating in surface attachment, these appendages confer modes of surface motility that promote exploration and colonization. During these periods of surface motility, P. aeruginosa is generally in a state with low levels of intracellular c-di-GMP1,2 even though these bacteria may be present at high cell density and experiencing sufficiently elevated levels of acyl homoserine lactone quorum-sensing signals. Thus, these quorum sensing-induced populations are spreading as groups and are not yet producing the matrix exopolysaccharides Pel and Psl that have been demonstrated to promote antibiotic tolerance by biofilms.3-7
During the surface exploration mode termed swarming, P. aeruginosa use their flagella to move in high cell density groups over semi-solid surfaces. Optimal swarming also requires self-production of rhamnolipid, which is regulated through the rhl quorum-sensing cascade. Several studies have shown that swarming P. aeruginosa cells exhibit tolerance to antibiotics by upregulation of several factors in addition to those exhibited by sessile biofilm cells.8-13
We recently found that while the antibiotics tobramycin and carbenicillin both elicit a swarm phenotype, the responses are quite distinct.14 In connection with these swarm phenotype differences, we found that production of alkyl quinolone (AQ) molecules varied in response to tobramycin but not carbenicillin. There are over 50 known P. aeruginosa AQs that fall within three primary subclasses: (1) 2-alkyl-4(1H)-quinolones such as 2-heptyl-4(1H)-quinolone (HHQ) and 2-nonyl-4(1H)-quinolone (NHQ); (2) 2-alkyl-3-hydroxy-4(1H)-quinolones, such as 2-heptyl-3-hydroxy-4(1H)-quinolone (Pseudomonas quinolone signal; PQS) and 2-heptyl-3-nonyl-4(1H)-quinolone (C9-PQS); and (3) 2-alkyl-4-hydroxyquinoline N-oxides (AQNOs) such as 2-heptyl-4-hydroxyquinoline N-oxide (HQNO) and 2-nonyl-4-hydroxyquinoline N-oxide (NQNO).15 PQS, specifically, is a known quorum-sensing molecule for P. aeruginosa, for which the signaling cascade has interconnection with the las and rhl cascades.16-18
While previous reports have indicated that PQS may be part of a universal stress response elicited by P. aeruginosa,19,20 we recently found that tobramycin elicited a PQS response while carbenicillin did not.14 We employed a multiplexed chemical imaging strategy to show that P. aeruginosa secretes markedly distinct PQS and AQNO profiles in response to tobramycin exposure and influences these swarms on different spatial scales. The distribution of AQs varied by several orders of magnitude within the same swarm. More notably, our results suggest that multiple intercellular signals acting on different spatial scales are possible from one common cue.
Here, we detail more general AQ signatures of P. aeruginosa during the transition to surface growth and also between motile swarms to stationary biofilm communities. These experiments were conducted entirely in the absence of antibiotic exposure. We find that PQS signatures are abundant and readily available within stationary biofilm communities within 24 hours, while AQNO signatures can be used to distinguish active surface swarming cells from both planktonic and biofilm phenotypes.
Materials and Methods
Bacterial strains and culturing conditions
Pseudomonas aeruginosa PA14 was used for all experiments. Overnight planktonic cultures were grown by inoculation of isolated bacterial colonies from Lysogeny Broth (LB) agar into 6 mL of modified FAB minimal medium supplemented with 30 mM glucose (<16-20 hours) at 37°C, 240 r/min. Overnight cultures were normalized to an OD600 nm of 0.5 in FAB broth (no glucose), and 1 µL spots were used to inoculate swarm and biofilm assays.
Swarm/biofilm assays
Swarm motility or surface biofilm plate assays were performed in 60-mm diameter petri dishes containing 7.5 mL of modified FAB culture media supplemented with 12 mM glucose and solidified with 0.45% agar for swarm assays and 1.5% agar for biofilm assays (Noble agar, Sigma, St. Louis, MO).21 Assay plates were incubated inverted at 30°C, 85% RH. Whole plate light images of all plates were acquired using a Nikon D3300 camera (Nikon, Melville, NY) with an 18-55 mm zoom f/3.5-5.6G VR II lens.
