Abstract
Most organisms contain multiple soluble protein‐based redox carriers such as members of the ferredoxin (Fd) family, that contain one or more iron–sulfur clusters. The potential redundancy of Fd proteins is poorly understood, particularly in connection to the ability of Fd proteins to deliver reducing equivalents to members of the “radical SAM,” or S‐adenosylmethionine radical enzyme (ARE) superfamily, where the activity of all known AREs requires that an essential iron–sulfur cluster bound by the enzyme be reduced to the catalytically relevant [Fe4S4]1+ oxidation state. As it is still unclear whether a single Fd in a given organism is specific to individual redox partners, we have examined the five Fd proteins found within Thermotoga maritima via direct electrochemistry, to compare them in a side‐by‐side fashion for the first time. While a single [Fe4S4]‐cluster bearing Fd (TM0927) has a potential of −420 mV, the other four 2x[Fe4S4]‐bearing Fds (TM1175, TM1289, TM1533, and TM1815) have potentials that vary significantly, including cases where the two clusters of the same Fd are essentially coincident (e.g., TM1175) and those where the potentials are well separate (TM1815).
Keywords: redox homeostasis, iron–sulfur cluster, redox potential, ferredoxin, S‐adenosylmethionine, thiomethylation
Introduction
Ferredoxins are ubiquitous iron–sulfur cluster containing proteins that are often thought as playing essential roles as redox‐relays.1 While associated with the fermentative chemistry of anaerobic organisms,2 ferredoxin (Fd) proteins are now found in many essential biochemical pathways.1 Any given organism that contains at least one Fd tends to contain multiple Fd proteins, although the specific biological chemistry of these proteins is often unclear. In a similar way, Fds have been shown to support the required step of electron delivery required by the AdoMet radical enzyme (ARE) superfamily. Each member of the family contains at least one [4Fe–4S] cluster, and canonical activity of the ARE superfamily includes reduction of the [4Fe–4S]2+ state to a 1+ oxidation state in order to reductively cleave AdoMet, generating a powerful 5′‐dA• radical species that then participates in further chemistry.3, 4, 5 As AREs are widely distributed in bacterial and archaeal organisms,4 the potential native electron donors are also diverse, including flavodoxins and Fds. Fds are often invoked as a potential electron donor in this case, on the basis of redox potential, which can be often found in the literature as an un‐measured quantity, but approximated to ∼ −400 mV, where it is often assumed that various Fd proteins are equivalent to one another. The ARE superfamily illustrates how the same electron donor may have variable successes in supporting enzymatic catalysis: pyruvate formate–lyase activating enzyme (PFL‐AE) from Escherichia coli, for example, has been successful reduced by E. coli flavodoxin,6 as has the activase for the anaerobic ribonucleotide reductase,7, 8, 9 biotin synthase,10 lipoyl synthase,11 and RlmN12 by supplying reducing equivalents from NADPH via ferredoxin (flavodoxin): NADP+ reductase. Yet, in the case of AREs of the methylthiotransferase (MTTase) subfamily, MiaB and RimO, only poor catalytic properties could be observed using flavodoxin or Fd.13, 14, 15, 16
In our related paper,17 we have demonstrated that in terms of potential native electron donors for AREs in Thermotoga maritima, there are five candidate Fds, but no flavodoxins. In this context, we have realized that T. maritima provides an opportunity to examine the redox potentials of all five single‐domain Fd proteins in a comparative fashion, in hopes of rationalizing the reactivity of Fd in support of MiaB based catalysis. The Fds of T. maritima include TM0927, which is known to possess a single [4Fe–4S] cluster, and which has been crystallographically characterized.18 The other four (TM1175, TM1289, TM1533, and TM1815) are all predicted contain two [4Fe–4S] clusters (2x[4Fe–4S] Fds) with unknown redox potentials, and no structural data.17 In our related work,17 we found that all five of the potential candidate Fds proteins are variably successful at supporting the MTTase chemistry of MiaB, where the single‐cluster bearing TM0927 was generally most proficient, and that the majority of the 2‐cluster Fds were noticeably worse, with one exception being TM1175.17 While the apparent kinetics and yield of methylthiolation depended upon the identity of the Fd in question, there appeared to be a correlation between the support of MiaB activity and those Fds that could be readily reduced by either an external NADPH‐supported reductase system or by direct pre‐reduction with dithionite. Here, we pursue the question of the importance of the redox characteristics of these ferredoxin proteins by directly assessing their redox potentials and the temperature and proton‐dependencies thereof, illuminating the requisite Fd traits that can be required to support catalytic systems of varying thermodynamic needs.
Results
Protein film electrochemistry
As described in the related manuscript, five ferredoxins were identified in Tm and characterized to determine their ability to serve as the native electron donor for MiaB (as described previously).17 To directly correlate reduction potentials for the ferredoxins to the potentials of MiaB, protein film electrochemistry (PFE) of the Fds was performed at pH 8. TM0927 is the only ferredoxin examined in this report containing a single [4Fe–4S] cluster compared to all other ferredoxins that are predicted to be and have the redox traits of 2 [4Fe–4S] Fds. Figure 1 displays the resulting voltammogram of TM0927, with a single reversible feature with a midpoint potential of −420 mV (vs. SHE). The relationship of the cathodic and anodic peak heights with respect to scan rate shows a linear relationship, confirming an adsorbed species (Fig. 1, inset).
Figure 1.
