Abstract
The complexity of the human sense of smell is increasingly reflected in complex and high-dimensional data, which opens opportunities for data-driven approaches that complement hypothesis-driven research. Contemporary developments in computational and data science, with its currently most popular implementation as machine learning, facilitate complex data-driven research approaches. The use of machine learning in human olfactory research included major approaches comprising 1) the study of the physiology of pattern-based odor detection and recognition processes, 2) pattern recognition in olfactory phenotypes, 3) the development of complex disease biomarkers including olfactory features, 4) odor prediction from physico-chemical properties of volatile molecules, and 5) knowledge discovery in publicly available big databases. A limited set of unsupervised and supervised machine-learned methods has been used in these projects, however, the increasing use of contemporary methods of computational science is reflected in a growing number of reports employing machine learning for human olfactory research. This review provides key concepts of machine learning and summarizes current applications on human olfactory data.
Keywords: bioinformatics, data-driven research, data science, human olfaction, machine learning
Introduction
Human olfaction comprises the chemosensory modality dedicated to detecting low concentrations of airborne and volatile chemical substances (Ache and Young 2005). It is based on a complex molecular network of approximately 400 G-protein-coupled olfactory receptors (Niimura 2009) enabling detecting and discriminating thousands of odorants (Buck and Axel 1991; Menashe and Lancet 2006) by pattern recognition (Holley et al. 1974; Malnic et al. 1999; Ache and Young 2005), and many further structures such as enzymes, ion channels, protein kinases, neurotrophins, and further G-protein-coupled receptors (Lin et al. 2004). Information from the olfactory epithelium in the nasal cavity is transmitted via the olfactory bulbs to a range of projection areas in the brain, e.g., the piriform, entorhinal and orbito-frontal cortices, the amygdalae, and the thalamus (Gottfried 2006). Modulators of human olfactory function include genetics (Keller et al. 2007), sex and hormonal status (Doty and Cameron 2009), age (Doty, Shaman, Applebaum, et al. 1984), environmental factors (Schwartz et al. 1989) including drugs (Doty and Bromley 2004; Lötsch et al. 2012, 2015), and neurological diseases (Serby et al. 1985; Quinn et al. 1987; Peters et al. 2003; Albers et al. 2006; Lutterotti et al. 2011).
While different facets of human olfaction have been accessible for successful classical biomedical research approaches, its complexity is increasingly reflected in high-dimensional data, which opens opportunities for data-driven approaches that complement hypothesis-driven research. Contemporary developments in computational and data science (President’s Information Technology Advisory 2005) facilitate complex data-driven research approaches. Machine learning is currently the most popular application of artificial intelligence (AI) and can be referred to as a set of methods, that can automatically detect patterns in data and then use the uncovered patterns to predict or classify future data, to observe structures such as subgroups in the data or to extract information from the data suitable to derive new knowledge (Murphy 2012; Dhar 2013; Chollet and Allaire 2018).
Together with (bio-)statistics, machine learning aims at learning from data. However, while statistics can be regarded as a branch of mathematics, machine learning has developed from computer science and aims at learning from data without the immediate necessity of previous knowledge. In contrast to statistics which focuses at analyzing the probabilities of observations given a known underlying data distribution, machine learning focuses more on the optimization and performance of algorithms (Shalev-Shwartz and Ben-David 2014). The present review introduces some key concepts of machine learning to an audience of biomedical researchers interested in the human sense of smell. Several methods that have been applied in this field will be summarized, however, without aiming at replacing textbooks of machine learning (Murphy 2012; Dhar 2013).
Human olfactory research involving machine learning
A literature search was conducted in PubMed at https://www.ncbi.nlm.nih.gov/pubmed on August, 2018, for “(machine-learn* OR machine learn* OR support* vector machines OR svm OR naive bayes OR bayes OR random forest* OR knn OR k nearest neighbor* OR k-nearest neighbor OR adaptive boosting OR boosting OR boosted tree* OR decision tree* OR deep learning OR artificial neural network*) AND (smell OR olfact*) AND (human OR patient OR volunt*) NOT review[Publication Type]”. After eliminating papers mentioning smell but either not further addressing it or using olfaction related information only as a technical sample data set, and after removing a report of a study in mice mentioning the translational possibility without providing it, this search produced 18 results published between 2005 and 2018 (Table 1). Further publications were obtained by explicit searches for computer aided research in human olfaction, such as the use of self-organizing maps in 2003 (Madany Mamlouk et al. 2003) or an attempt to determine the interaction of volatile molecules with human olfactory receptors (Sanz et al. 2008).
Table 1.
Reports of human olfactory research where machine-learned methods were used
| Olfactory context | Analyzed problem | Machine-learning methods of main data analysis | Ref. |
|---|---|---|---|
| Physiology of pattern-based odor detection and recognition | Relationship between odorant response and mutations in olfactory receptors | Naive Bayes, neural networks, SVM, kNN, meta learning, decision trees (CART) | (Gromiha et al. 2012) |
| Odor recognition and discrimination | Gnostic fields as a sub-neuronal network derived algorithm | (Kanan 2013) | |
| Ordering olfactory stimuli according to descriptors of odor perception | Multidimensional scaling and self-organizing maps | (Madany Mamlouk et al. 2003) | |
| Prediction of the affective component of an odor from EEG-derived responses to control olfactory stimulation | Principal component analysis-based feature selection, linear discriminant analysis-based classifier | (Lanata et al. 2016) | |
| Prediction of personalized olfactory perception | RF classifier followed by Pearson’s correlation | (Li et al. 2018) | |
| Prediction of the activity of chemicals for a given odorant receptor | SVM algorithm | (Bushdid et al. 2018) | |
| Pattern recognition inolfactory phenotypes | Prediction of olfactory diagnosis and underlying etiologies from olfactory subtest results | Emergent self-organized (Kohonen) maps (ESOM) | (Lötsch et al. 2016) |
| Determination of pattern and types of odor affected in Alzheimer’s disease | RF with recursive feature elimination algorithm (RF-RFE) | (Velayudhan et al. 2015) | |
| Pattern recognition by construction of an olfactory bionic model and a 3-layered cortical model, mimicking the main features of the olfactory system | Three artificial neural networks (ANNs): back-propagation network, SVM classifier, and a radial basis function classifier | (Gonzalez et al. 2010) | |
| Olfactory acuity as diagnostic biomarker | Early Parkinson diagnosis from multimodal features including olfaction | Naive Bayes classifier, logistic regression, adaptive boosted trees, RFs, and SVM | (Prashanth et al. 2016) (Prashanth et al. 2014) |
| Diagnosis of Parkinson’s disease from olfactory phenotypes | SVM, linear discriminant analysis | (Gerkin et al. 2017) | |
| Feature selection for predicting rapid progression of Parkinson’s disease using public data from the Parkinson’s Progression Markers Initiative, including olfactory parameters | Feature selection using the so-called wrapper approach with decision tree and naive Bayes based methods, classification using the C4.5 decision tree algorithm | (Tsiouris et al. 2017) | |
| Odor identification as diagnostic tool for Parkinson’s disease. | Random-forests classifier | (Casjens et al. 2013) | |
| Odor recognition from physico-chemical properties of volatile molecules (electronic noses) | Prediction of odors from molecular properties of odorant molecular separated by means of gas chromatography | Feature selection using RF, classification using RF, SVM, extreme learning machines | (Shang et al. 2017) |
| Prediction of olfactory perception from chemical features of odor molecules | Regularized linear models, random-forests | (Keller et al. 2017) | |
| Relationships between molecular structure and perceived odor quality of ligands for a human olfactory receptor. | 3D pharmacophore-based molecular modeling techniques | (Sanz et al. 2008) | |
| Evaluation of the applicability of composite odors | ANN | (Wagner et al. 2006) | |
| Application of Sensory Evaluation by an electronic nose for quality detection in citrus fruits | RF-based on bootstrap sampling | (Qiu and Wang 2015) | |
| Knowledge discovery in publicly available big databases. | Biological roles exerted by the genes expressed in the human olfactory bulb | Over-representation analysis | (Lötsch et al. 2014) |
The list has been obtained from a PubMed search at https://www.ncbi.nlm.nih.gov/pubmed on 21 September 2018, for “(machine-learn* OR machine learn* OR support* vector machines OR svm OR naive bayes OR bayes OR random forest* OR knn OR k nearest neighbor* OR k-nearest neighbor* OR adaptive boosting OR boosting OR boosted tree* OR decision tree* OR deep learning OR artificial neural network*) AND (smell OR olfact*) AND (human OR patient OR volunt*) NOT review[Publication Type]. The search obtained 57 hits, followed by data cleaning for reports where the focus was non-olfactory research such as using olfactory data as an example for method validation.