CRM imaging and analysis
Confocal Raman microspectroscopy (CRM) imaging was performed as previously described.14,21 Briefly, Raman images were acquired using a 40× objective (NA = 0.6), with a full Raman spectrum being obtained at each image pixel (150 × 150, 100 × 100, or 80 × 80 pixel) over a selected region on the swarm sample with an integration time of 100 ms per spectrum. The AQ standards were dissolved in either high-performance liquid chromatography (HPLC)-grade ethanol or methanol (Sigma, St. Louis, MO), then deposited and air-dried on clean Si wafers for CRM analysis. MATLAB was used to perform principal component analysis (PCA) using previously described custom scripts22 to detect chemical variations in the samples. Cellular components were identified by the presence of the thymine ring stretch in DNA (745 cm−1), and to the C-N stretch in proteins, C-O stretch in lipids (1127 cm−1), amide III stretch from proteins (1311 cm−1), and ring stretch in adenine and guanine attributed to presence of DNA/RNA and amide II stretch in proteins (1583 cm−1).23,24 In biological samples, the PQS subclass is associated with features at 1157, 1372, 1466, and 1654 cm−1, and the AQNO subclass with features in 715, 1205, 1359, and 1508 cm−1.14,21,24 Spectral positions are reproducible to ±3 cm−1.
Results and Discussion
We find an abundance of AQs in surface-growing cultures of P. aeruginosa and detail distinct signatures between swarms and stationary biofilm communities. Using nondestructive CRM, we obtained spatial chemical image profiles of P. aeruginosa growing on nutrient agar using our established protocols.14 Here, biofilm assays were performed by inoculating P. aeruginosa on 1.5% agar, which exhibited minimal spreading and promoted development of a stationary biofilm phenotype. Swarm assays were performed by inoculating P. aeruginosa on 0.45% agar, which promoted characteristic swarming motility over time. The same minimal medium (FAB-glucose) was used for all experiments, thus nutrient composition was not a factor for discerning phenotype in our results.
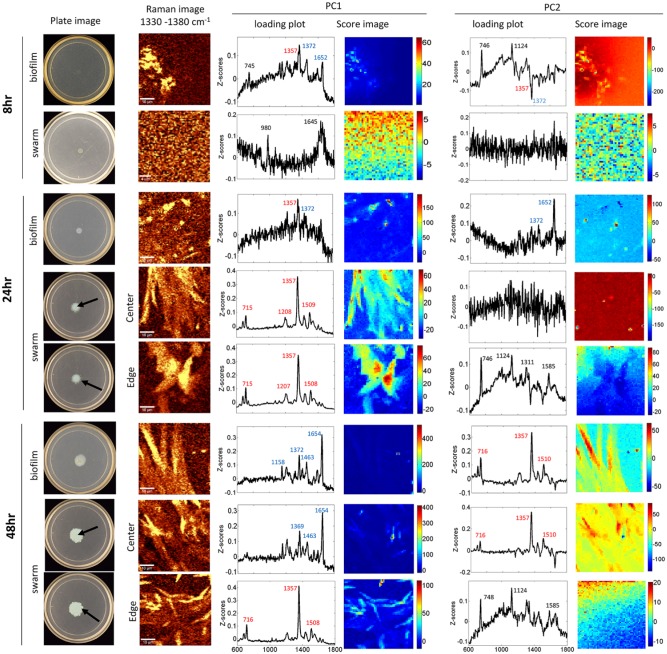
Confocal Raman microspectroscopy imaging of both 0 hour swarm and 0 hour biofilm samples (not shown) exhibit features, e.g. at 745, 1137, and 1311 cm−1, characteristic of planktonic bacterial cells. By 8 hours post-inoculation, the biofilm spectra show features of both PQS (1372 cm−1) and AQNO (1357 cm−1), as evidenced by both the first (PC1) and second (PC2) principal component loading plots (Figure 1), along with features assigned to planktonic cells (745 cm−1). The relatively low z-score values, compared to later time points, reflect the low concentration of AQ molecules and the heterogeneous environment occupied by bacterial cells at this early stage of development. Confocal Raman microspectroscopy images of swarm samples at 8 hours post-inoculation do not exhibit features of AQs, instead being dominated by medium-derived signals, such as those at 980 and 1645 cm−1.
Figure 1.
Pseudomonas aeruginosa biofilms and swarms present different chemical signals at different time points. The plate images shows the swarming motility and biofilm colonies on agar-based assays incubated for 8, 24, and 48 hours. Plate assays were analyzed directly by combining CRM (Raman image includes both PQS and AQNO subclasses) and PCA analysis (loading plots and score images for PC1 and PC2) to identify chemical features within the samples. Representative CRM results (n ⩾ 3) collected from biofilm samples and swarm regions both near and away from inoculation center are shown. Scale bars on Raman images represent 10 µm. Loading plots for PC1 and PC2 include features corresponding to tabulated features from Raman spectra of cellular and matrix components (black), PQS/C9-PQS (blue), and AQNOs (HQNO/NQNO; red). Score images of PC1 and PC2 show the distribution of each of the principal components.