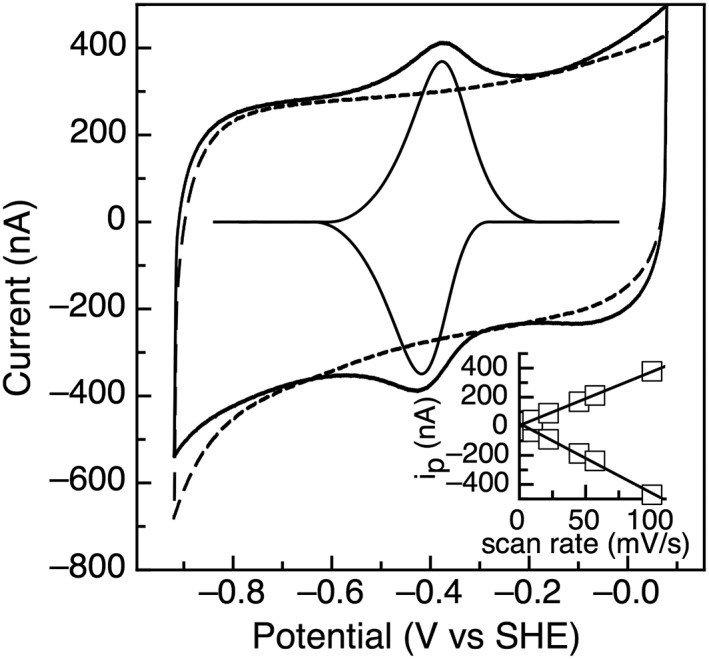
Cyclic voltammetry of TM0927 with the baseline response of the electrode shown as the dashed line. Data was collected at 100 mV/s, pH 8.0, 200 mM NaCl, and 10°C. The inset shows the dependence of peak height (i p) as a function of scan rate.
TM1175 has two [4Fe‐4S] clusters that are close (but not identical) in redox potential, with potentials of −395 and − 490 mV at pH 8 [Fig. 2(A)]. Following the established naming convention for clusters in 2 [4Fe–4S] Fds, the cluster with the higher potential is cluster A. Here, across a range of scan rates, the magnitude of the signal for cluster A is larger than that for cluster B. This is unexpected, as the electroactive coverage for the clusters should be the same presuming that the cluster loading of each site is identical. TM1289 also contains two [4Fe–4S] clusters; however, here the clusters have a larger separation in potential and display reversible peaks at −380 and − 560 mV [Fig. 2(B)]. Both of these peaks are consistent with one‐electron centers with peak widths at half height of ~ 110 mV for both clusters.19
Figure 2.
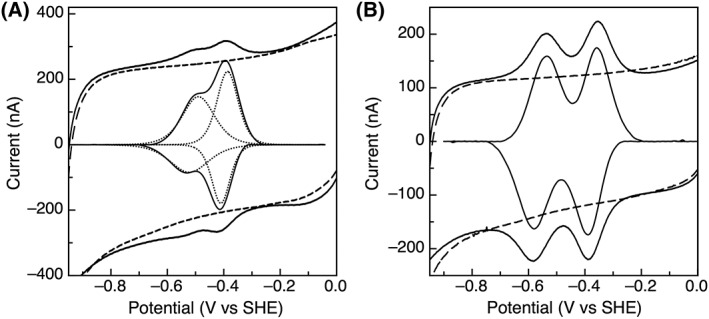
(A) Cyclic voltammetry of TM1175 with the baseline response of the electrode shown in the dashed lines and the dotted lines representing the fit for 2 one‐electron centers. (B) TM1289 on a PGE electrode with the baseline response of the electrode shown as dashed lines. Data was collected at 100 mV/s, pH 8, and 10°C.
TM1533 and TM1815 display unusually low potentials for a 2 [4Fe–4S] Fd. Cluster A of TM1533 has a midpoint potential of −415 mV and Cluster B is very low in potential at −710 mV [Fig. 3(A)]. Similarly, Cluster A of TM1815 has a reduction potential of −320 mV and Cluster B has an unusually low potential of −725 mV [Fig. 3(B)]. For both clusters in TM1533 and TM1815, the features are representative of one‐electron centers as the peak‐width at half height are ~100 mV, consistent with a one electron process. Potentials in this range have only been observed by direct electrochemistry for either the [4Fe–4S]2+/1+ couple of BtrN,20 or the unusual two‐electron, [3Fe–4S]0/−2 couple found for [3Fe–4S],[4Fe–4S]‐containing ferredoxins, which would be half as narrow in peak‐width and four‐times in height, as described by theory.19
Figure 3.
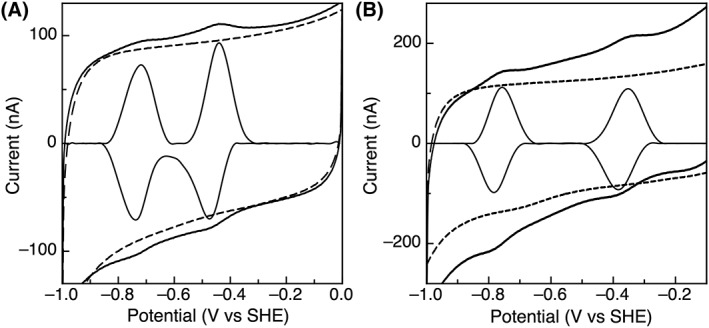
PFE response of (A) TM1533 and (B) TM1815 on PGE electrodes. Data was collected at 100 mV/s, pH 8, 200 mM NaCl, and 10°C.
Temperature dependence of reduction potentials
As T. maritima is a hyperthermophile, the impact of temperature on the reduction potential was examined. Figure 4(A) shows the temperature dependence of the midpoint potential of TM0927, which is linear up to 40°C with a slope of −0.702 mV/K. The slope of this linear fit can be related to the entropy change for the redox reaction, S rc/nF, yielding a value of −67.7 J/mol K. A similar interpretation of the Gibbs–Helmholtz plot [Fig. 4(B)] gives the H rc for the redox process, yielding a value of −28.4 kJ/mol. Over the temperature range examined, the midpoint potential for cluster A of TM1289 is linear with a slope of −0.444 mV/K, yielding an entropy change for the redox reaction of −42.8 J/mol K [Fig. 4(C)]. Figure 4(D) is the Gibbs–Helmholtz plot giving H rc of −24.5 kJ/mol. Cluster B of TM1289 does not exhibit a discernible temperature dependence, with midpoint potentials of −560 ± 3 mV over the entire temperature range examined. Only TM0927 and TM1289 had films stable enough at elevated temperatures to allow for this analysis. Even when a new film was generated for each temperature with TM1175, TM1533, and TM1815, signals were not observed for temperatures >30°C, indicating rapid desorption.
