Abstract
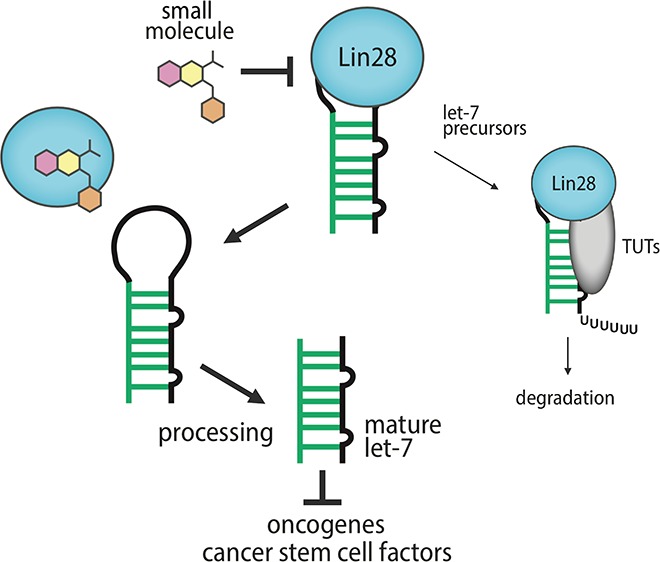
Abnormal function of RNA-binding proteins can lead to dysregulation of RNA function, causing a variety of disease states. Thus, developing small-molecule modulators of protein–RNA interactions is one of the key challenges in chemical biology. Herein, we performed a high-throughput screening of chemical libraries using a Förster resonance energy transfer-based Lin28–let-7 interaction assay to identify a potent small-molecule inhibitor of the protein–microRNA interaction, as it is an important target implicated in stem cell-like phenotypes in cancer cells. The new inhibitor KCB3602 selectively restored cellular let-7 microRNA levels, decreased the expression of a panel of oncogenes responsible for cancer stem cell maintenance, and showed potential anticancer activities. We expect that our Lin28–let-7 interaction inhibitor will provide a good starting point for pharmacological eradication of cancer stem cells.
Keywords: microRNA modulator, protein−RNA interaction, Lin28−let-7 axis, cancer stem cell, anticancer agent
RNA-binding proteins (RBPs) play crucial roles in the post-transcriptional regulation of coding and noncoding RNAs.1 Aberrant expression or mutation in RBPs often results in a variety of diseases including cancer, cardiovascular diseases, and inflammatory diseases.2−4 Therefore, finding small-molecule modulators of various types of protein–RNA interactions (PRIs) is one of the emerging challenges in chemical biology and drug discovery. For example, a group of small molecules that specifically bind oncogenic microRNA (miRNA) precursors have been successfully used to block their biogenesis in cells and animal models.5−7
Lin28–let-7 interaction is an example of highly interesting therapeutic targets. Lin28a and Lin28b (collectively referred to as Lin28) are aberrantly expressed in many cancer cells and act by binding to let-7 microRNA precursors, which blocks the Drosha- and Dicer-mediated maturation of tumor-suppressor let-7 miRNAs.8,9 Moreover, Lin28-bound let-7 precursors can be oligouridylated by terminal uridyltransferases (TUTs), which leads to the miRNA degradation (Figure 1).10,11 In particular, Lin28 is a stem cell factor whose expression confers stem cell-like properties to cancer cells.9 Indeed, there are many evidence showing that Lin28 plays important roles in the formation of cancer stem cells (CSCs), and the protein is now being considered as a biomarker for CSC.9 At the same time, it has been reported that let-7 depletion drives CSC-like phenotypes in cancers.12−14 On the contrary, forced let-7 expression could block the progression of CSCs.13 Therefore, small-molecule inhibitors of the Lin28–let-7 interaction would restore let-7 biogenesis and hence be developed as potential anticancer agents, especially for eradicating CSCs, which are responsible for tumor recurrence and resistance to conventional therapies.9
Figure 1.