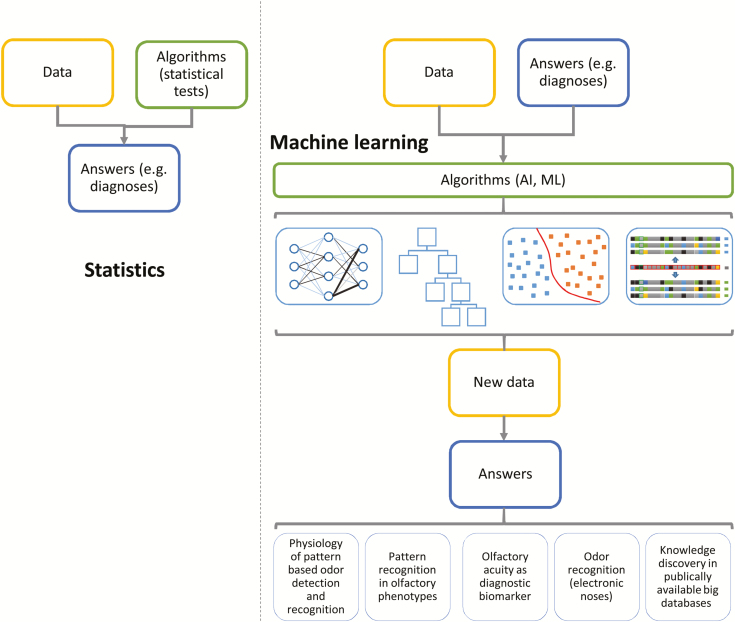
The use of machine learning in human olfactory research included several major approaches (Figure 1) comprising 1) the study of the physiology of pattern-based odor detection and recognition processes, 2) pattern recognition in olfactory phenotypes, 3) the development of complex disease biomarkers including olfactory features, 4) odor prediction from physico-chemical properties of volatile molecules such as electronic noses, and 5) knowledge discovery in publicly available big databases. A limited set of machine-learned methods has been used in these projects, which will be shortly described in the following.
Figure 1.
Overview about approaches to data processing, pursed either by statistics (left part) or by machine learning (right part). As explained previously (Chollet and Allaire 2018), statistics applies preselected rules, mathematical methods, or statistical algorithms to data with the aim to obtain an answer about a preformulated hypothesis such as a difference in a parameter among diagnostic groups of subjects. By contrast, in typical machine-learning tasks data are provided together with the answers such as the membership in a diagnostic group with the aim to obtain rules or algorithms that can provide the diagnosis group membership from new data where this is unknown yet. Such artificial intelligence or machine learning–based algorithms can take several different forms. The icons in the third line of the right part of the figure symbolize respective typical machine-learning methods, i.e., multilayer neuronal networks, decision tree–based algorithms, algorithms such as SVM that separate the classes by placing a hyperplane between them, or prototype-based algorithms such as k nearest neighbors that compare the feature vectors carried by a case with those carried by other cases and assign the class on the basis of the classes to which cases with most similar feature vectors belong. In human olfactory research, machine-learned algorithms have been applied to obtain answers that can be classified into 5 groups (bottom line of the right part of the figure; Table 1).
Preliminary knowledge of machine-learning algorithms
Machine learning evolved from computer science, with the start perhaps marked by Turing who proposed an experiment where 2 players, who can either be human or artificial, try to convince a human third player, that they are also humans (Turing 1950). If the, human, third player cannot name the AI among his/her counter players, then the AI has passed the so-called Turing test. From there, AI and machine learning, which is the currently most popular subtype of AI, have developed at increasing speed. Major steps included the creation of the first neural network called the perceptron (Rosenblatt 1958), the proposal of the nearest neighbors-based classification (Cover and Hart 1967), the development of self-organizing topologically correct feature maps (Kohonen 1982) or the description of the random forest (RF) ensemble learning algorithm (Ho 1995; Breiman 2001).
Although machine learning and statistics work in concert, they differ fundamentally. As elaborated elsewhere (Chollet and Allaire 2018), in statistics, data together with rules to analyze them are processed to obtain answers about hypotheses, such as significant differences of a diagnostic marker between clinical diagnoses (Figure 1). By contrast, in machine learning, data and answers, the latter being for example clinical diagnoses, are typically analyzed to obtain rules or trained algorithms, and these rules or trained algorithms are then applied to new data to obtain the correct answer or diagnosis (Figure 1). This describes a main use of machine-learning technique for 1) classification, i.e., the prediction of a class membership of a case from collected information. An example is diagnosing Parkinson’s disease by assigning a subject to the healthy or the patient class from the individual performance in olfactory tests.
Machine-learning techniques are further used for 2) data structure detection aiming for example at finding a group structure of distinct phenotypes among a cohort. In contrast to classification, a specific and already known membership or diagnosis plays no immediate role in this task. By contrast, it is the task of data structure detection to provide hints at possible novel classes or diagnostic groups. Machine-learning techniques can also be used for 3) knowledge discovery. This can be used in 2 different scenarios. The most frequent application of machine-learned knowledge discovery is the discovery of new knowledge from large data sets stored in knowledge bases. For example, combining the genes expression pattern in the olfactory bulb with the worldwide acquired knowledge about the function of all genes can create new knowledge, e.g., can point at neurogenesis in the human olfactory bulb (Lötsch et al. 2014). However, differing from this use, the classification ability of machine-learned algorithms is increasingly used for data analysis and knowledge discovery rather than for building diagnostic tools. The idea behind this use is that if an algorithm can be trained with information, consisting for example of clinical symptoms or markers, to provide a more accurate classification, or clinical diagnosis, than obtained by guessing, then the acquired parameters are relevant to the disease (Kringel et al. 2018). This complements classical statistical approaches to the data.
Machine learning can be applied in a supervised or unsupervised fashion. Supervised learning means that while an algorithm is being trained to learn a correct class assignment from a set of parameters, such as to make the correct diagnosis from clinical and laboratory information, its success is supervised as the information about the correct diagnose is available. Subsequently, the trained algorithm will be able to perform the correct class assignment, e.g., to make the correct diagnosis in new and formerly unseen cases, if it is provided with clinical or laboratory data similar to those on which it had been trained. Thus, the data space addressed in supervised learning is split into an input space that comprises vectors with different parameters (variables, features (Guyon et al. 2003)) acquired from cases (subjects), which belong to the output classes that can be a clinical phenotype, an olfactory diagnosis or any other group criterion applied to a subject.
Supervised machine learning has been used in human olfactory research with a preference for a few main methods. Among them, naive Bayesian classifiers provide the probability of a data point being assigned to a specific class calculated by application of the Bayes’ theorem (Bayes and Price 1763). Frequently, decision tree–based methods have been implemented in different ways. In decision tree methods (Loh 2014), an algorithm is created with conditions on variables (parameters) as vertices and classes (e.g., diagnoses) as leaves. Tree structured rule-based classifiers analyze ordered variables , such as the results of olfactory subtests, by recursively splitting the data at each node into children nodes, starting at the root node (Quinlan 1986). During learning, the splits are modified such that misclassification is minimized. By contrast, in RF (Ho 1995; Breiman 2001), which also uses decision trees, the splits are set to minimize misclassification but just set randomly along the range of the respective parameters. Many of the randomly split simple trees are created from the so-called RF. The classification is obtained as the majority vote for class membership provided by many decision trees, i.e., RF uses collective decisions or ensemble learning for classification.
Prototype-based classifiers are based on the learning of typical properties of members of a class such as sets of clinical or laboratory parameters. For example, the k-nearest neighbor (kNN) classifier (Cover and Hart 1967) belongs to the most frequently used algorithms in data science while it is one of the most basic methods in machine learning. During kNN classifier training, the entire labeled training data set, i.e., data for which the class membership is known, is stored. A test case, i.e., a new unlabeled case for which the class membership is unknown, is placed in the feature space near the training cases at the smallest high distance in the high-dimensional space. The test case receives the class label according to the majority vote of the class labels of the k training cases closest to it.
A component early introduced in machine-learning algorithms are so-called artificial neurons, which can be arranged to form neuronal networks (Rosenblatt 1958). A perceptron is built from artificial neurons arranged in one to several successive layers. The neurons are provided with several inputs, a processing level and an output level, which connects them to other artificial neurons. Each neuron sums up its weighed inputs determining its response. The combined responses of all neurons in the network determine the class association of a data point. During learning, weights are adapted from initial random values in a way that the activation is shifted toward the desired output, i.e., the learning of a perceptron takes place by adjusting the weighting functions of each neuron. By contrast, in support vector machines (SVM) geometrical and statistical approaches are employed for finding an optimum decision surface in the high-dimensional feature space, i.e., a hyperplane, that can separate the neurons carrying data of one class from those carrying data belonging to another class (Cortes and Vapnik 1995).
While in supervised learning the goal of the training is to enable the algorithm to find the correct class association of a case, such as the right diagnosis, i.e., to map input information to output classes, in unsupervised learning the output classes are missing and hence, the goal of the training cannot be enabling the algorithm to find a correct class assignment. The data space addressed in supervised learning consists only of the input space that comprises vectors with different parameters (variables, features (Guyon et al. 2003)) acquired from cases (subjects). Output classes to which the cases belong are not known or not immediately relevant in unsupervised learning. Instead, unsupervised methods aim at finding “interesting” structures, which are structures that according to topical experts merit further exploration. Such structures can, for example, hint at subgroups among the subjects not hypothesized before (clusters) or can be associated with clinical phenotypes.