Figure 4.
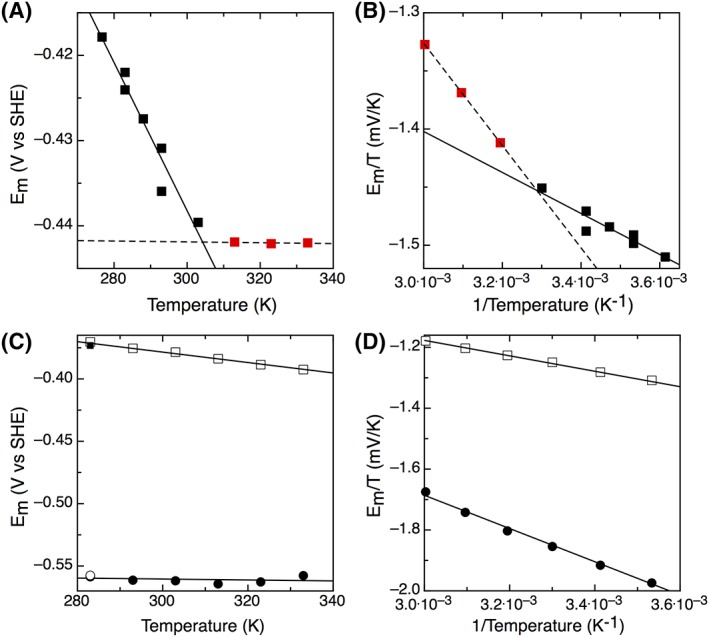
(A) Temperature dependence of the midpoint potential of TM0927 and (B) the resulting Gibbs–Helmholtz plot. The data appeared biphasic, requiring separate fitting for the two phases in both panels and A and B, where the black squares (and solid line) represent the first temperature‐dependent phase, and the red squares (and dashed line) represent the second phase of the temperature‐dependence in the panels. Data were collected at 100 mV/s and pH 8. (C) Temperature dependence of the midpoint potential of TM1289. Data were collected at 100 mV/s and pH 8 and the closed square and open circle represent Clusters A and B, respectively, for the film cooled back to 283 K. (D) The resulting Gibbs–Helmholtz plot.
Proton‐coupled electron transfer with protein film voltammetry
The possibility of proton‐coupled electron transfer for these ferredoxins was investigated by monitoring the midpoint potentials over a pH range of 4–10. Based on electrochemical theory, the pH‐dependence for a one‐electron: one‐proton process would be −56 mV/pH unit at 10°C. None of the T. maritima Fds studied exhibits a slope consistent with proton coupled electron transfer. Very modest, essentially pH invariant slopes of −6 mV/pH unit to −13 mV/pH unit were observed for the FeS clusters in TM0927, TM1289, and TM1815 (data not shown). While cluster A of TM1175 has a modest slope of −6 mV/pH unit, Cluster B displays a slope of −31 mV/pH unit (Fig. 5) due to closely spaced values of pKox (6.4) and pKred (7.3).
Figure 5.
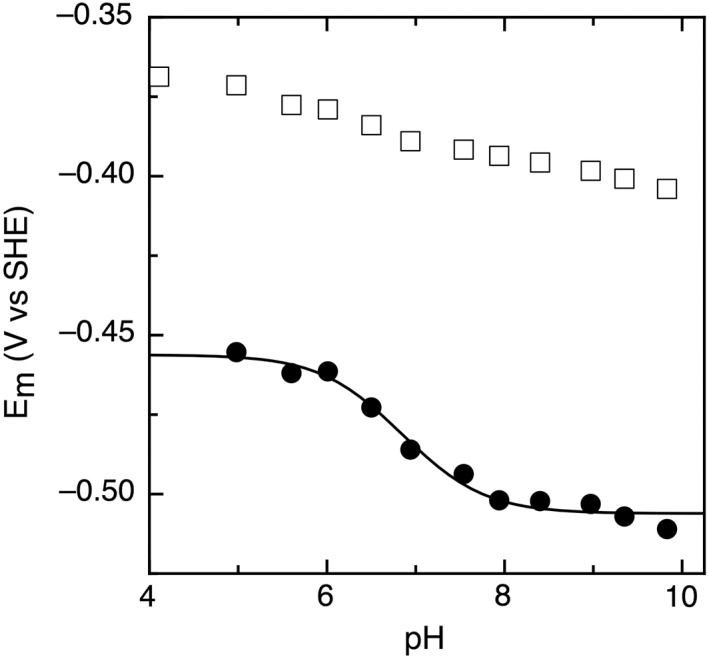
Pourbaix diagram of TM1175 with Cluster A shown in open squares and Cluster B shown in closed circles. Data were collected at 100 mV/s, 10°C.
Discussion
Tuning Fd redox potentials by alterations of sequence
The monocluster [4Fe–4S] ferredoxin from Thermotoga maritima, TM0927, was examined with protein film electrochemistry and was determined to have a reduction potential of −420 mV (Fig. 1). This is slightly lower in potential than a previous study, which measured a potential of −388 mV using differential pulse voltammetry.21 Both potentials are in the expected range of −280 to −715 mV for a [4Fe–4S]+2/+1 cluster.21, 22, 23, 24, 25 There are relatively few single [4Fe–4S] cluster Fd redox potentials reported to date. Table 1 lists the reduction potentials for characterized [4Fe–4S] Fds, where the potentials range narrowly from −370 mV to −426 mV.26, 27, 28, 29 The highest potential reported thus far comes from a Pyrococcus furiosus Fd that possesses unusual ligation of the [4Fe–4S] cluster with three cysteines and one aspartic acid ligand. The potential for this unusual [4Fe–4S] cluster is −370 mV and when the aspartic acid was changed to the canonical cysteine, the potential of the cluster decreased to −426 mV.28 This value is very close to that measured for TM0927, which does share a 50% sequence identity with the D14C variant of P. furiosus Fd.