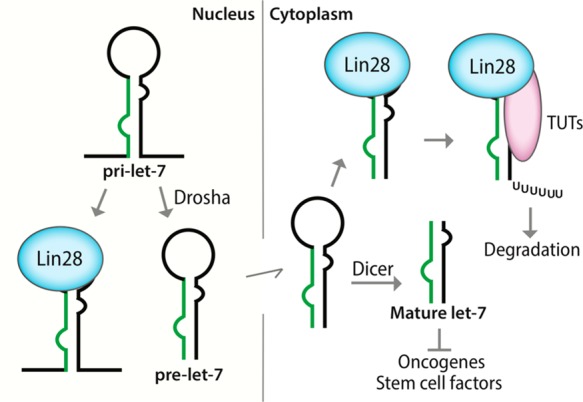
Schematic of the biogenesis of let-7 miRNAs. Lin28 binds both pri-let-7 and pre-let-7 to block their Drosha- and Dicer-mediated processing.
Previously, we have reported a Förster resonance energy transfer (FRET)-based Lin28–let-7 binding assay that can be applied to high-throughput screening of compound libraries (Figure S1a).15 The assay was successfully employed in this study to screen a large collection of drug-like small molecules, which resulted in the identification of a new class of small-molecule inhibitor of the Lin28–let-7 interaction. We investigated potent and selective cellular activities as well as mode of action of the new inhibitor. Especially, we found that the inhibitor blocks the expression of a panel of let-7 target oncogenes, which are closely related with CSC proliferation. We also investigated anticancer activities of the new inhibitor in the context of CSC-like phenotypes.
Given that our FRET-based binding assay is simple, homogeneous, and robust, we envisioned the high-throughput screening of an 8,400-member Korea Chemical Bank (KCB) representative compound library. A few inhibitors of the Lin28–let-7 interaction reported thus far suffer from low potency in cells.15−17 Therefore, new chemical entities targeting this interaction are highly desirable for the development of anticancer agents, especially for targeting stem cell-like features of cancer cells. Compounds showing inhibitory activities higher than 30% were selected as hit candidates from this high-throughput screening, and they were retested by using an independent gel-based electrophoretic mobility shift assay (EMSA) to exclude false positives and measure accurate inhibitory effects. We used the Lin28-binding module “pre-element (preE)” within long let-7 precursor sequences8 in this assay. The miRNA was fluorescently labeled at its 5′ end to enable direct visualization and quantification of protein-bound and unbound fractions.
We first identified KCB3566 as an initial hit with an IC50 of 11 μM in EMSA (Figures 2a and S1b). To identify compounds with enhanced potency, we performed a structure–activity relationship study by searching for analogues of KCB3566 within the entire 430,000-member KCB compound libraries and evaluating them by EMSA. Among these analogues, KCB3602 exhibited the highest potency with an IC50 at 4.8 μM and was confirmed as a nonpromiscuous binder. Therefore, KCB3602 was selected as the final hit (Figures 2a and S2a,b). KCB3602 showed similar activities in an EMSA using a different member of the let-7 family (PreE-let-7a-1, S2c). It also exhibited dose-dependent inhibitory activity in our FRET assay (S2d).
Figure 2.
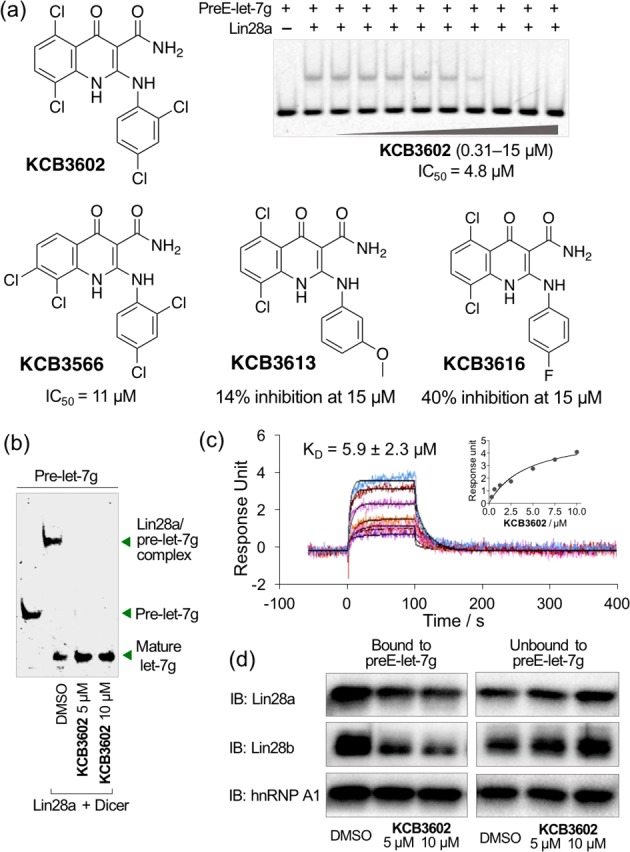
In vitro hit validation. (a) Structures and inhibitory activities of the hit analogues. EMSA was performed by using Lin28a–preE-let-7g pair. (b) KCB3602 promoted let-7 maturation at the Dicer assay. (c) SPR analysis to detect direct binding event between Lin28a and KCB3602 at various concentrations ranging from 0.31 to 10 μM. Inset shows the dose–response curve. Calculated Rmax is 43. (d) RNA pull-down assay using 5′-biotinylated preE-let-7g and proteome from JAR human choriocarcinoma cells to check the target engagement at the cellular context.