Unsupervised machine-learning methods have been used for structure detection in human olfactory research data (Table 1). While principal component analysis is occasionally named among them, it dates back to 1901 which is too early when timing the beginning of machine learning at the emergence of computers science in the middle of last century (Turing 1950). Among unsupervised methods further addressed in the present review of machine-learned methods in olfactory research figure self-organizing maps (SOM) of artificial neurons (Kohonen 1982, 1995). SOM are based on a projection of high-dimensional data points onto a 2-dimensional self-organizing network consisting of a grid of neurons in a way that large or small distances in the high-dimensional space are preserved as large or small distances, respectively, on the map. Each neuron holds, in addition to a position vector on the 2-dimensional grid, a further vector carrying “weights” of the same dimensions as the dimensions of the input data. The weights are initially randomly drawn from the range of the data variables During the learning phase of the SOM, the weights are adapted to the data so that finally neurons on the grid carry data vectors that are most similar to a case’s data vector. To obtain clusters, the so-called U-matrix (Ultsch 2003) can be added that displays the distances between neurons in the high-dimensional space as a third dimension onto the 2-dimensional SOM projection grid. Then, large heights indicate a large gap in the data space, whereas low U-heights indicate that the points are close to each other in the data space, indicating structure in the data set. The corresponding visualization technique is a topographical map with a coloring chosen analogously to a geographical physical map facilitating the recognition of cluster structures in the data. Large “heights” in brown and white colors represent large distances between data, green “valleys” and blue “lakes” represent clusters of similar data (Figure 2).
Figure 2.
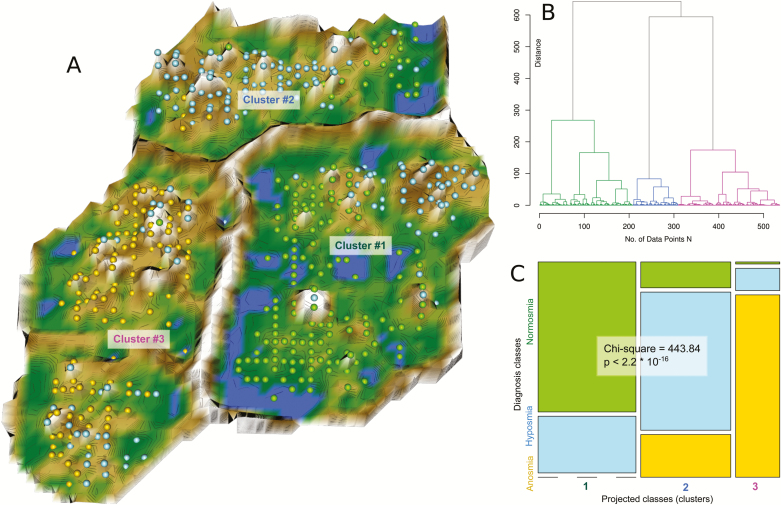
An example of unsupervised machine learning applied on data related to human olfaction. Representation of the olfactory subtest results pattern obtained using a projection of the data points onto a toroid neuronal grid. The data originate from a previous analysis of pattern in olfactory subtests acquired in 10,714 subjects (Lötsch et al. 2016). For the present graphical demonstration, a subset of 5% of these data had been randomly drawn in a class-proportional manner, i.e., preserving the relative numbers of subjects with normosmia, hyposmia, or anosmia. (A) The projection was obtained using a parameter-free polar swarm, Pswarm consisting of so-called DataBots (Thrun 2018) which are self-organizing artificial “life forms” that carry vectors of the individual olfactory subtest results. During the learning phase, the DataBots were allowed adaptively adjusting their location on the grid close to DataBots, according to the Euclidean distance, carrying data with similar features, with successively decreasing search radius. When the algorithm ended, the DataBots became projected points. To enhance the emergence of data structures on this projection, an U-matrix (Ultsch 2003; Lötsch and Ultsch 2014) displaying the distance in the high-dimensional space was added as a third dimension. It was colored in a geographical map analogy with brown snow-covered heights and green valleys with blue lakes. Watersheds indicate borderlines between different groups of subjects suggesting 3 clusters. (B) Hierarchical clustering of the projected data also indicated 3 clusters, supporting the machine-leaned results shown in A. (C) Mosaic plot representing a contingency table of the olfactory diagnoses versus the machine-learned clusters of olfactory subtest results. The size of the cells is proportional to the number of subjects included. The calculations and figure creation were performed using the R software package (version 3.4.3 for Linux; http://CRAN.R-project.org/; R Development Core Team 2008), in particular the libraries “DatabionicSwarm” (https://cran.r-project.org/package=DatabionicSwarm [Thrun 2018]). The figure reproduces results of a previous analysis of the same data set (Lötsch et al. 2016), however, using a different unsupervised machine-learning method for non-redundancy.
Machine-learned approaches to the physiology of odor recognition
Important features of odorants relate to their physico-chemical properties and to their perception. The physiological basis for the relation between physico-chemical properties of odorants and their perception is laid by the approximately 400 olfactory receptor (OR) types in humans (Axel 1995; Niimura et al. 2014). Within the olfactory epithelium, there appears to be a topographical distribution of OR (Ressler et al. 1993; Vassar et al. 1993; Strotmann et al. 1994) which may also exist in humans, at least in terms of the hedonic aspects of odor perception in humans (Lapid et al. 2011). Molecules bind to a large range of OR (Zhao et al. 1998; Duchamp-Viret et al. 1999; Malnic et al. 1999; Saito et al. 2009) which ultimately results in an odorant-specific pattern of activation at the next level of processing. Their physico-chemical properties determine their affinity to receptors and thus—to a certain degree—their perceptual characteristics (Khan et al. 2007; Secundo et al. 2014; Sinding et al. 2017).
Machine-learning methods may be suitable for knowledge discovery approaches to the physiology of odor perception and recognition. For example, the consequences of mutation-caused amino acid exchanges in olfactory receptors for the potency of their ligands was addressed (Gromiha et al. 2012) using various methods of supervised machine learning including naive Bayes classifiers, neural networks, regression analyses, SVM, k-nearest neighbors, and decision tree–based algorithms. The classifiers were trained with physico-chemical properties of both, amino acids and receptor ligands and in addition with experimentally obtained data of EC50, odorant response and cAMP acquired from goldfish, mouse, and human olfactory receptors. The task was to discriminate mutant receptors with enhanced or reduced EC50 values for their ligands, which was successful at an accuracy of > 90% (Gromiha et al. 2012).
Machine-learning concepts have also been used for the implementation of so-called gnostic fields, which aim at modeling the cerebral processes of object recognition from sensory information including that of odors (Kanan 2013), based on a theoretical model in which competing sets of “gnostic” neurons sitting atop sensory processing hierarchies enabled stimuli to be robustly categorized (Konorski 1967). A transformation of the theoretical model into a computational one was applied to the classification of olfactory stimuli (Kanan 2013). The model was applied on 16-dimensional time varying signal recordings from an electronic nose during odor sensing. Following transformation of electronic-nose features from a stimulus into a single vector, SVM classifiers and gnostic fields were used for an odor classification task, with the result that the gnostic model approached 90% correct classification and outperforming SVM-based predictors.
Self-organizing maps and multidimensional scaling have been applied onto a data set (Aldrich 1996) comprising 871 olfactory stimuli that had been described using 278 descriptors (Madany Mamlouk et al. 2003). The relationship between the odor descriptors and the corresponding stimuli was examined to determine whether they might reveal an underlying structure in human odor perception. As a result, relationships between odors were projected onto a topology conserving map and strong support was found for a high-dimensionality of human olfactory perception space. In addition, an ordering of similar odors and corresponding perceptions was found where the resulting maps also allowed the authors to order odor perception in a way not previously possible. For example, a map was produced on which the perceptions were grouped into clusters. The authors highlight that the odor perception of, e.g., apple, banana, and cherry might be ordered, indicating that apple and cherry are perceived more alike than cherry and banana.
A comparative analysis of olfactory neuronal activity between humans and rats was performed proposing a machine-learning approach for predicting perceptual similarities of odors in humans from glomerular activity patterns of rats (Soh et al. 2014). Features extracted from glomerular activity patterns of the olfactory bulb of rats were compared with results of perceptual similarity experiments of odorants in humans. The latter had first been presented with a standard odorant and subsequently, with 6 different but alike odorants, with the task to identify those most similar to the standard. The similarities between rat olfactory bulb signals during sensing similar odors and human responses were analyzed by means of a log-linearized Gaussian mixture network (Tsuji et al. 1999) that expands the Gaussian mixture model to a neural network. The authors concluded their results as satisfactory. Although the correct rate of classification varied from 33.3% to 92.9%, they support the feasibility of linking the glomerular responses of rats to human perception (Soh et al. 2014).
A machine-learning approach was developed to recognize features extracted from EEG signals recorded during perception of either pleasant or unpleasant olfactory stimuli in a group of subjects exposed to a controlled olfactory stimulation experiment (Lanata et al. 2016). Subjects were asked to rate the hedonic properties of the smell of benzaldehyde or isovaleric acid using the Self-Assessment Manikin questionnaire (Bradley and Lang 1994). The EEG response to olfactory stimulation was acquired from 19 electrodes placed according to the international 10/20 system and transformed into power spectra by means of fast Fourier transformation (Cooley and Tukey 1965). Subsequently, the feature space obtained as EEG power spectra was mapped to the output space obtained as hedonic questionnaire outcomes. For this purpose, a linear discriminant classifier was trained repeatedly on data from n – 1 subjects, i.e., one subject was left out for training. Subsequently, the classifier was applied to that subject. This leave-one out scenario was repeated 32 times and the finally obtained classifier achieved 75% classification accuracy of pleasant versus unpleasant stimuli from EEG signal-based features (Lanata et al. 2016).