Table 1.
Reduction Potentials of [4Fe–4S] Ferredoxins
The four additional Fds from T. maritima were examined next, all of the 2x[4Fe–4S] clusters composition. These Fds display unusual sequence and electrochemical properties compared to the two classical ferredoxin subfamilies exemplified by clostridial Fd (CpFd) and Alvin Fd, indicating that the ferredoxin family is more diverse than previously believed. Typically, bacterial 2x[4Fe–4S] Fds are small proteins approximately 55 amino acids in length with two CXXCXXCXXXCP binding motifs separated by 18 amino acids.21, 24, 25, 30, 31, 32, 33 The clostridial‐like Fds feature isopotential clusters, i.e., both clusters display very similar, if not identical potentials. Here, all of our Fds from T. maritima have discernible different redox potentials. In contrast, the Alvin‐like subfamily of Fds display two distinct redox potentials, and possess a primary sequence characterized by a six amino acid insertion between the second and third cysteines in the second binding motif (shown in red in Fig. 6). This insertion is believed to impart the unique low potential of ~ −650 mV observed for these Fds, resulting in an ~ 200 mV difference in reduction potential between the two clusters.34 Just as the T. maritima 2x[4Fe–4S] Fds do not appear be like the clostridal Fds, they also do not contain the six amino acid insertion in the second cluster‐binding motif.
Figure 6.

Sequence alignment of the four 2[4Fe–4S] Fds examined from T. maritima with the canonical CpFd and Alvin Fd. The cysteines binding the first cluster are highlighted in green, while the cysteines binding the second cluster are in cyan. The proline of the cluster‐binding motif is highlighted in yellow, where appropriate.
The reduction potentials for the clusters in TM1175 are −390 mV and − 495 mV for Cluster A and Cluster B, respectively [Fig. 2(A)]. Unexpectedly, in all scans the signal observed for Cluster A is larger than that of Cluster B, despite both centers being one electron centers, which should have equal coverage. This behavior could be due to the orientation of the protein on the electrode, with Cluster A positioned closer to the electrode, as the non‐covalent adsorption of TM1175 to the electrode results in a distribution of orientations. TM1175 shares 49% and 45% sequence identity with the Fds from Clostridium pasteurianum and Chlorobium tepidum (Ct). In comparison with these Fds, TM1175 seem contains more positively charged residues (shown in orange in Fig. 6), particularly close to the cluster binding motifs, which may help stabilize the negative charge on the clusters. At the same time, TM1175 also contains more hydrophobic residues (such as an extra Phe that sits in the second cluster binding motif (bold)) which are believed to be responsible for lower reduction potentials of FeS clusters. Previous reports have indicated that the residue before the final cysteine of the binding motif is important for tuning the potential, with bulky, hydrophobic residues resulting in lower potentials.34, 35, 36, 37 For the two cluster‐binding motifs in TM1175, the residues are Phe and Ile (also in bold), in contrast to the glutamic acid and valine for the motifs in both CpFd and CtFd. These bulkier, hydrophobic residues likely contribute to the lower potential (−495 mV) measured for TM1175.
The two clusters of TM1289 display reduction potentials similar to those of Alvin‐like Fds (−380 mV and − 560 mV), despite lacking the six amino acid insertion in the second cluster‐binding motif (red in Fig. 6). Similar to TM1175, TM1289 also has more/bulkier hydrophobic residues and more positively charged residues than the clostridial Fds, notably a series of basic residues (largely Arg, navy in Fig. 6). Again, the presence of the positively charged residues may contribute to the low potential (−560 mV) measured by stabilizing the oxidized state. The final amino acid before the fourth cysteine of the binding motifs for the first and second clusters are a gain hydrophobic, Val and Ala, additionally contributing to the low potential for cluster B.
TM1533 and TM1815 also reveals unusually low potentials for the 2[4Fe–4S] Fds. TM1533 displays potentials of −415 mV and − 710 mV with peak widths at half height of ~100 mV, consistent with 2 one‐electron centers [Fig. 3(A)]. TM1533 does not contain the Alvin‐type six amino acid insertion in the second cluster‐binding motif, which could account for the −710 mV potential of cluster B. However, TM1533 does feature a binding motif with three inserted amino acids, which may shield the cluster, resulting in the low potential.
Similar to TM1533, TM1815 has a higher potential cluster [−315 mV, Fig. 3(B)], but also a cluster of very low redox potential, −725 mV. All electrochemical features are consistent with one‐electron redox couples, but the lower potential feature is the lowest potential reported for a 2x[4Fe–4S] Fd to date. Despite having unusual electrochemical properties, TM1815 shares 41% sequence identity with CpFd. One distinct feature is that the first binding motif in TM1815 features the canonical proline residue replaced with a glycine (see the absence of the yellow highlight in Fig. 6). Proline replacement has only been observed for a few sequences of bacterial ferredoxins, including Desulfovibrio africanus FdIII, D. vulgaris Miyazaki Fd, and Rhodobacter capsulatus FdI.38, 39, 40 Previous studies on CpFd have examined the impact of the proline residue on the reduction potential of the cluster and found that when changed to lysine, asparagine, methionine, threonine, or lysine, the potential shifted by no more than 20 mV, indicating that while this residue does affect the potential, it likely is not a major contributor to tuning the potential.31
In further examining the sequences of 2x[4Fe–4S] Tm Fds, TM1175, TM1533, and TM1815 all have non‐canonical binding motifs with additional amino acids inserted in the cluster‐binding motifs. The impact of inserted amino acids in the cluster‐binding motif has been previously assessed for Rhodobacter capsulatus (Rc) FdI (FdxN) involved in the nitrogen fixation pathway. Rc FdI is an Alvin‐type 2[4Fe–4S] Fd, containing a six amino acid insertion in the cluster binding motif. For the native protein, both clusters exhibited redox potentials of −490 mV, but when the eight amino acid sequence was changed to glycine and alanine to give the more conventional two amino acid length loop, the potential for one of the clusters shifted positive in potential to −430 mV (a shift of + 60 mV). Along with structural modeling, it was proposed that the inserted amino acids form a loop‐out region, which shields the cluster, thus lowering the potential.41 The amino acids inserted in the cluster‐binding motifs for these proteins likely are not solely responsible for the redox properties observed, as TM1289 has the canonical Cp Fd binding motifs, but still displays unusual redox properties. However, the cluster‐binding motifs for these proteins contain more, and bulkier, hydrophobic residues, which may act to prevent surface accessibility to the clusters, resulting in the lower reduction potential. Additionally, the N‐terminal and C‐terminal extensions found in the Tm Fds may further shield the clusters; however, further structural analysis is required for this conclusion.