To check whether the inhibition of Lin28–let-7 interaction would lead to increased let-7 biogenesis, we performed in vitro Dicer assay with KCB3602. In the presence of Lin28, pre-let-7 binds to Lin28 and Dicer could not fully convert pre-let-7 to mature let-7. However, KCB3602 was able to block the engagement of pre-let-7 with Lin28, and this blocking resulted in the complete formation of mature let-7 miRNA (Figure 2b). It is important to note, however, that KCB3602 did not affect the activity of Dicer itself (Figure S3).
Next, we elucidated the mode of action of KCB3602 through a biophysical study using surface plasmon resonance (SPR) analysis and showed its binding to full length Lin28a (Figure 2c). Lin28 has two distinct RNA-binding domains—cold shock domain (CSD) and zinc knuckle domain (ZKD)—and both of them are responsible for high-affinity binding to let-7 miRNA precursors.8 SPR analysis using each of these domains showed that KCB3602 exclusively binds to CSD, not to ZKD (Figure S4). Therefore, disrupting the binding event between let-7 precursors and CSD itself was sufficient enough to dissociate let-7 precursors from Lin28 and rescue let-7 biogenesis.
We then proceeded to verify whether KCB3602 is active at the cellular context. In a let-7 miRNA pull-down assay, biotinylated preE-let-7g was immobilized on streptavidin beads and incubated with the proteome from JAR human choriocarcinoma cells expressing Lin28a and Lin28b. As expected, both Lin28a and Lin28b were efficiently pulled down by the RNA. In addition, the Lin28–let-7 binding event was clearly blocked by KCB3602 in a dose-dependent manner (Figure 2d). Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), another well-known let-7 precursor-binding protein,18 was also efficiently pulled down by the immobilized miRNA. However, their interaction was not disrupted by KCB3602, which clearly demonstrated its selectivity toward Lin28 proteins (Figure 2d).
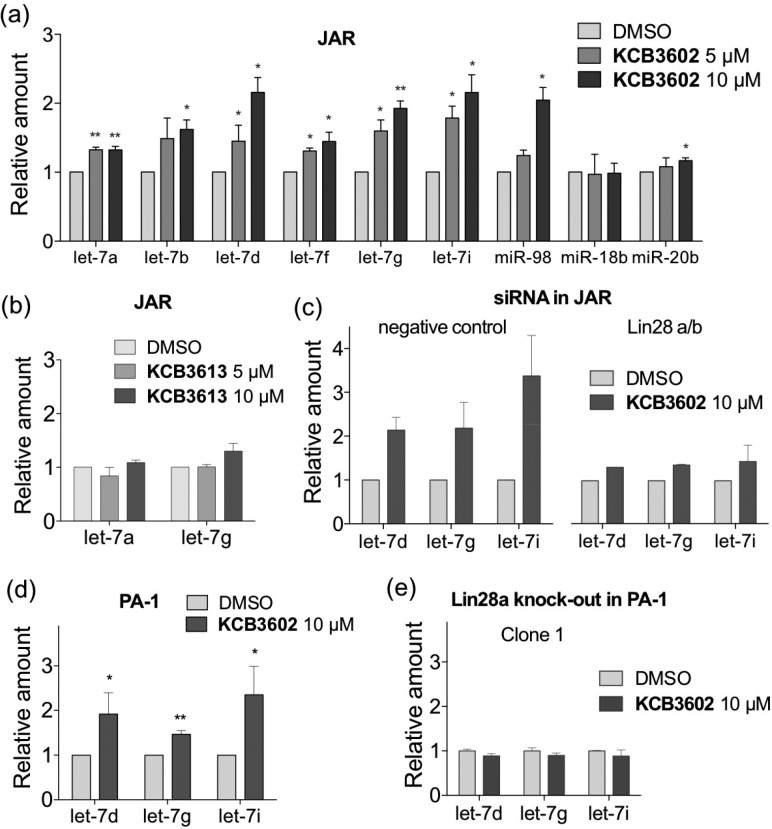
The previously reported inhibitors of Lin28 exhibited cellular activities only at higher concentrations.15−17 In contrast, 10 μM of KCB3602 was enough to induce a sufficient increase in the cellular let-7 levels in Lin28-expressing JAR cells, and this increase was dose-dependent (Figure 3a). Moreover, the cellular amount of all let-7 family members tested in this study was increased, while the level of unrelated miRNAs (miR-18b and miR-20b) remained largely unchanged. We also found that KCB3613, a structurally similar analogue of KCB3602 with quite low potency in EMSA, exhibited significantly reduced activity in cells (Figure 3b). In order to confirm that the increase in cellular let-7 levels was the outcome of Lin28-specific