By 24 hours post-inoculation, the AQ attributes are evident in both swarms and biofilms. Biofilm spectra at this time point exhibit quinolone stretch features indicative of PQS at 1372 cm−1 and a co-secreted peptide-related band at 1652 cm−1 in both the first (PC1) and second (PC2) principal components (Figure 1). By 24 hours, the swarming phenotype is readily apparent at the macroscale, and we observe strong signal features at 715, 1205, 1359, and 1508 cm−1, characteristic of the N-oxide quinolones AQNO (both C7 and C9 congeners), in PC1 at both the swarm edge and the swarm center at this time point. PC2 features for 24 hours swarms are not indicative of AQs but are attributable to features of individual P. aeruginosa cells14,24 at the swarm edge. Importantly, the 1372 cm−1 feature characteristic of PQS appears in PC1 of the biofilm, but not the swarm. At 24 hours post-inoculation, AQNO are produced by both biofilm and swarming cells. Interestingly, when comparing the 24 hours biofilm and swarm samples, the production of AQNO and PQS is consistent across the entire biofilm. However, the swarm sample, already at 24 hours, shows clear differences between the center and edge positions. The center is dominated by AQNO features, while the edge (PC2) still shows a significant contribution from planktonic cells.
At 48 hours post-inoculation, the biofilm spectra exhibit multiple features—1158, 1372, 1463, and 1654 cm−1, in the PC1 loading plot that can be attributed to PQS (Figure 1). The biofilm sample at 48 hours also shows features of AQNO (PC2). The presence of both PQS and AQNO is confirmed across the entire biofilm sample. Interestingly, CRM of the swarm center at 48 hours strongly resembles the AQ profile of the biofilm with features characteristic of PQS and the co-secreted peptide in PC1 and features of AQNO in PC2. In contrast, the swarm edge continues to exhibit features of AQNO in PC1 and cellular components in PC2. We interpret these results as an indication that bacteria, in the center of the swarm zone, have ceased surface motility and have transitioned to a stationary biofilm state while cells at the swarm edge continue to expand over the swarm plate surface.
Overall, we find that surface-growing P. aeruginosa is readily distinguished from planktonic cultures by probing for the AQs. Planktonic cultures inoculated onto agar that promoted stationary biofilm colony growth exhibit both PQS and AQNO signatures within 8 hours incubation even when minimal growth can be observed by eye at the macroscale. After 24 hours, when biofilm colonies are apparent on plate assays, both PQS and AQNO signatures continue to be detected, with additional features of PQS becoming apparent. When robust growth is apparent by 48 hours, the PQS signature of these biofilm colonies is dominant by our spatial CRM analysis.
Swarming cells also showed a mixture of PQS and AQNO features over the 48-hours sampling period; however, areas containing the most active swarming cells showed features solely attributable to AQNO or to individual cells. While cells inoculated onto swarm agar that do not exhibit a robust swarm phenotype (8 hours) do not exhibit an AQ signature, by 24 hours when swarm tendrils have formed, these groups exhibit multiple features of AQNOs. This expression of AQNO continues through the 48 hours time point for these swarms. However, at this time point, the swarm center and swarm edge can be distinguished by the presence of PQS features in PC1 of the swarm center—indicating that these swarm center cells have begun (or completed) their transition to a stationary biofilm state.
It was readily apparent that all surface-growing cultures could be readily distinguished from planktonic cultures by the increase of AQs generally. It is tempting to oversimplify these results to conclude that PQS indicates stationary biofilms and AQNO indicates surface motile swarming. However, all biofilm samples analyzed by CRM also exhibited characteristic AQNO features. In addition, our swarm results are somewhat in contrast to a prior report by Ha et al25 that showed HHQ as an inhibitor of swarm motility during growth on arginine. It is likely that the modulation of AQNO on surfaces response to multiple environmental cues, only a few of which have been cataloged. Certainly, further work is required to refine the spatial scale(s) of AQ production by P. aeruginosa growing on surfaces with the intent of further understanding the transition from planktonic to surface-colonized growth and the interpretation of these surface-growing cells to their surrounding environment.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was completed with support from the National Institutes of Health (grant no.: R01AI113219) to J.D.S. and P.W.B.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: NMS and TC contributed equally to this manuscript.