To further examine the sequence and structure of these low potential Fds, threading models of TM1175, TM1289, TM1533, and TM1815 were constructed (see Fig. 6, Ref. 17). While such models can be useful for illustrating the localization of hydrophobic residues, the N‐ and C‐termini of the T. maritima Fds differ with respect to most search models, and the models do not accurately depict the immediate environment around both clusters. For example, CpFd can be used as the basis for a threaded model, but it is only 50 amino acids long, where homology exists only in the middle of the TM1815 sequence (Fig. 6).
Table 2 summarizes the potentials measured for the 2[4Fe–4S] Fd clusters of the five ferredoxins of T. maritima along with the potentials for other 2[4Fe–4S] Fds. The potentials of the T. maritima 2[4Fe–4S] Fds mostly lie in the −330 to −675 mV range reported for other 2[4Fe–4S] Fds.25, 31, 32, 33, 34, 41, 42, 43 In terms of the native electron donor for MiaB, all of the ferredoxins from T. maritima examined contain at least one cluster with a potential capable of reducing MiaB, based on the potentials for the clusters in Bt MiaB (−390, −450 mV) determined previously.
Table 2.
Reduction Potentials of 2[4Fe–4S] Fds
| Ferredoxin | E m Cluster A | E m Cluster B | Reference |
|---|---|---|---|
| Clostridium pasteurianum Fd | −380 | −380 | 31 |
| Chlorobium tepidum FdI | −514 | −514 | 32 |
| Chlorobium tepidum FdII | −584 | −584 | 32 |
| Chromatium vinosum Fd | −460 | −655 | 25 |
| Allochromatium vinosum FdIII | −486 | −644 | 34 |
| Thauera aromatica Fd | −431 | −587 | 43 |
| Azotobacter vinelandii Fix Fd | −440 | −440 | 24 |
| Escherichia coli Fd | −418 | −675 | 34 |
| Pseudomonas aeruginosa Fd | −475 | −655 | 33 |
| Entamoeba histolytica Fd | −333 | −333 | 21 |
| Rhodobacter capsulatus FdI, FdxN | −490 | −490 | 47 |
| Rhodobacter capsulatus FdI, FdxNGA | −490 | −430 | 47 |
| TM1175 | −395 | −490 | This Study |
| TM1289 | −380 | −570 | This Study |
| TM1533 | −415 | −710 | This Study |
| TM1815 | −320 | −725 | This Study |
Temperature dependence of reduction potentials
As T. maritima is a hyperthermophilic organism with a growth optimum of ~80°C, the temperature dependence of the reduction potential for the ferredoxins was investigated. Only TM0927 and TM1289 were suitable for this analysis; TM1175, TM1533, and TM1815, all had films that were unstable at elevated temperatures, preventing measurement of the reduction potentials. TM0927 and TM1289 both showed a decrease in reduction potential as the temperature increased (Fig. 4). TM0927 displayed temperature dependence up to 40°C. In contrast, only Cluster A of TM1289 shows clear temperature dependence. The slope of these plots can be used to determine the entropic component of the redox reaction. For TM0927, the S rc is −67.7 J/mol K, whereas the value for Cluster A of TM1289 is −42.8 J/mol K. These relatively large negative entropy values could be attributed to conformation and/or solvation effects, as seen as a more rigid reduced protein or charge neutralization in reduced protein with more ordered waters.
Proton coupled electron transfer with protein film voltammetry
PFV was used to examine whether the Fds from T. maritima display proton‐coupled electron transfer. The clusters in TM0927, TM1289, and TM1815 all were essentially pH invariant with very modest slopes of −6 mV/pH to −13 mV/pH unit over a range of pH 4–10. These are much less than the slope of −56 mV/pH unit expected for a one‐proton: one‐electron coupled process (at 10°C). Cluster A of TM1175 also displays a modest slope of −6 mV/pH unit; however, Cluster B has a slope of −31 mV/pH unit across physiological pH (Fig. 5). Currently, the significance of the slope for cluster B is not clear; it may be attributed to protonation events further away, and is evocative of BtrN, another ARE that bears a proton‐dependent [4Fe–4S]2+/1+ redox couple, though in the case of BtrN the cluster is the AdoMet‐binding active site cluster.20 Cluster A of TM1533 displayed modest pH‐dependence of −12 mV/pH unit, indicating that it is not coupled to a proton. However, Cluster B showed more complicated pH‐dependence data: the reversibility of the signal is dependent on scan rate and pH (data not shown). At slow scan rates, the voltammetry appears reversible due to the protonation and deprotonation occurring on the timescale of the potential sweep. As the scan rate increases at low pH, the oxidation peak of Cluster B becomes less defined and disappears as the cluster is trapped in the protonated form. This phenomenon has been extensively studied in the case of Azotobacter vinelandii FdI, where rates could be measured and an aspartic acid residue was identified as being important in the proton gating observed.44 Unfortunately, the overall low electroactive coverage displayed here prevents the further comparable ‘fast‐scan’ analysis in order to determine the rates and site of protonation for Cluster B of TM1533.