targeting, we knocked down both Lin28a and Lin28b by siRNA in JAR cells and investigated the effect of KCB3602. As shown in Figures 3c and S5a, the increase in cellular let-7 levels was clearly attenuated by Lin28 knock-down. In addition, let-7 levels were substantially restored in PA-1 human ovarian teratocarcinoma cells that express Lin28a (Figure 3d).19 On the contrary, we were able to see minimal changes in cellular let-7 levels upon treatment with KCB3602 in MCF7 human breast adenocarcinoma cells that barely express endogenous Lin28 (Figure S6a).20,21 Furthermore, we performed clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated genome editing22 in PA-1 cells and obtained three clonal cell lines in which the Lin28a gene was completely knocked out (Figure S5b). Treatment of the Lin28a-knocked out PA-1 cell lines with KCB3602 caused no increase in cellular let-7 levels, thus reconfirming that KCB3602 acts by targeting Lin28 (Figures 3e and S5c).
Figure 3.
Quantification of mature let-7 levels by qRT-PCR to confirm cellular activity and Lin28-specificity of KCB3602. Changes in miRNA levels after treatment of JAR cells with (a) KCB3602 (≥3 biological replicates) and (b) KCB3613 (3 biological replicates) for 20 h. (c) Effect of KCB3602 on let-7 levels in Lin28 knocked-down JAR cells. Cells were treated with KCB3602 for 20 h (2 biological replicates). (d) Changes in let-7 levels after 20 h treatment of PA-1 human ovarian teratocarcinoma cells with KCB3602 (4 biological replicates). (e) Effect of KCB3602 on let-7 levels in PA-1 cells where Lin28a was knocked out by CRISPR/Cas9-mediated genome editing. Cells were treated with KCB3602 for 20 h (2 technical replicates). Error bars represent standard deviation (*P < 0.05, **P < 0.01, paired two-tailed t test was performed when n ≥ 3). U6 snRNA was used as an endogenous control in all experiments.
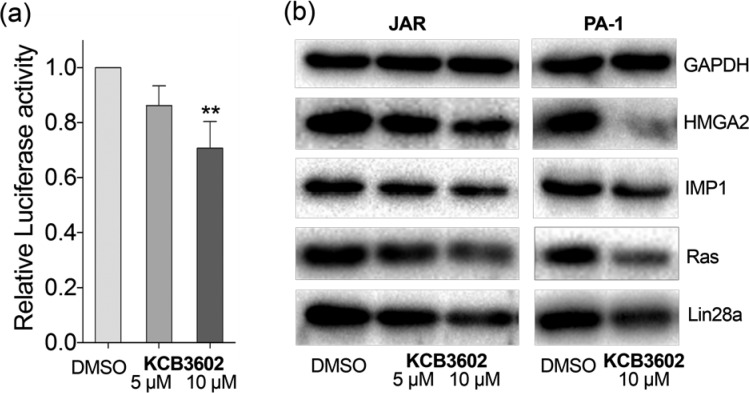
Based on these results, we then investigated whether KCB3602 could block the expression of let-7 target genes. First, a dual luciferase assay was performed by transfecting JAR cells with a reporter construct (wt) where the luciferase gene was fused with 3′ untranslated region (UTR) of high-mobility group AT-hook2 (HMGA2) gene that contains seven let-7 complementary sites.23 At the same time, cells were transfected with a control construct (m7) in which all let-7 target sites were mutated to block their association with let-7.23 The expression of the target luciferase gene was dose-dependently decreased when compared to that of the control luciferase gene upon KCB3602 treatment (Figure 4a), indicating that increase in let-7 miRNA levels apparently leads to the suppression of its target gene expression.