Machine-learned pattern recognition in olfactory phenotypes
Three dimensions are frequently attributed to the human sense of smell and separately tested in clinical diagnostics, namely 1) odor identification performance addressing the ability to name or associate an odor (Cain 1979; Doty, Shaman, Kimmelman, et al. 1984), 2) odor threshold addressing the lowest concentration of a selected odorant at which it is still perceived (Cain et al. 1988), and 3) odor discrimination performance addressing the ability to distinguish different smells (Cain and Krause 1979). However, there are numerous other dimensions, like pleasantness/liking, familiarity, and edibility (e.g., Fournel et al. 2016). Currently, these dimensions receive little attention on a clinical level and are not included in the most widely distributed clinical olfactory tests, like the Sniffin’ Sticks or the UPSIT, so that, in the following, the 3 dimensions referred to are odor identification, odor discrimination and odor threshold sensitivity.
A distinct importance of the 3 main components of the sense of smell has been challenged by the suggestion that the 3 olfactory subtests measure a common source of variance (Doty et al. 1994); nevertheless it has remained an active research topic owing to the desire of deeper understanding of the pathological mechanisms via which various different etiologies may cause olfactory dysfunction. Two reports explicitly employed machine learning to address these problems in a large data set comprising olfactory subtest results (threshold, discrimination, identification) acquired in subjects whose olfactory performance ranged from normosmia, i.e., olfactory test scores similarly to those found in large samples of subjects who indicated an intact sense of smell (Doty, Shaman, and Dann 1984; Hummel et al. 1997) to anosmia, i.e., the absence of olfactory function. Olfactory performance was associated with 9 different etiologies, i.e., healthy, sinunasal disease, congenital, neurodegenerative diseases, idiopathic, postinfectious, trauma, toxic, and brain tumor/stroke. This data set was assessed for structures, e.g., clusters, that coincided with the olfactory diagnoses or the underlying etiologies, using unsupervised machine learning implemented as emergent self-organizing feature maps (ESOM) applied in combination with the use of the U-matrix (Ultsch 2003). On a trained ESOM, the distance structure in the high-dimensional feature space was visualized using a geographical map analogy with data clusters represented as “valleys” separated by “mountain ridges”. This structure reflected the olfactory diagnoses into normosmia, hyposmia, and anosmia while no consistent overlap with the etiologies underlying the individual olfactory functions could be proven (see Figure 3 in Lötsch et al. 2016).
To demonstrate this method, in the following experiment an alternative data projection method was used for non-redundancy (Lötsch et al. 2016). Specifically, the topographic mapping was implemented as swarm intelligence instead of self-organizing map, i.e., as an algorithm guided by the flocking behavior of numerous independent but cooperating so-called “DataBots” (Ultsch 2000), which are self-organizing artificial “life forms” identified with single data objects (subjects tested for olfactory function). These “DataBots” can move on a 2-dimensional grid, and their movements are either random or follow the attractive or repulsive forces proportionally to the (dis-)similarities of neighboring “DataBots”. A subset of 5% of the original data was randomly drawn for this demonstration. As this method is computationally more demanding than ESOM, the data space was again explored for distance-based structures, presently using a parameter-free projection method of a polar swarm (Thrun 2018). Following successful swarm learning, “DataBots” carrying items with similar features were located in groups on the projection grid. After calculation of a U-matrix (Ultsch and Sieman 1990; Lötsch and Ultsch 2014), 3 clusters emerged (Figure 2) among subjects with 3 unevenly distributed olfactory diagnosss, i.e., the classification of subjects with respect to the diagnosis of “normosmia”, “hyposmia”, or “anosmia” coincided with the observed clusters at a statistical significance level of P < 2.2 · 10–16 (χ2 test), which reproduced previously obtained results of an analysis using an emergent self-organizing map (Lötsch et al. 2016). This indicated that unsupervised machine learning is suitable to detect group structures in olfaction related data that reflect a known grouping of subjects and supports the utility of the method for further exploration of clinical subgroups in similar data.
Machine learning–derived complex biomarkers with olfactory and non-olfactory features
Olfactory loss is a symptom of several neurological conditions including neurodegenerative diseases such as Parkinson’s disease (Doty et al. 1988), Alzheimer’s disease (Murphy et al. 1990), or multiple sclerosis (Hawkes 1996), in which an impaired sense of smell can be both, an early symptom and a marker of disease progression. However, complete olfactory loss has an estimated prevalence of 5% in the population (Murphy et al. 2002; Brämerson et al. 2004; Landis et al. 2004) and therefore, specific patterns of features are needed to associate it with a neurological disease.
To create this kind of complex biomarker for Parkinson progression (Prashanth et al. 2014, 2016), non-motor parameters (results of the 40-item University of Pennsylvania smell identification test (UPSIT [Doty, Shaman, and Dann 1984]), ratings in the REM sleep behavior disorder screening questionnaire [Stiasny-Kolster et al. 2007]), cerebrospinal fluid related markers (Aβ1−42, T-tau, and P-tau181 and their ratios) and markers obtained by means of dopaminergic imaging (striatal binding ratio [SBR] obtained SPECT imaging using 123I-Ioflupane) available from patients with Parkinson disease (disease stage 1 or 2 according to (Hoehn and Yahr 1967)) and healthy subjects were explored for their utility as an early diagnostic of Parkinson’s disease. Using 100 times cross-validation with randomly splitting the data set into training (70% of the data) and test (30%) subsets, 5 different machine-learned classifiers were tested with respect to their performance to obtain the correct diagnosis of early Parkinson’s disease (naive Bayes classifier, logistic regression, adaptive boosted trees, RF, and SVM). Using the 13 above-mentioned different features, classifiers were compared with the results that SVM performed best in diagnostic accuracy, SVM, and logistic regression performed best when using the area under the ROC curve as performance measure, RF and SVM provided the best diagnostic sensitivity, and SVM performed best in diagnostic specificity. From this, the SVM-based complex biomarker was chosen that provided 96.40% diagnostic accuracy for Parkinson’s disease, with 97.03% sensitivity and 95.01% specificity. Comparative analysis of the feature importance, accessible via RF for example as mean decrease in classification accuracy when the respective feature was omitted from forest building, indicated that the UPSIT score figured as the third important marker among the 13 components of the early-Parkinson biomarker (see Figure 5 in Prashanth et al. 2016). However, this was not further used for feature selection, i.e., for eliminating features contributing only minimally to the diagnosis and thus reducing the efforts to acquire the biological markers necessary to use the classifier for diagnostics.
Machine-learned marker selection was, however, employed to extract the most informative features for predicting rapid progression of Parkinson’s disease from public data provided by the Parkinson’s Progression Markers Initiative (http://www.ppmi-info.org/) (Tsiouris et al. 2017). Several markers acquired in patients diagnosed with Parkinson’s disease less than 2 years before including 3 Movement Disorder Society MDS Unified Parkinson’s Disease Rating Scales (MDS-UPDRS (Goetz et al. 2008)), the Montreal Cognitive Assessment scale (Nasreddine et al. 2005), the Epworth Sleeping Scale (Johns 1991), REM Sleep Behavior Disorder Screening Questionnaire (Stiasny-Kolster et al. 2007), neurological examination, and the UPSIT olfactory test (Doty, Shaman, and Dann 1984). Using the so-called wrapper approach, which searches all possible subsets of features for the optimal feature subset achieving the best possible performance with a particular learning algorithm on a particular training set (Kohavi and John 1997), a feature set was created that subsequently was submitted to the C4.5 decision tree algorithm (Quinlan 1986) to identify patients likely to display rapid disease progression already at the baseline patient evaluation. This was successful at an accuracy of almost 90% when using the algorithm to identify the top 5% of the worsening patients (Tsiouris et al. 2017). Among the features used to predict patients with a rapid progress of Parkinson’s disease speech problems, olfactory dysfunctions, high rigidity, and affected leg muscle reflexes (Tsiouris et al. 2017) have been identified. Similarly, the UPSIT-based olfactory phenotype was used together with basic demographic parameters including age and sex to build an SVM and linear discriminant analysis-based classifier for the diagnosis of Parkinson’s disease (Gerkin et al. 2017). Specifically, both clinical and postmortem neuropathological data were available for deceased individuals who had taken at least one UPSIT. The obtained classifier was judged by the authors to perform well and in addition, they observed that the responses to single UPSIT test items provided a more suitable basis for classifier building than using just the UPSIT sum score (Gerkin et al. 2017).
Machine-learned odor recognition from physico-chemical properties of volatile molecules
Chromatographic analyses of volatile molecules have been employed for automated detection of odors with medical applications as odor-based biomarkers or to predict the smell of molecules in other context. These efforts may possibly be summarized as development and application of so-called “electronic noses”. A specific search on 5 February 2018, for “(electronic nose) AND (algorithm OR machine) NOT review[Publication Type]”, identified 228 items. Reviews about activities in electronic nose developments have been published previously (Yan et al. 2015; Wojnowski et al. 2017). As the present review focuses on the study on biomedical topics involving the human sense of smell rather than technical implementations, only a few examples will be provided, mainly those reports that explicitly named machine learning and had been found already in the main search (see above).