Attempts to reconcile biochemical function
While the redox potentials of the four 2x[Fe4–S4] T. maritima Fds all suggest that they could reduce MiaB or other ARE family members, we considered genome context and similarity as one way to provide additional clues regarding potential function. Intriguingly three of the Fds (TM1289, TM1533, 1819) appear to have connections to the low‐potential Fds that might be used for nitrogen fixation pathways, although T. maritima does not fix nitrogen: TM1289 appears to be related to NifB for example (in Azotobacter vinelandii, there is a ferredoxin located next to nifB and the two genes are co‐transcribed).42, 45 While specific nif genes are lacking, TM1288 does contain the signature CXXCXXC motif of an ARE family member and TM1291 is annotated as another putative FeS cluster binding protein. TM1533 has strong sequence similarity to ferredoxins of the fix nitrogen fixation pathway (FixX), with 49% sequence identity with the FixX protein from A. vinelandii. Further examination of the genomic region surrounding TM1533 shows that TM1530, TM1531, and TM1532 are FixA, FixB, and FixC, respectively, completing the FixABCX nitrogen fixation operon, though this may be an unused pathway in Thermotoga. TM1815 is most similar to FdxN, ferredoxin III, a ferredoxin from nitrogen fixation regions of nitrogen‐fixing bacteria. While there does not seem to be a nif operon, TM1816 is a predicted dinitrogenase iron–molybdenum cofactor biosynthesis protein, which is a Nif X,Y‐like protein, although of unknown function in this system. Collectively the current biological data for Thermotoga does not appear to help resolve the potential functions of the four additional 2x[Fe4–S4] cluster Fds, suggesting that new efforts in proteomics or transcriptomics may be necessary to address the biological function of these diverse electron carriers.
Conclusion
In our preceding work, we had observed that only two of the Fd proteins of Tm substantially support the catalysis of the native Tm MiaB enzyme, the single‐cluster bearing TM0927 and to a lesser degree, the two‐cluster bearing TM1175. Here, we have found that on the basis of thermodynamics alone, each of the Fd protein from Tm should be reduced by any of the reducing systems used previously, at least to the extent of a single cluster being reduced. Thus, it is clear that additional factors of kinetic reactivity with a reducing system must be at work (whether based on dithionite or flavodoxin/ferredoxin reductase), and are yet to be elucidated. However, these studies set the stage for further inquiry into the specificity of Fd proteins as redox carriers sought after in key biological pathways.
Materials and Methods
Protein film voltammetry
Experiments were performed anaerobically in an MBraun Labmaster glovebox using a PGSTAT 12 potentiostat (EcoChemie). A three‐electrode configuration was used with a standard calomel reference electrode, a platinum wire counter electrode, and a pyrolytic graphite edge (PGE) working electrode in a water‐jacketed glass cell. The electrochemical cell was thermostated using a circulating water bath and the reference electrode was maintained at room temperature in the course of the experiments. Potentials reported are relative to the standard hydrogen electrode.
Baseline measurements were collected using the PGE electrode polished with 1 M alumina, rinsed, and placed into the cell containing a 10 °C mixed buffer solution composed of 10 mM MES, CHES, TAPS, HEPES, at pH 8.0 with 200 mM NaCl. A 3 L sample of protein (ranging in concentration from 196 M to 1.135 mM) was applied directly to the polished PGE electrode surface, the protein sample was removed after 3 min, and the electrode was immediately placed back into the cell solution. Non‐turnover electrochemical signals were analyzed by correction of the non‐Faradaic component of the current from the raw data using the SOAS package.
Temperature dependence of the reduction potential
The entropic component of the redox reaction was determined using the slope of a plot of E m versus T (S rc = nF(dE m/dT)). The data were collected using a cell where the reference calomel electrode was maintained at a constant temperature (293 ± 0.5 K), while the cell containing the working electrode was varied. The enthalpic component was similarly determined by a Gibbs–Helmholtz plot (E m/T vs. 1/T).
Proton‐coupled electron transfer with protein film voltammetry. For measuring pH dependent voltammetry, the above buffer system was used, with the buffer pH adjusted with dilute KOH or HCl as needed.
Acknowledgments
This work has been supported by the NIH (GM‐101957 (S.J.B. and S.J.E.), GM‐122595 (S.J.B.) and GM‐120283 (S.J.E.)) and the Howard Hughes Medical Institute (S.J.B.).