Figure 4.
(a) Luciferase reporter assay with constructs containing HMGA2 3′ UTR. Cells were treated with KCB3602 for 20 h (4 biological replicates, standard deviation, **P < 0.01 from paired two-tailed t test). The expression of the reporter gene (wt) was normalized to that of the control gene (m7). (b) Western blot analysis in JAR cells and PA-1 cells to observe the effect of KCB3602 on the expression of endogenous let-7 target oncogenes. Cells were treated with KCB3602 for 24 h.
As shown in Figures 4b and S7a, KCB3602 decreased the amount of endogenous let-7 target gene products as well. These include HMGA2, IGF2 mRNA-binding protein 1 (IMP1), Ras, and Lin28a itself, all of which are well-known oncogenes.9,24 This result shows that restoring let-7 biogenesis is an efficient way to block the function of a panel of oncogenes simultaneously. More importantly, those proteins have been reported to play functional roles for the maintenance of CSCs.9,25−28 For example, HMGA2 up-regulation is associated with stem cell signatures in human intestinal cancer and gastric cancer.26,28 IMP1 promotes stem cell-like phenotype including drug resistance and increased metastasis in triple-negative breast carcinoma, colorectal cancer, and hepatocellular carcinoma.27 In addition, K-Ras has been proposed to activate CSCs during colorectal tumorigenesis,25 and CSC inhibitor salinomycin targets K-Ras signaling.29 Therefore, small molecule-based modulation of Lin28–let-7 axis could be an effective way to modulate the proliferation of CSCs that are hard to remove by conventional chemotherapy.30 We also checked the effect of the hit compound on the expression of the above oncogenes in PA-1 and MCF7 cells. In line with its effect on let-7 levels, the oncogenes were clearly down-regulated by KCB3602 in Lin28a-expressing PA-1 cells (Figures 4b and S7b). On the contrary, KCB3602 did not affect the expression of those oncogenes in MCF7 cells that barely express endogenous Lin28 (Figures S6b,c). Moreover, expression of these genes were largely unchanged by KCB3613 (Figure S8). These results verified that KCB3602 selectively blocks the expression of those oncogenes by inhibiting Lin28 and restoring cellular let-7 levels.
Because KCB3602 down-regulated a panel of CSC-related oncogene products, we hypothesized that the Lin28 inhibitor would block the stem cell-like phenotypes of cancer cells. To prove this, we tested whether KCB3602 can relieve drug-resistance which is one of distinctive features of CSCs. First, we enriched drug-tolerant persister cells31 from JAR cell line by treating the cells with cytotoxic concentration of paclitaxel and regrowing them in the absence of the drug. When these paclitaxel-resistant cells were pretreated with KCB3602, the IC50 value of paclitaxel was clearly reduced (Figures 5a and S9a). In fact, IMP1 protein, a let-7 target and CSC factor,27 has been known to bind and stabilize the mRNA of multidrug resistance 1 (MDR1)32 which is overexpressed in many CSCs.33,34 Our mechanistic investigation revealed that MDR1 protein was down-regulated and thereby paclitaxel became more potent because the let-7-enhancing KCB3602 decreased IMP1 levels in the drug-resistant cells (Figure 5b). A previously reported benzopyranylpyrazole-based Lin28 inhibitor,15 which we named as SB1301, also relieved drug-resistance by the same mechanism (Figure 5a,b). In addition, KCB3602 clearly blocked the sphere-like growth of cancer cells which is one of the representative characteristics of CSCs (Figure 5c,d).35 Finally, we investigated the effect of KCB3602 on viabilities of a couple of cell lines. Even though the compound preferentially blocked the proliferation of the target cancer cell lines (JAR and PA-1), it exhibited the marginal growth arrest of normal cell lines (C2C12 and C8-D1A) at a high concentration (Figure S9b). More extensive structure–acitvity relationship studies will be performed in further studies for identifying compounds with enhanced potency as well as improved selectivity.