The use of volatile molecules emitted from patient materials for clinical diagnostics offers the promise of non-invasive and rapid biomarkers. Variations in small molecular weight volatile organic compounds, i.e., odorants in urine were assessed as possible biomarkers for lung cancer in mice, which could be used to discriminate between odors of mice with and without experimental tumors (Matsumura et al. 2010). Following this establishment of the principal utility of odorants from urine as cancer biomarkers, urinary volatile compounds were analyzed using solid-phase-microextraction, followed by gas chromatography coupled with mass spectrometry. From the typical total ion chromatograms, unsupervised machine learning implemented as principal component analysis established the separation of the odorants into clusters, which encouraged the use of these data for class association, which was subsequently implemented as supervised machine learning in the form of SVM. The diagnostic accuracy of the SVM classifiers was 95% and above, supporting the utility of this approach and encouraging further translational research (Matsumura et al. 2010). Further odor-based disease biomarkers have been proposed for colorectal cancer (de Meij et al. 2014) and other gastrointestinal and liver diseases (for reviews, see Probert et al. 2009; Chan et al. 2016), various types of cancer (reviewed in Zhou et al. 2017; Kabir and Donald 2018), pulmonary diseases (Pizzini et al. 2018), renal diseases (Liu et al. 2018), and many other medical conditions.
Chromatographic analyses of volatile molecules have also been used to automatically predict the emitted odor to replace human assessors. For example, aiming at reducing subjective components of the sensory assessment of smells by panelists, which have been described to be influenced by many factors such as the testing environment, experimental bias, assessor sensitivity, assessor selection, and training (Delahunty et al. 2006), the physico-chemical molecular parameters of several flavors and fragrances obtained by means of gas chromatography coupled with mass spectrometry were used to train supervised machine-learned classifiers (Shang et al. 2017). The algorithms were provided with the GC/MS derived molecular characteristics and database-derived information about the molecular parameters of 1026 different odorants annotated with 160 different odor descriptors (the 20 most frequently occurring being: sweet, green, fruity, floral, meaty, wine-like, apple, fatty, woody, herbaceous, sulfurous, ethereal, nutty, spicy, oily, earthy, pineapple, waxy, creamy, and rose). Machine-learning algorithms including SVM, RF, and extreme learning machine were used and their prediction results were compared. The best classifier was based on extreme machine learning, which is a special form of neuronal networks with single or multiple layers of hidden nodes with the advantage over other network approaches that the parameters of hidden nodes do not need to be tuned, which, as the authors pointed out, makes them faster than other implementations of neural networks (Huang et al. 2006). ELM provided the best identification accuracy (97.53%), followed by SVM (97.19%) and RF (92.79%). However, when applied to 30 primary volatile organic compounds from Golden Delicious apples that had not been included in the analyses, they predicted only 70% of compounds accurately. Although the authors concluded that this does not suffice to replace panelists, it represents a proof-of-concept that machine learning is probably suitable for automatic odor detection (Shang et al. 2017).
Finally, the prediction of whether or not a given molecule will induce the perception of smell or what kind of olfactory perception it will produce was addressed by means of machine learning, i.e., algorithms were trained to predict sensory attributes of molecules based on their chemo-informatic features (Keller et al. 2017). A large psychophysical data set collected from 49 individuals who profiled 476 structurally and perceptually diverse molecules (Keller and Vosshall 2016) was supplied with 4884 physico-chemical features of each of the molecules smelled by the subjects, including atom types, functional groups, and topological and geometrical properties. A RF-based classifier successfully predicted 8 among 19 rated semantic descriptors (“garlic”, “fish”, “sweet”, “fruit,” “burnt”, “spices”, “flower”, “sour”). Alternative approaches at the relationship between the chemical structure of odorant molecules and their interaction with human olfactory receptors used 3D-modeling cheminformatics techniques (Sanz et al. 2008) based on the pharmacophore concept, which is regarded as the ensemble of steric and electronic features that is necessary to ensure the optimal interactions with a specific biological target and to trigger or block the biological response (Guner 2002).
Knowledge discovery in publicly available big databases
Computational methods, publicly available databases, and data mining tools provide a contemporary basis to combine the knowledge about the biological roles of genes, or about the interactions of chemicals including drugs with proteins, with the acquired knowledge about higher-level organization of gene products into biological pathways (Hu et al. 2007). In genetic research, the gold-standard is the Gene Ontology (GO) knowledge base (Ashburner et al. 2000), which has been used to study the biological functions of the genes expressed in the human olfactory bulb (Lötsch et al. 2014). Specifically, a set intersection of n = 1427 genes identified to be expressed at protein level in the human olfactory bulb (Fernandez-Irigoyen et al. 2012)) with n = 669 genes found to be expressed in human olfactory bulbs at mRNA level provided a set of n = 231 genes that can be regarded to be supported by 2 independent analyses as expressed in the human olfactory bulb. The biological processes including functional subcategories covered by these genes were queried from the Gene Ontology knowledge base which provides a dynamic, controlled vocabulary (GO terms) capturing the acquired knowledge about gene product attributes. The GO terms are connected with each other by “is-a”, “part-of”, “regulates”, and “subclass of” relationships (Camon et al. 2003, 2004) forming a polyhierarchy organized in a directed acyclic graph (DAG, Thulasiraman and Swamy 1992). In this polyhierarchy, the particular biological roles covered by the n = 231 genes were identified by means of over-representation analysis, which compared the biological processes annotated to the expressed genes with the occurrence of processes in the set of all human genes and determined the statistical significance of the deviation from change by means of hypergeometric tests, that annotated the resulting GO terms with P-values p(Ti), applying Bonferroni α correction (Bonferroni 1936) and a heuristically determined P-value threshold of 10–4. This analysis resulted in a polyhierarchy of 94 GO terms with a significantly over-represented subset of genes with respect to all annotated human genes. Among the GO terms to which the genes expressed in the human olfactory bulb were statistically significantly annotated the authors figured a subset of particular remarkable terms, that served as headlines describing the biological functions of the analyzed genes as so-called functional areas (Ultsch and Lötsch 2014). These areas included “nervous system development” and “neuron development”, of which an associated term in the GO polyhierarchy was neurogenesis. Indeed, neurogenesis emerged as a significant GO term (expected number of genes in this category: 8.3, found: 34, P = 1.14 × 10–9); hence, 2 GO categories provided primary support for the existence of neurogenesis in the human olfactory bulb contributing a positive finding to the ongoing scientific discussion about the occurrence of postnatal neurogenesis in the human olfactory bulb (Bergmann et al. 2012).
Conclusions and outlook
Recent literature supports that the complexity of human olfaction (Buck and Axel 1991; Menashe and Lancet 2006) is reflected in high-dimensional data increasingly resulting from laboratory and clinical research. Contemporary computational science allows the extraction of information and the generation of knowledge from these data (President’s Information Technology Advisory 2005) facilitating research approaches structured as “DIKW” hierarchies (data – information – knowledge – wisdom). These data-driven approaches complement classical hypothesis-driven approaches. However, an advantage lies in the assessment of big data, i.e., data not only large in sample size but also complex and high-dimensional. Machine learning is better suited for data mining and exploratory data analyses than classical statistics, as it does not require prior hypotheses while it can be used to generate hypotheses.
Moreover, machine learning has been shown to outperform classical methods in task such as clustering. For example, while classical Ward hierarchical clustering failed in several artificial or biomedical data sets to obtain the correct, i.e., known cluster structures and even suggested subgroups in clearly unstructured data, machine learning implemented as emergent self-organizing map provided always the correct cluster structure or indicated correctly the absence of subgroups (Ultsch and Lötsch 2017). Similarly, neuronal networks can be shown to outperform principal component analysis, which has been introduced more than 100 years ago (Pearson 1901) and it is still used as a standard method for comparative data projections although it is sensitive to the scaling of the data and correlation structures and the results may be inferior to those obtained with machine learning.
A difficulty for employing the full power of machine learning in olfactory research is recruiting large numbers of subjects. While machine-learned methods have been successfully applied to data sets comprising 100 subjects, a strength of these methods lies in the analysis of large data sets where classifiers can be trained with many samples. A further potential weakness of machine learning is its vulnerability to overfitting. In this case, the algorithm learns to perfectly assign the classes in the training sample but fails to correctly predict new cases, i.e., the algorithm has learned the classification “by heart”. Several measures have been proposed to be taken against overfitting such as building the classifier on a training data set and testing its performance on a test data set, obtained in a separate experiment or via splitting the available data, and/or by employing cross-validation using the creation of data subsets by random resampling from the original data set. Furthermore, machine learning may be fooled by data sets containing dominant but irrelevant features, such as in the parable where neural networks were trained to automatically detect camouflaged tanks on photos of tanks in trees and photos of trees without tanks (Dreyfus and Dreyfus 1992).
The application of machine learning for (human) olfactory research has been mentioned explicitly in scientific publications since 2012. Among many machine-learning methods (Murphy 2012; Dhar 2013), a subset has so far been applied to problems related to human olfactory research, of which SVM, regression models and several kinds of decision tree algorithms have so far most frequently been mentioned. Machine learning has been successfully applied to 1) obtain a better understanding of the complex signaling processes that enable the recognition of odors from patterns of sensory input signals, 2) to allow a deeper knowledge discovery in complex data of olfactory phenotypes, or to develop biomarkers that 3) include olfactory information, or 4) provide a diagnostic tool based on the detection of volatile substances emitted by patients suffering from a certain disease, or to perform knowledge discovery in big data such as the GeneOntology database addressing the biological roles of olfaction relevant genes. For example, the idiopathic Parkinson’s Syndrome is characterized by olfactory loss as an early symptom, but the diagnosis cannot be concluded solely from this; therefore, olfactory-related features have been successfully included in more complex biomarkers that allow a satisfactory diagnosis of the disease at early stages and enable predictions of its future course.