References
- 1. Atkinson J, Campbell I, Bennett G, Silberg J (2016) Cellular assays for ferredoxins: a strategy for understanding electron flow through protein carriers that link metabolic pathways. Biochemistry 55:7047–7064. [DOI] [PubMed] [Google Scholar]
- 2. Mortenson L, Valentine R, Carnahan J (1962) An electron transport factor from Clostridium pasteurianum . Biochem Biophys Res Commun 7:448–452. [DOI] [PubMed] [Google Scholar]
- 3. Frey PA, Booker SJ (2001) Radical mechanisms of S‐adenosylmethionine‐dependent enzymes. Advan Protein Chem 58:1–45. [DOI] [PubMed] [Google Scholar]
- 4. Sofia HJ, Chen G, Hetzler BG, Reyes‐Spindola JF, Miller NE (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res 29:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dowling DP, Vey JL, Croft AK, Drennan CL (2012) Structural diversity in the AdoMet radical enzyme superfamily. Biochim Biophys Acta 1824:1178–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knappe J, Blaschkowski HP, Gröbner P, Schmitt T (1974) Pyruvate formate‐lyase of Escherichia coli: the acetyl‐enzyme intermediate. Eur J Biochem 50:253–263. [DOI] [PubMed] [Google Scholar]
- 7. Harder J, Eliasson R, Pontis E, Ballinger MD, Reichard P (1992) Activation of the anaerobic ribonucleotide reductase from Escherichia coli by S‐adenosylmethionine. J Biol Chem 267:25548–25552. [PubMed] [Google Scholar]
- 8. Bianchi V, Eliasson R, Fontecave M, Mulliez E, Hoover DM, Matthews RG, Reichard P (1993) Flavodoxin is required for the activation of the anaerobic ribonucleotide reductase. Biochem Biophys Res Commun 197:792–797. [DOI] [PubMed] [Google Scholar]
- 9. Bianchi V, Reichard P, Eliasson R, Pontis E, Krook M, Jornvall H, Haggard‐Ljungquist E (1993) Escherichia coli ferredoxin NADP+ reductase: activation of E. coli anaerobic ribonucleotide reduction, cloning of the gene (fpr), and overexpression of the protein. J Bacteriol 175:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ifuku O, Koga N, Haze S, Kishimoto J, Wachi Y (1994) Flavodoxin is required for conversion of dethiobiotin to biotin in Escherichia coli . Eur J Biochem 224:173–178. [DOI] [PubMed] [Google Scholar]
- 11. Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu‐Gogonea C, Souder MG, Tu L, Booker SJ (2004) Lipoyl synthase requires two equivalents of S‐adenosyl‐L‐methionine to synthesize one equivalent of lipoic acid. Biochemistry 43:6378–6386. [DOI] [PubMed] [Google Scholar]
- 12. Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ (2011) A radically different mechanism for S‐adenosylmethionine‐dependent methyltransferases. Science 332:604–607. [DOI] [PubMed] [Google Scholar]
- 13. Pierrel F, Hernandez HL, Johnson MK, Fontecave M, Atta M (2003) MiaB protein from Thermotoga maritima. Characterization of an extremely thermophilic tRNA‐methylthiotransferase. J Biol Chem 278:29515–29524. [DOI] [PubMed] [Google Scholar]
- 14. Pierrel F, Douki T, Fontecave M, Atta M (2004) MiaB protein is a bifunctional radical‐S‐adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J Biol Chem 279:47555–47563. [DOI] [PubMed] [Google Scholar]
- 15. Lee KH, Saleh L, Anton BP, Madinger CL, Benner JS, Iwig DF, Roberts RJ, Krebs C, Booker SJ (2009) Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily. Biochemistry 48:10162–10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landgraf BJ, Arcinas AJ, Lee KH, Booker SJ (2013) Identification of an intermediate methyl carrier in the radical S‐adenosylmethionine methylthiotransferases RimO and MiaB. J Am Chem Soc 135:15404–15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arcinas AJ, Maiocco SJ, Elliott SJ, Booker SJ (2018) Ferredoxins as interchangeable redox components in support of MiaB, a radical S‐adenosylmethionine methylthiotransferase. Protein Sci 28:267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Macedo‐Ribeiro S, Darimont B, Sterner R, Huber R (1996) Small structural changes account for the high thermostability of 1[4Fe–4S] ferredoxin from the hyperthermophilic bacterium Thermotoga maritima . Structure 4:1291–1301. [DOI] [PubMed] [Google Scholar]
- 19. Leger C, Bertrand P (2008) Direct electrochemistry of redox enzymes as a tool for mechanistic studies. Chem Rev 108:2379–2438. [DOI] [PubMed] [Google Scholar]
- 20. Maiocco SJ, Grove TL, Booker SJ, Elliott SJ (2015) Electrochemical resolution of the [4Fe‐4S] centers of the AdoMet radical enzyme BtrN: evidence of proton coupling and an unusual, low‐potential auxiliary cluster. J Am Chem Soc 137:8664–8667. [DOI] [PubMed] [Google Scholar]
- 21. Smith ET, Blamey JM, Zhou ZH, Adams MW (1995) A variable‐temperature direct electrochemical study of metalloproteins from hyperthermophilic microorganisms involved in hydrogen production from pyruvate. Biochemistry 34:7161–7169. [DOI] [PubMed] [Google Scholar]
- 22. Matsubara H, Saeki K (1992) Structural and functional diversity of ferredoxins and related proteins. Advan Inorgan Chem 38:223–280. [Google Scholar]
- 23. Iwasaki T, Wakagi T, Isogai Y, Tanaka K, Iizuka T, Oshima T (1994) Functional and evolutionary implications of a [3Fe–4S] cluster of the dicluster‐type ferredoxin from the thermoacidophilic Archaeon, Sulfolobus Sp strain‐7. J Biol Chem 269:29444–29450. [PubMed] [Google Scholar]
- 24. Gao‐Sheridan HS, Pershad HR, Armstrong FA, Burgess BK (1998) Discovery of a novel ferredoxin from Azotobacter vinelandii containing two [4Fe–4S] clusters with widely differing and very negative reduction potentials. J Biol Chem 273:5514–5519. [DOI] [PubMed] [Google Scholar]
- 25. Kyritsis P, Hatzfeld OM, Link TA, Moulis JM (1998) The two [4Fe‐4S] clusters in Chromatium vinosum ferredoxin have largely different reduction potentials. Structural origin and functional consequences. J Biol Chem 273:15404–15411. [DOI] [PubMed] [Google Scholar]