Figure 5.
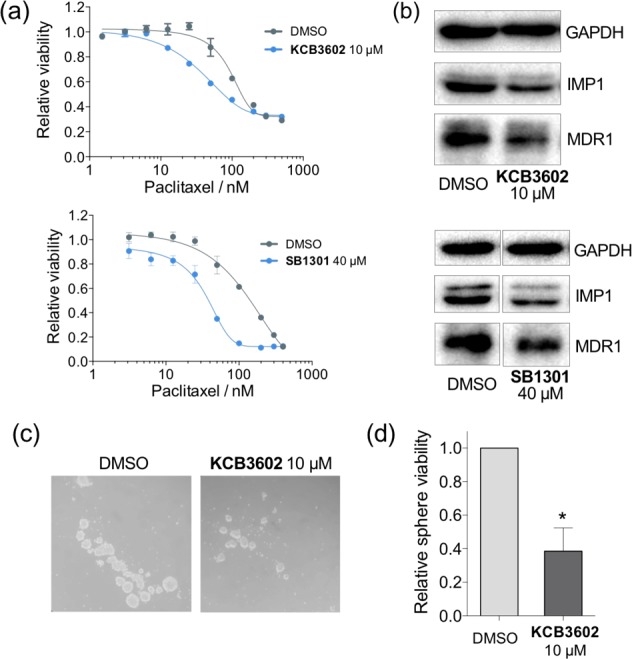
(a) Water-soluble tetrazolium (WST) assay to measure the viability of paclitaxel-resistance JAR cells. Cells were pretreated with KCB3602 or SB1301 for 30 h, and then treated with paclitaxel for 72 h in the absence of those compounds. (b) Effect of KCB3602 or SB1301 on the expression of IMP1 and MDR1 which are associated with drug resistance. Cells were treated with the compound for 30 h. (c) Sphere formation assay to observe the effect of KCB3602 on stem-cell like growth of cancer cells. JAR cells were pretreated with KCB3602 for 24 h, dissociated to single cells, and the same number of viable cells were incubated in sphere forming media for 12 days. Degree of sphere formation was observed under a microscope or (d) measured by WST assay (3 biological replicates, standard deviation, *P < 0.05 from paired two-tailed t test).
In summary, we have discovered a new class of Lin28 inhibitor, KCB3602, with significantly improved potency in cellular systems. This Lin28-selective inhibitor exerted its action by increasing cellular let-7 levels and subsequently decreasing the expression of diverse oncogenes which have important roles for the maintenance of CSCs. On the basis of these results, we expected that KCB3602 would block stem cell-like features of cancer cells and proved that it relieved the drug-resistance and inhibited the sphere-like growth in a cancer model. Accordingly, we believe that small-molecule inhibitors of Lin28–let-7 interaction will provide new tools for the removal of CSCs which are responsible for tumor relapse, metastasis, and drug resistance.30 Meanwhile, we have shown that PRIs, which were once regarded undruggable, can be viable therapeutic targets for small-molecule therapeutics. We expect that chemical modulators of different types of PRIs would be developed for expanding the repertoire of drug candidates having novel modes of action.
Acknowledgments
We thank Korea Chemical Bank of Korea Research Institute of Chemical Technology for providing compounds for screening and biological assays.
Glossary
ABBREVIATIONS
- RBP
RNA-binding protein
- PRI
protein–RNA interaction
- miRNA or miR
microRNA
- TUT
terminal uridyltransferase
- CSC
cancer stem cell
- FRET
Förster resonance energy transfer
- EMSA
electrophoretic mobility shift assay
- preE
pre-element
- SPR
surface plasmon resonance
- CSD
cold shock domain
- ZKD
zinc knuckle domain
- hnRNP A1
heterogeneous nuclear ribonucleoprotein A1
- CRISPR
clustered regularly interspaced short palindromic repeats
- UTR
untranslated region
- HMGA2
high-mobility group AT-hook2
- IMP1
IGF2 mRNA-binding protein 1
- MDR1
multidrug resistance 1
- WST
water-soluble tetrazolium
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00323.