Machine learning has started to be acknowledged in olfactory research and first results can be observed. It promises benefits for this field of research; however, it may also be reasonable to consider Amara’s law that states that one tends to overestimate an effect of a technology in the short run while underestimating its effect in the long run (https://en.wikipedia.org/wiki/Roy_Amara), which is illustrated in the so-called Gartner hype cycle (https://www.gartner.com/technology/research/methodologies/hype-cycle.jsp) where the expectations quickly inflate followed by a period of disillusionment that finally equilibrates in a plateau where the method is productive. When raising expectations, topics of olfactory research so far unresolved such as 1) fitting of ligands onto olfactory receptors, for which initial attempts at a machine-learned solution have been made (Sanz et al. 2008; Keller et al. 2017), and 2) a deeper understanding of the combinatorial code of olfaction, i.e., how receptors act in concert to create the odor percept. Indeed, pattern recognition and network analysis are important domains of machine learning, qualifying it for future applications in (human) olfactory research. It will broaden the scientific armamentarium of human olfactory research and provides new and intriguing solutions for complex questions.
Funding
This work has been funded by the Landesoffensive zur Entwicklung wissenschaftlich-ökonomischer Exzellenz (LOEWE), LOEWE-Zentrum für Translationale Medizin und Pharmakologie (J.L.). We also would like to thank the Deutsche Forschungsgemeinschaft for support (DFG HU 441/18-1 to T.H. and DFG Lo 612/10–1 to J.L.). Further support was gathered from the European Union Seventh Framework Programme (FP7/2007 - 2013) under grant agreement no. 602919 (“GLORIA”, J.L.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors have declared that no further conflicts of interest exist.
Acknowledgment
A short form of this review was presented as an oral presentation on at the “human CHEMOSENSATION – the lab meeting”, at Dresden, Germany, on 24 February 2018.
References
- Ache BW, Young JM. 2005. Olfaction: diverse species, conserved principles. Neuron. 48:417–430. [DOI] [PubMed] [Google Scholar]
- Albers MW, Tabert MH, Devanand DP. 2006. Olfactory dysfunction as a predictor of neurodegenerative disease. Curr Neurol Neurosci Rep. 6:379–386. [DOI] [PubMed] [Google Scholar]
- Aldrich E. 1996. Flavor and fragrances catalog. Milwaukee (WI): Company SAC. [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R. 1995. The molecular logic of smell. Sci Am. 273:154–159. [DOI] [PubMed] [Google Scholar]
- Bayes M, Price M. 1763. An essay towards solving a problem in the doctrine of chances. By the Late Rev. Mr. Bayes, F. R. S. communicated by Mr. Price, in a letter to John Canton, A. M. F. R. S. Philosophical Transactions. 53:370–418. [Google Scholar]
- Bergmann O, Liebl J, Bernard S, Alkass K, Yeung MS, Steier P, Kutschera W, Johnson L, Landén M, Druid H, et al. 2012. The age of olfactory bulb neurons in humans. Neuron. 74:634–639. [DOI] [PubMed] [Google Scholar]
- Bonferroni CE. 1936. Teoria statistica delle classi e calcolo delle probabilita. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 8:3–62. [Google Scholar]
- Bradley MM, Lang PJ. 1994. Measuring emotion: the self-assessment manikin and the semantic differential. J Behav Ther Exp Psychiatry. 25:49–59. [DOI] [PubMed] [Google Scholar]
- Brämerson A, Johansson L, Ek L, Nordin S, Bende M. 2004. Prevalence of olfactory dysfunction: the Skövde population-based study. Laryngoscope. 114:733–737. [DOI] [PubMed] [Google Scholar]
- Breiman L. 2001. Random forests. Mach. Learn. 45:5–32. [Google Scholar]
- Buck L, Axel R. 1991. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 65:175–187. [DOI] [PubMed] [Google Scholar]
- Bushdid C, de March CA, Fiorucci S, Matsunami H, Golebiowski J. 2018. Agonists of G-protein-coupled odorant receptors are predicted from chemical features. J Phys Chem Lett. 9:2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain WS. 1979. To know with the nose: keys to odor identification. Science. 203:467–470. [DOI] [PubMed] [Google Scholar]
- Cain WS, Gent JF, Goodspeed RB, Leonard G. 1988. Evaluation of olfactory dysfunction in the Connecticut Chemosensory Clinical Research Center. Laryngoscope. 98:83–88. [DOI] [PubMed] [Google Scholar]
- Cain WS, Krause RJ. 1979. Olfactory testing: rules for odor identification. Neurol Res. 1:1–9. [DOI] [PubMed] [Google Scholar]
- Camon E, Magrane M, Barrell D, Binns D, Fleischmann W, Kersey P, Mulder N, Oinn T, Maslen J, Cox A, et al. 2003. The Gene Ontology Annotation (GOA) project: implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 13:662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R. 2004. The Gene Ontology Annotation (GOA) database: sharing knowledge in Uniprot with Gene ontology. Nucleic Acids Res. 32:D262–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Eckert A, Woitalla D, Ellrichmann G, Turewicz M, Stephan C, Eisenacher M, May C, Meyer HE, Brüning T, et al. 2013. Diagnostic value of the impairment of olfaction in Parkinson’s disease. PLoS One. 8:e64735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DK, Leggett CL, Wang KK. 2016. Diagnosing gastrointestinal illnesses using fecal headspace volatile organic compounds. World J Gastroenterol. 22:1639–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet F, Allaire JJ. 2018. Deep learning with R. Shelter Island (NY): Manning Publications Co. [Google Scholar]
- Cooley JW, Tukey JW. 1965. An algorithm for the machine calculation of complex Fourier series. Math Comput. 19:297–301. [Google Scholar]
- Cortes C, Vapnik V. 1995. Support-vector networks. Mach Learn. 20:273–297. [Google Scholar]
- Cover T, Hart P. 1967. Nearest neighbor pattern classification. IEEE Trans Inf Theor. 13:21–27. [Google Scholar]
- Delahunty CM, Eyres G, Dufour JP. 2006. Gas chromatography-olfactometry. J Sep Sci. 29:2107–2125. [DOI] [PubMed] [Google Scholar]
- de Meij TG, Larbi IB, van der Schee MP, Lentferink YE, Paff T, Terhaar Sive Droste JS, Mulder CJ, van Bodegraven AA, de Boer NK. 2014. Electronic nose can discriminate colorectal carcinoma and advanced adenomas by fecal volatile biomarker analysis: proof of principle study. Int J Cancer. 134:1132–1138. [DOI] [PubMed] [Google Scholar]
- Dhar V. 2013. Data science and prediction. Commun ACM. 56:64–73. [Google Scholar]
- Doty RL, Bromley SM. 2004. Effects of drugs on olfaction and taste. Otolaryngol Clin North Am. 37:1229–1254. [DOI] [PubMed] [Google Scholar]
- Doty RL, Cameron EL. 2009. Sex differences and reproductive hormone influences on human odor perception. Physiol Behav. 97:213–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty RL, Deems DA, Stellar S. 1988. Olfactory dysfunction in Parkinsonism: a general deficit unrelated to neurologic signs, disease stage, or disease duration. Neurology. 38:1237–1244. [DOI] [PubMed] [Google Scholar]
- Doty RL, Shaman P, Applebaum SL, Giberson R, Siksorski L, Rosenberg L. 1984. Smell identification ability: changes with age. Science. 226:1441–1443. [DOI] [PubMed] [Google Scholar]
- Doty RL, Shaman P, Dann M. 1984. Development of the University of Pennsylvania Smell Identification Test: a standardized microencapsulated test of olfactory function. Physiol Behav. 32:489–502. [DOI] [PubMed] [Google Scholar]
- Doty RL, Shaman P, Kimmelman CP, Dann MS. 1984. University of Pennsylvania smell identification test: a rapid quantitative olfactory function test for the clinic. Laryngoscope. 94:176–178. [DOI] [PubMed] [Google Scholar]
- Doty RL, Smith R, McKeown DA, Raj J. 1994. Tests of human olfactory function: principal components analysis suggests that most measure a common source of variance. Percept Psychophys. 56:701–707. [DOI] [PubMed] [Google Scholar]
- Dreyfus HL, Dreyfus SE. 1992. What artificial experts can and cannot do. AI & Society. 6:18–26. [Google Scholar]
- Duchamp-Viret P, Chaput MA, Duchamp A. 1999. Odor response properties of rat olfactory receptor neurons. Science. 284:2171–2174. [DOI] [PubMed] [Google Scholar]
- Fernandez-Irigoyen J, Corrales FJ, Santamaría E. 2012. Proteomic atlas of the human olfactory bulb. J Proteomics. 75:4005–4016. [DOI] [PubMed] [Google Scholar]