- 26. Stombaugh NA, Burris RH, Ormejohn W (1973) Ferredoxins from Bacillus‐polymyxa – low potential iron‐sulfur proteins which appear to contain single 4‐iron, 4‐sulfur centers accepting a single electron on reduction. J Biol Chem 248:7951–7956. [PubMed] [Google Scholar]
- 27. Asso M, Mbarki O, Guigliarelli B, Yagi T, Bertrand P (1995) Epr and redox characterization of ferredoxin‐I and ferredoxin‐Ii from Desulfovibrio vulgaris miyazaki. Biochem Biophys Res Commun 211:198–204. [DOI] [PubMed] [Google Scholar]
- 28. Brereton PS, Verhagen MFJM, Zhou ZH, Adams MWW (1998) Effect of iron‐sulfur cluster environment in modulating the thermodynamic properties and biological function of ferredoxin from Pyrococcus furiosus . Biochemistry 37:7351–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li B, Elliott SJ (2016) The catalytic bias of 2‐oxoacid: ferredoxin oxidoreductase in CO2: evolution and reduction through a ferredoxin‐mediated electrocatalytic assay. Electrochim Acta 199:349–356. [Google Scholar]
- 30. Iismaa SE, Vazquez AE, Jensen GM, Stephens PJ, Butt JN, Armstrong FA, Burgess BK (1991) Site‐directed mutagenesis of Azotobacter vinelandii ferredoxin I. Changes in [4Fe–4S] cluster reduction potential and reactivity. J Biol Chem 266:21563–21571. [PubMed] [Google Scholar]
- 31. Quinkal I, Davasse V, Gaillard J, Moulis JM (1994) On the role of conserved proline residues in the structure and function of Clostridium pasteurianum 2[4Fe‐4S] ferredoxin. Protein Engin 7:681–687. [DOI] [PubMed] [Google Scholar]
- 32. Yoon KS, Bobst C, Hemann CF, Hille R, Tabita FR (2001) Spectroscopic and functional properties of novel 2[4Fe–4S] cluster‐containing ferredoxins from the green sulfur bacterium Chlorobium tepidum . J Biol Chem 276:44027–44036. [DOI] [PubMed] [Google Scholar]
- 33. Giastas P, Pinotsis N, Efthymiou G, Wilmanns M, Kyritsis P, Moulis JM, Mavridis IM (2006) The structure of the 2[4Fe–4S] ferredoxin from Pseudomonas aeruginosa at 1.32‐Å resolution: comparison with other high‐resolution structures of ferredoxins and contributing structural features to reduction potential values. J Biol Inorg Chem 11:445–458. [DOI] [PubMed] [Google Scholar]
- 34. Saridakis E, Giastas P, Efthymiou G, Thoma V, Moulis JM, Kyritsis P, Mavridis IM (2009) Insight into the protein and solvent contributions to the reduction potentials of [4Fe‐4S]2+/+ clusters: crystal structures of the Allochromatium vinosum ferredoxin variants C57A and V13G and the homologous Escherichia coli ferredoxin. J Biol Inorg Chem 14:783–799. [DOI] [PubMed] [Google Scholar]
- 35. Stephens PJ, Jollie DR, Warshel A (1996) Protein control of redox potentials of iron–sulfur proteins. Chem Rev 96:2491–2514. [DOI] [PubMed] [Google Scholar]
- 36. Kümmerle R, Gaillard J, Kyritsis P, Moulis JM (2001) Intramolecular electron transfer in [4Fe–4S] proteins: estimates of the reorganization energy and electronic coupling in Chromatium vinosum ferredoxin. J Biol Inorg Chem 6:446–451. [DOI] [PubMed] [Google Scholar]
- 37. Chen K, Jung YS, Bonagura CA, Tilley GJ, Prasad GS, Sridhar V, Armstrong FA, Stout CD, Burgess BK (2002) Azotobacter vinelandii ferredoxin I: a sequence and structure comparison approach to alteration of [4Fe–4S]2+/+ reduction potential. J Biol Chem 277:5603–5610. [DOI] [PubMed] [Google Scholar]
- 38. Bovierlapierre G, Bruschi M, Bonicel J, Hatchikian EC (1987) Amino‐acid‐sequence of Desulfovibrio africanus ferredoxin. 3. A unique structural feature for accommodating iron‐sulfur clusters. Biochim Biophys Acta 913:20–26. [Google Scholar]
- 39. Okawara N, Ogata M, Yagi T, Wakabayashi S, Matsubara H (1988) Amino acid sequence of ferredoxin I from Desulfovibrio vulgaris Miyazaki. J Biochem 104:196–199. [DOI] [PubMed] [Google Scholar]
- 40. Schatt E, Jouanneau Y, Vignais PM (1989) Molecular cloning and sequence analysis of the structural gene of ferredoxin I from the photosynthetic bacterium Rhodobacter capsulatus . J Bacteriol 171:6218–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saeki K, Tokuda K, Fukuyama K, Matsubara H, Nadanami K, Go M, Itoh S (1996) Site‐specific mutagenesis of Rhodobacter capsulatus ferredoxin I, FdxN, that functions in nitrogen fixation ‐ Role of extra residues. J Biol Chem 271:31399–31406. [DOI] [PubMed] [Google Scholar]
- 42. Reyntjens B, Jollie DR, Stephens PJ, GaoSheridan HS, Burgess BK (1997) Purification and characterization of a fixABCX‐linked 2[4Fe–4S] ferredoxin from Azotobacter vinelandii . J Biol Inorgan Chem 2:595–602. [Google Scholar]
- 43. Boll M, Fuchs G, Tilley G, Armstrong FA, Lowe DJ (2000) Unusual spectroscopic and electrochemical properties of the 2[4Fe–4S] ferredoxin of Thauera aromatica . Biochemistry 39:4929–4938. [DOI] [PubMed] [Google Scholar]
- 44. Chen K, Hirst J, Camba R, Bonagura CA, Stout CD, Burgess BK, Armstrong FA (2000) Atomically defined mechanism for proton transfer to a buried redox centre in a protein. Nature 405:814–817. [DOI] [PubMed] [Google Scholar]
- 45. Rodriquez‐Quiñones F, Bosch R, Imperial J (1993) Expression of the nifBfdxNnifOQ region of Azotobacter vinelandii and its role in nitrogenase activity. J Bacteriol 175:2926–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Asso M, Mbarki O, Guigliarelli B, Yagi T, Bertrand P (1995) EPR and redox characterization of ferredoxins I and II from Desulfovibrio vulgaris Miyazaki. Biochem Biophys Res Commun 211:198–204. [DOI] [PubMed] [Google Scholar]
- 47. Jouanneau Y, Meyer C, Gaillard J, Forest E, Gagnon J (1993) Purification and characterization of a novel dimeric ferredoxin (FdIII) from Rhodobacter capsulatus . J Biol Chem 268:10636–10644. [PubMed] [Google Scholar]