ORCID iDs: Tianyuan Cao  https://orcid.org/0000-0002-5854-1972
https://orcid.org/0000-0002-5854-1972
Joshua D. Shrout  https://orcid.org/0000-0001-9509-2187
https://orcid.org/0000-0001-9509-2187
References
- 1. Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:8165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chung IY, Choi KB, Heo YJ, Cho YH. Effect of PEL exopolysaccharide on the wspF mutant phenotypes in Pseudomonas aeruginosa PA14. J Microbiol Biotechnol. 2008;18:1227-1234. [PubMed] [Google Scholar]
- 4. Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of Psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ma L, Lu H, Sprinkle A, Parsek MR, Wozniak DJ. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J Bacteriol. 2007;189:8353-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Starkey M, Hickman JH, Ma LY, et al. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191:3492-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Allison DG, Matthews MJ. Effect of polysaccharide interactions on antibiotic susceptibility of Pseudomonas aeruginosa. J Appl Bacteriol. 1992;73:484-488. [DOI] [PubMed] [Google Scholar]
- 8. Overhage J, Bains M, Brazas MD, Hancock RE. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol. 2008;190:2671-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lai S, Tremblay J, Déziel E. Swarming motility: a multicellular behaviour conferring antimicrobial resistance. Environ Microbiol. 2009;11:126-136. [DOI] [PubMed] [Google Scholar]
- 10. Yeung ATY, Torfs ECW, Jamshidi F, et al. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol. 2009;191:5592-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zimelis VM, Jackson GG. Activity of aminoglycoside antibiotics against Pseudomonas aeruginosa: specificity and site of calcium and magnesium antagonism. J Infect Dis. 1973;127:663-669. [DOI] [PubMed] [Google Scholar]
- 12. Butler MT, Wang Q, Harshey RM. Cell density and mobility protect swarming bacteria against antibiotics. Proc Nat Acad Sci U S A. 2010;107:3776-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Merritt JH, Brothers KM, Kuchma SL, O’Toole GA. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol. 2007;189:8154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morales-Soto N, Dunham SJB, Baig NF, et al. Spatially dependent alkyl quinolone signaling responses to antibiotics in Pseudomonas aeruginosa swarms. J Biol Chem. 2018;293:9544-9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lépine F, Milot S, Déziel E, He J, Rahme LG. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Amer Soc Mass Spec. 2004;15:862-869. [DOI] [PubMed] [Google Scholar]
- 16. Pesci EC, Milbank JB, Pearson JP, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Nat Acad Sci U S A. 1999;96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wade DS, Calfee MW, Rocha ER, et al. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005;187:4372-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diggle SP, Winzer K, Chhabra SR, Worrall KE, Camara M, Williams P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50:29-43. [DOI] [PubMed] [Google Scholar]
- 19. Häussler S, Becker T. The Pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 2008;4:e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cummins J, Reen FJ, Baysse C, Mooij MJ, O’Gara F. Subinhibitory concentrations of the cationic antimicrobial peptide colistin induce the pseudomonas quinolone signal in Pseudomonas aeruginosa. Microbiology. 2009;155:2826-2837. [DOI] [PubMed] [Google Scholar]
- 21. Baig NF, Dunham SJ, Morales-Soto N, Shrout JD, Sweedler JV, Bohn PW. Multimodal chemical imaging of molecular messengers in emerging Pseudomonas aeruginosa bacterial communities. Analyst. 2015;140:6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ahlf DR, Masyuko RN, Hummon AB, Bohn PW. Correlated mass spectrometry imaging and confocal Raman microscopy for studies of three-dimensional cell culture sections. Analyst. 2014;139:4578-4585. [DOI] [PubMed] [Google Scholar]
- 23. Maquelin K, Kirschner C, Choo-Smith LP, et al. Identification of medically relevant microorganisms by vibrational spectroscopy. J Microbiol Meth. 2002;51:255-271. [DOI] [PubMed] [Google Scholar]
- 24. Masyuko RN, Lanni EJ, Driscoll CM, Shrout JD, Sweedler JV, Bohn PW. Spatial organization of Pseudomonas aeruginosa biofilms probed by combined matrix-assisted laser desorption ionization mass spectrometry and confocal Raman microscopy. Analyst. 2014;139:5700-5708. [DOI] [PubMed] [Google Scholar]
- 25. Ha D-G, Merritt JH, Hampton TH, et al. 2-Heptyl-4-Quinolone, a precursor of the Pseudomonas quinolone signal molecule, modulates swarming motility in Pseudomonas aeruginosa. J Bacteriol. 2011;193:6770-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]