- Fournel A, Ferdenzi C, Sezille C, Rouby C, Bensafi M. 2016. Multidimensional representation of odors in the human olfactory cortex. Hum Brain Mapp. 37:2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerkin RC, Adler CH, Hentz JG, Shill HA, Driver-Dunckley E, Mehta SH, Sabbagh MN, Caviness JN, Dugger BN, Serrano G, et al. 2017. Improved diagnosis of Parkinson’s disease from a detailed olfactory phenotype. Ann Clin Transl Neurol. 4:714–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz CG, Tilley BC, Shaftman SR, Stebbins GT, Fahn S, Martinez-Martin P, Poewe W, Sampaio C, Stern MB, Dodel R, et al. ; Movement Disorder Society UPDRS Revision Task Force. 2008. Movement Disorder Society-sponsored revision of the Unified Parkinson’s Disease Rating Scale (MDS-UPDRS): scale presentation and clinimetric testing results. Mov Disord. 23:2129–2170. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Liljenström H, Ruiz Y, Li G. 2010. A biologically inspired model for pattern recognition. J Zhejiang Univ Sci B. 11:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried JA. 2006. Smell: central nervous processing. Adv Otorhinolaryngol. 63:44–69. [DOI] [PubMed] [Google Scholar]
- Gromiha MM, Harini K, Sowdhamini R, Fukui K. 2012. Relationship between amino acid properties and functional parameters in olfactory receptors and discrimination of mutants with enhanced specificity. BMC Bioinformatics. 13 (Suppl 7):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guner OF. 2002. History and evolution of the pharmacophore concept in computer-aided drug design. Curr Top Med Chem. 2:1321–1332. [DOI] [PubMed] [Google Scholar]
- Guyon I, Elisseeff A. 2003. An introduction to variable and feature selection. J Mach Learn Res. 3:1157–1182. [Google Scholar]
- Hawkes CH. 1996. Assessment of olfaction in multiple sclerosis. Chem Senses. 21:486. [Google Scholar]
- Ho TK. 1995. Random decision forests. In: M. Kavanaugh and P. Storms, editors. Proceedings of the Third International Conference on Document Analysis and Recognition. p. 278–282. New York: IEEE Computer Society Press. [Google Scholar]
- Hoehn MM, Yahr MD. 1967. Parkinsonism: onset, progression and mortality. Neurology. 17:427–442. [DOI] [PubMed] [Google Scholar]
- Holley A, Duchamp A, Revial MF, Juge A. 1974. Qualitative and quantitative discrimination in the frog olfactory receptors: analysis from electrophysiological data. Ann N Y Acad Sci. 237:102–114. [DOI] [PubMed] [Google Scholar]
- Hu P, Bader G, Wigle DA, Emili A. 2007. Computational prediction of cancer-gene function. Nat Rev Cancer. 7:23–34. [DOI] [PubMed] [Google Scholar]
- Huang G-B, Zhu Q-Y, Siew C-K. 2006. Extreme learning machine: theory and applications. Neurocomputing. 70:489–501. [Google Scholar]
- Hummel T, Sekinger B, Wolf SR, Pauli E, Kobal G. 1997. ‘Sniffin’ sticks’: olfactory performance assessed by the combined testing of odor identification, odor discrimination and olfactory threshold. Chem Senses. 22:39–52. [DOI] [PubMed] [Google Scholar]
- Johns MW. 1991. A new method for measuring daytime sleepiness: the Epworth sleepiness scale. Sleep. 14:540–545. [DOI] [PubMed] [Google Scholar]
- Kabir KMM, Donald WA. 2018. Cancer breath testing: a patent review. Expert Opin Ther Pat. 28:227–239. [DOI] [PubMed] [Google Scholar]
- Kanan C. 2013. Recognizing sights, smells, and sounds with gnostic fields. PLoS One. 8:e54088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Gerkin RC, Guan Y, Dhurandhar A, Turu G, Szalai B, Mainland JD, Ihara Y, Yu CW, Wolfinger R, et al. ; DREAM Olfaction Prediction Consortium. 2017. Predicting human olfactory perception from chemical features of odor molecules. Science. 355:820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Vosshall LB. 2016. Olfactory perception of chemically diverse molecules. BMC Neurosci. 17:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. 2007. Genetic variation in a human odorant receptor alters odour perception. Nature. 449:468–472. [DOI] [PubMed] [Google Scholar]
- Khan RM, Luk CH, Flinker A, Aggarwal A, Lapid H, Haddad R, Sobel N. 2007. Predicting odor pleasantness from odorant structure: pleasantness as a reflection of the physical world. J Neurosci. 27:10015–10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohavi R, John GH. 1997. Wrappers for feature subset selection. Artificial Intelligence. 97:273–324. [Google Scholar]
- Kohonen T. 1982. Self-organized formation of topologically correct feature maps. Biol Cybernet. 43:59–69. [Google Scholar]
- Kohonen T. 1995. Self-organizing maps. Berlin: Springer. [Google Scholar]
- Konorski J. 1967. Integrative activity of the brain. Chicago (IL): University of Chicago Press. [Google Scholar]
- Kringel D, Geisslinger G, Resch E, Oertel BG, Thrun MC, Heinemann S, Lötsch J. 2018. Machine-learned analysis of the association of next-generation sequencing-based human TRPV1 and TRPA1 genotypes with the sensitivity to heat stimuli and topically applied capsaicin. Pain. 159:1366–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanata A, Guidi A, Greco A, Valenza G, Di Francesco F, Scilingo EP. 2016. Automatic recognition of pleasant content of odours through ElectroEncephaloGraphic activity analysis. Conf Proc IEEE Eng Med Biol Soc. 2016:4519–4522. [DOI] [PubMed] [Google Scholar]
- Landis BN, Konnerth CG, Hummel T. 2004. A study on the frequency of olfactory dysfunction. Laryngoscope. 114:1764–1769. [DOI] [PubMed] [Google Scholar]
- Lapid H, Shushan S, Plotkin A, Voet H, Roth Y, Hummel T, Schneidman E, Sobel N. 2011. Neural activity at the human olfactory epithelium reflects olfactory perception. Nat Neurosci. 14:1455–1461. [DOI] [PubMed] [Google Scholar]
- Li H, Panwar B, Omenn GS, Guan Y. 2018. Accurate prediction of personalized olfactory perception from large-scale chemoinformatic features. GigaScience. 7. doi:10.1093/gigascience/gix127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DM, Yang YH, Scolnick JA, Brunet LJ, Marsh H, Peng V, Okazaki Y, Hayashizaki Y, Speed TP, Ngai J. 2004. Spatial patterns of gene expression in the olfactory bulb. Proc Natl Acad Sci USA. 101:12718–12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Zhao N, Wang M, Pi X, Feng Y, Wang Y, Tong H, Zhu L, Wang C, Li E. 2018. Urine volatile organic compounds as biomarkers for minimal change type nephrotic syndrome. Biochem Biophys Res Commun. 496:58–63. [DOI] [PubMed] [Google Scholar]
- Loh W-Y. 2014. Fifty years of classification and regression trees. Int Stat Rev. 82:329–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötsch J, Geisslinger G, Hummel T. 2012. Sniffing out pharmacology: interactions of drugs with human olfaction. Trends Pharmacol Sci. 33:193–199. [DOI] [PubMed] [Google Scholar]
- Lötsch J, Hummel T, Ultsch A. 2016. Machine-learned pattern identification in olfactory subtest results. Sci Rep. 6:35688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötsch J, Knothe C, Lippmann C, Ultsch A, Hummel T, Walter C. 2015. Olfactory drug effects approached from human-derived data. Drug Discov Today. 20:1398–1406. [DOI] [PubMed] [Google Scholar]
- Lötsch J, Schaeffeler E, Mittelbronn M, Winter S, Gudziol V, Schwarzacher SW, Hummel T, Doehring A, Schwab M, Ultsch A. 2014. Functional genomics suggest neurogenesis in the adult human olfactory bulb. Brain Struct Funct. 219:1991–2000. [DOI] [PubMed] [Google Scholar]
- Lötsch J, Ultsch A. 2014. Exploiting the structures of the U-matrix. In: Villmann T, Schleif F-M, Kaden M, Lange M, editors. Advances in intelligent systems and computing. Heidelberg: Springer; p. 248–257. [Google Scholar]
- Lutterotti A, Vedovello M, Reindl M, Ehling R, DiPauli F, Kuenz B, Gneiss C, Deisenhammer F, Berger T. 2011. Olfactory threshold is impaired in early, active multiple sclerosis. Mult Scler. 17:964–969. [DOI] [PubMed] [Google Scholar]
- Madany Mamlouk A, Chee-Ruiter C, Hofmann UG, Bower JM. 2003. Quantifying olfactory perception: mapping olfactory perception space by using multidimensional scaling and self-organizing maps. Neurocomputing. 52–54: 591–597. [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. 1999. Combinatorial receptor codes for odors. Cell. 96:713–723. [DOI] [PubMed] [Google Scholar]
- Matsumura K, Opiekun M, Oka H, Vachani A, Albelda SM, Yamazaki K, Beauchamp GK. 2010. Urinary volatile compounds as biomarkers for lung cancer: a proof of principle study using odor signatures in mouse models of lung cancer. PLoS One. 5:e8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I, Lancet D. 2006. Variations in the human olfactory receptor pathway. Cell Mol Life Sci. 63:1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KP. 2012. Machine learning: a probabilistic perspective. Cambridge (MA): The MIT Press. [Google Scholar]
- Murphy C, Gilmore MM, Seery CS, Salmon DP, Lasker BR. 1990. Olfactory thresholds are associated with degree of dementia in Alzheimer’s disease. Neurobiol Aging. 11:465–469. [DOI] [PubMed] [Google Scholar]
- Murphy C, Schubert CR, Cruickshanks KJ, Klein BE, Klein R, Nondahl DM. 2002. Prevalence of olfactory impairment in older adults. JAMA. 288:2307–2312. [DOI] [PubMed] [Google Scholar]