Experimental procedures, supporting figures, and spectroscopic data (PDF)
Author Contributions
∥ These authors contributed equally. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by the National Creative Research Initiative Grant (NRF-2014R1A3A2030423) and the Bio & Medical Technology Development Program (2012M3A9C4048780) through the National Research Foundation of Korea (NRF) funded by the Korean Government (Ministry of Science and ICT). W.G.B is grateful for the BK21 Fellowship Program.
The authors declare no competing financial interest.
Supplementary Material
References
- Gerstberger S.; Hafner M.; Tuschl T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E.; Harris M. E. Specificity and nonspecificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2015, 16, 533–544. 10.1038/nrm4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B.; Billaud M.; Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer 2017, 3, 506–528. 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- Li Y.; Kowdley K. V. MicroRNAs in common human diseases. Genomics, Proteomics Bioinf. 2012, 10, 246–253. 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Cameron M. D.; Haga C. L.; Rosenberg L. H.; Lafitte M.; Duckett D. R.; Phinney D. G.; Disney M. D. Design of a small molecule against an oncogenic noncoding RNA. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 5898–5903. 10.1073/pnas.1523975113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney M. D.; Angelbello A. J. Rational Design of small molecules targeting oncogenic noncoding RNAs from sequence. Acc. Chem. Res. 2016, 49, 2698–2704. 10.1021/acs.accounts.6b00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Luo Y.; Tran T.; Haniff H. S.; Nakai Y.; Fallahi M.; Martinez G. J.; Childs-Disney J. L.; Disney M. D. Defining RNA-small molecule affinity landscapes enables design of a small molecule inhibitor of an oncogenic noncoding RNA. ACS Cent. Sci. 2017, 3, 205–216. 10.1021/acscentsci.7b00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y.; Chen C.; Gregory R. I.; Chou J. J.; Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011, 147, 1080–1091. 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzeau J.; Menezes M. R.; Cao S. Y.; Hagan J. P. The Lin28/let-7 pathway in cancer. Front. Genet. 2017, 8, 1–16. 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle C. R.; Walleshauser J.; Joshua-Tor L. Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis. Nat. Struct. Mol. Biol. 2017, 24, 658–665. 10.1038/nsmb.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. F.; Nam Y.; Lee A. K.; Yu C. X.; Roth K.; Chen C.; Ransey E. M.; Sliz P. Lin28 zinc knuckle domain is required and sufficient to induce let-7 oligouridylation. Cell Rep. 2017, 18, 2664–2675. 10.1016/j.celrep.2017.02.044. [DOI] [PubMed] [Google Scholar]
- Jin B.; Wang W.; Meng X. X.; Du G.; Li J.; Zhang S. Z.; Zhou B. H.; Fu Z. H. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer 2016, 16, 863. 10.1186/s12885-016-2904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Liu J.; Xu C.; Tang S. C.; Ren H. The insights of Let-7 miRNAs in oncogenesis and stem cell potency. J. Cell. Mol. Med. 2016, 20, 1779–1788. 10.1111/jcmm.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F.; Li T. T.; Wang K. L.; Xiao G. Q.; Wang J. H.; Zhao H. D.; Kang Z. J.; Fan W. J.; Zhu L. L.; Li M.; Cui B.; Zheng F. M.; Wang H. J.; Lam E. W.; Wang B.; Xu J.; Liu Q. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017, 8, e2569 10.1038/cddis.2016.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.; Byun W. G.; Koo J. Y.; Park H.; Park S. B. Discovery of a small-molecule inhibitor of protein-microRNA interaction using binding assay with a site-specifically labeled Lin28. J. Am. Chem. Soc. 2016, 138, 13630–13638. 10.1021/jacs.6b06965. [DOI] [PubMed] [Google Scholar]
- Roos M.; Pradere U.; Ngondo R. P.; Behera A.; Allegrini S.; Civenni G.; Zagalak J. A.; Marchand J. R.; Menzi M.; Towbin H.; Scheuermann J.; Neri D.; Caflisch A.; Catapano C. V.; Ciaudo C.; Hall J. A small-molecule inhibitor of Lin28. ACS Chem. Biol. 2016, 11, 2773–2781. 10.1021/acschembio.6b00232. [DOI] [PubMed] [Google Scholar]