- Nasreddine ZS, Phillips NA, Bédirian V, Charbonneau S, Whitehead V, Collin I, Cummings JL, Chertkow H. 2005. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 53:695–699. [DOI] [PubMed] [Google Scholar]
- Niimura Y. 2009. Evolutionary dynamics of olfactory receptor genes in chordates: interaction between environments and genomic contents. Hum Genomics. 4:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K. 2014. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 24:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson K. 1901. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dubl. Phil. Mag. 2:559–572. [Google Scholar]
- Peters JM, Hummel T, Kratzsch T, Lötsch J, Skarke C, Frölich L. 2003. Olfactory function in mild cognitive impairment and Alzheimer’s disease: an investigation using psychophysical and electrophysiological techniques. Am J Psychiatry. 160:1995–2002. [DOI] [PubMed] [Google Scholar]
- Pizzini A, Filipiak W, Wille J, Ager C, Wiesenhofer H, Kubinec R, Blaško J, Tschurtschenthaler C, Mayhew CA, Weiss G, et al. 2018. Analysis of volatile organic compounds in the breath of patients with stable or acute exacerbation of chronic obstructive pulmonary disease. J Breath Res. 12:036002. [DOI] [PubMed] [Google Scholar]
- Prashanth R, Dutta Roy S, Mandal PK, Ghosh S. 2016. High-accuracy detection of early Parkinson’s disease through multimodal features and machine learning. Int J Med Inform. 90:13–21. [DOI] [PubMed] [Google Scholar]
- Prashanth R, Roy SD, Mandal PK, Ghosh S. 2014. Parkinson’s disease detection using olfactory loss and REM sleep disorder features. Conf Proc IEEE Eng Med Biol Soc. 2014:5764–5767. [DOI] [PubMed] [Google Scholar]
- President’s Information Technology Advisory Committee 2005. Report to the president: computational science: ensuring America’s competitiveness. Arlington, Virginia: National Coordination Office for Information Technology Research and Development. [Google Scholar]
- Probert CS, Ahmed I, Khalid T, Johnson E, Smith S, Ratcliffe N. 2009. Volatile organic compounds as diagnostic biomarkers in gastrointestinal and liver diseases. J Gastrointestin Liver Dis. 18:337–343. [PubMed] [Google Scholar]
- Qiu S, Wang J. 2015. Application of sensory evaluation, HS-SPME GC-MS, E-nose, and E-tongue for quality detection in citrus fruits. J Food Sci. 80:S2296–S2304. [DOI] [PubMed] [Google Scholar]
- Quinlan JR. 1986. Induction of decision trees. Mach Learn. 1:81–106. [Google Scholar]
- Quinn NP, Rossor MN, Marsden CD. 1987. Olfactory threshold in Parkinson’s disease. J Neurol Neurosurg Psychiatry. 50:88–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2008. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. 1993. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 73:597–609. [DOI] [PubMed] [Google Scholar]
- Rosenblatt F. 1958. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev. 65:386–408. [DOI] [PubMed] [Google Scholar]
- Saito YA, Strege PR, Tester DJ, Locke GR III, Talley NJ, Bernard CE, Rae JL, Makielski JC, Ackerman MJ, Farrugia G. 2009. Sodium channel mutation in irritable bowel syndrome: evidence for an ion channelopathy. Am J Physiol Gastrointest Liver Physiol. 296:G211–G218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz G, Thomas-Danguin T, Hamdani el H, Le Poupon C, Briand L, Pernollet JC, Guichard E, Tromelin A. 2008. Relationships between molecular structure and perceived odor quality of ligands for a human olfactory receptor. Chem Senses. 33:639–653. [DOI] [PubMed] [Google Scholar]
- Schwartz BS, Doty RL, Monroe C, Frye R, Barker S. 1989. Olfactory function in chemical workers exposed to acrylate and methacrylate vapors. Am J Public Health. 79:613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secundo L, Snitz K, Sobel N. 2014. The perceptual logic of smell. Curr Opin Neurobiol. 25:107–115. [DOI] [PubMed] [Google Scholar]
- Serby M, Corwin J, Conrad P, Rotrosen J. 1985. Olfactory dysfunction in Alzheimer’s disease and Parkinson’s disease. Am J Psychiatry. 142:781–782. [DOI] [PubMed] [Google Scholar]
- Shalev-Shwartz S, Ben-David S. 2014. Understanding machine learning: from theory to algorithms. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Shang L, Liu C, Tomiura Y, Hayashi K. 2017. Machine-learning-based olfactometer: prediction of odor perception from physicochemical features of odorant molecules. Anal Chem. 89:11999–12005. [DOI] [PubMed] [Google Scholar]
- Sinding C, Valadier F, Al-Hassani V, Feron G, Tromelin A, Kontaris I, Hummel T. 2017. New determinants of olfactory habituation. Sci Rep. 7:41047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh Z, Saito M, Kurita Y, Takiguchi N, Ohtake H, Tsuji T. 2014. A comparison between the human sense of smell and neural activity in the olfactory bulb of rats. Chem Senses. 39:91–105. [DOI] [PubMed] [Google Scholar]
- Stiasny-Kolster K, Mayer G, Schäfer S, Möller JC, Heinzel-Gutenbrunner M, Oertel WH. 2007. The REM sleep behavior disorder screening questionnaire–a new diagnostic instrument. Mov Disord. 22:2386–2393. [DOI] [PubMed] [Google Scholar]
- Strotmann J, Wanner I, Helfrich T, Beck A, Meinken C, Kubick S, Breer H. 1994. Olfactory neurones expressing distinct odorant receptor subtypes are spatially segregated in the nasal neuroepithelium. Cell Tissue Res. 276:429–438. [DOI] [PubMed] [Google Scholar]
- Thrun MC. 2018. Projection-based clustering through self-organization and swarm intelligence: combining cluster analysis with the visualization of high-dimensional data. Wiesbaden, Germany: Springer Fachmedien Wiesbaden. ISBN:978-3-658-20540-9. doi:10.1007/978-3-658-20540-9 [Google Scholar]
- Thulasiraman K, Swamy MNS. 1992. Graphs: theory and algorithms. New York (NY): Wiley. [Google Scholar]
- Tsiouris KM, Rigas G, Gatsios D, Antonini A, Konitsiotis S, Koutsouris DD, Fotiadis DI. 2017. Predicting rapid progression of Parkinson’s disease at baseline patients evaluation. Conf Proc IEEE Eng Med Biol Soc. 2017:3898–3901. [DOI] [PubMed] [Google Scholar]
- Tsuji T, Fukuda O, Ichinobe H, Kaneko M. 1999. A log-linearized Gaussian mixture network and its application to EEG pattern classification. IEEE Transact Syst Man Cybernet C Appl Rev. 29:60–72. [Google Scholar]
- Turing A. 1950. Computing machinery and intelligence. Mind LIX:433–460. doi:10.1093/mind/LIX.236.433 [Google Scholar]
- Ultsch A. 2000. Visualisation and classification with artificial life. In: Kiers HAL, Rasson J-P, Groenen PJF, Schader M, editors. Data analysis, classification, and related methods. Berlin (Heidelberg): Springer Berlin Heidelberg; p. 229–234. [Google Scholar]
- Ultsch A. 2003. Maps for visualization of high-dimensional data spaces. Kitakyushu, Japan: WSOM: 225–230. [Google Scholar]
- Ultsch A, Lötsch J. 2014. Functional abstraction as a method to discover knowledge in gene ontologies. PLoS One. 9:e90191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch A, Lötsch J. 2017. Machine-learned cluster identification in high-dimensional data. J Biomed Inform. 66:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch A, Sieman HP. 1990. Kohonen’s self organizing feature maps for exploratory data analysis. INNC’90, Int. Neural Network Conference Dordrecht (Netherlands): Kluwer; p. 305–308. [Google Scholar]
- Vassar R, Ngai J, Axel R. 1993. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 74:309–318. [DOI] [PubMed] [Google Scholar]
- Velayudhan L, Gasper A, Pritchard M, Baillon S, Messer C, Proitsi P. 2015. Pattern of smell identification impairment in Alzheimer’s disease. J Alzheimers Dis. 46:381–387. [DOI] [PubMed] [Google Scholar]
- Wagner M, Sudhoff H, Zamelczyk-Pajewska M, Kośmider J, Linder R. 2006. A computer-based approach to assess the perception of composite odour intensity: a step towards automated olfactometry calibration. Physiol Meas. 27:1–12. [DOI] [PubMed] [Google Scholar]
- Wojnowski W, Majchrzak T, Dymerski T, Gębicki J, Namieśnik J. 2017. Electronic noses: powerful tools in meat quality assessment. Meat Sci. 131:119–131. [DOI] [PubMed] [Google Scholar]
- Yan J, Guo X, Duan S, Jia P, Wang L, Peng C, Zhang S. 2015. Electronic nose feature extraction methods: a review. Sensors (Basel). 15:27804–27831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein S. 1998. Functional expression of a mammalian odorant receptor. Science. 279:237–242. [DOI] [PubMed] [Google Scholar]
- Zhou J, Huang ZA, Kumar U, Chen DDY. 2017. Review of recent developments in determining volatile organic compounds in exhaled breath as biomarkers for lung cancer diagnosis. Anal Chim Acta. 996:1–9. [DOI] [PubMed] [Google Scholar]