- Wang L.; Rowe R. G.; Jaimes A.; Yu C.; Nam Y.; Pearson D. S.; Zhang J.; Xie X.; Marion W.; Heffron G. J.; Daley G. Q.; Sliz P. Small-molecule inhibitors disrupt let-7 oligouridylation and release the selective blockade of let-7 processing by Lin28. Cell Rep. 2018, 23, 3091–3101. 10.1016/j.celrep.2018.04.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G.; Caceres J. F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010, 17, 1011–1018. 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S.; Maihle N. J.; Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 2010, 29, 2153–2159. 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- Zhong X.; Li N.; Liang S.; Huang Q.; Coukos G.; Zhang L. Identification of microRNAs regulating reprogramming factor Lin28 in embryonic stem cells and cancer cells. J. Biol. Chem. 2010, 285, 41961–41971. 10.1074/jbc.M110.169607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Li H.; Feng J.; Cui X.; Huang W.; Li Y.; Su F.; Liu Q.; Zhu J.; Lv X.; Chen J.; Huang D.; Yu F. Lin28 induces epithelial-to-mesenchymal transition and stemness via downregulation of let-7a in breast cancer cells. PLoS One 2013, 8, e83083 10.1371/journal.pone.0083083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A.; Hsu P. D.; Wright J.; Agarwala V.; Scott D. A.; Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C.; Hemann M. T.; Bartel D. P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007, 315, 1576–1579. 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch B.; Bley N.; Muller S.; Glass M.; Misiak D.; Lederer M.; Vetter M.; Strauss H. G.; Thomssen C.; Huttelmaier S. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Res. 2016, 44, 3845–3864. 10.1093/nar/gkw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon B. S.; Jeong W. J.; Park J.; Kim T. I.; Min do S.; Choi K. Y. Role of oncogenic K-Ras in cancer stem cell activation by aberrant Wnt/beta-catenin signaling. J. Natl. Cancer Inst. 2014, 106, djt373. 10.1093/jnci/djt373. [DOI] [PubMed] [Google Scholar]
- Madison B. B.; Jeganathan A. N.; Mizuno R.; Winslow M. M.; Castells A.; Cuatrecasas M.; Rustgi A. K. Let-7 represses carcinogenesis and a stem cell phenotype in the intestine via regulation of Hmga2. PLoS Genet. 2015, 11, e1005408 10.1371/journal.pgen.1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrauwe N.; Suva M. L.; Janiszewska M.; Riggi N.; Stamenkovic I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016, 30, 2459–2474. 10.1101/gad.287540.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.; Sun B.; Zhu D.; Zhao X.; Zhang Y.; Dong X.; Che N.; Li J.; Liu F.; Zhao N.; Zhang D.; Liu T.; Lin X. HMGA2 regulates CD44 expression to promote gastric cancer cell motility and sphere formation. Sci. Rep. 2017, 7, 260–274. [PMC free article] [PubMed] [Google Scholar]
- Najumudeen A. K.; Jaiswal A.; Lectez B.; Oetken-Lindholm C.; Guzman C.; Siljamaki E.; Posada I. M.; Lacey E.; Aittokallio T.; Abankwa D. Cancer stem cell drugs target K-ras signaling in a stemness context. Oncogene 2016, 35, 5248–5262. 10.1038/onc.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E.; Clevers H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- Hangauer M. J.; Viswanathan V. S.; Ryan M. J.; Bole D.; Eaton J. K.; Matov A.; Galeas J.; Dhruv H. D.; Berens M. E.; Schreiber S. L.; McCormick F.; McManus M. T. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 2017, 551, 247–250. 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B.; Park S. M.; Murmann A. E.; Gwin K.; Montag A. G.; Zillhardt M.; Hua Y. J.; Lengyel E.; Peter M. E. Let-7 modulates acquired resistance of ovarian cancer to Taxanes via IMP-1-mediated stabilization of multidrug resistance 1. Int. J. Cancer 2012, 130, 1787–1797. 10.1002/ijc.26190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah L. N.; Chow E. K. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013, 2, 3. 10.1186/2001-1326-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.; Huang Y. H.; Chen J. L. Understanding and targeting cancer stem cells: therapeutic implications and challenges. Acta Pharmacol. Sin. 2013, 34, 732–740. 10.1038/aps.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. C.; Ingram P. N.; Fouladdel S.; McDermott S. P.; Azizi E.; Wicha M. S.; Yoon E. High-throughput single-cell derived sphere formation for cancer stem-like cell identification and analysis. Sci. Rep. 2016, 6, 27301. 10.1038/srep27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.