Abstract
Glutathione transferase enzymes help plants to cope with biotic and abiotic stress. They mainly catalyze the conjugation of glutathione (GSH) onto xenobiotics, and some act as glutathione peroxidase. With X‐ray crystallography, kinetics, and thermodynamics, we studied the impact of oxidation on Arabidopsis thaliana glutathione transferase Phi 9 (GSTF9). GSTF9 has no cysteine in its sequence, and it adopts a universal GST structural fold characterized by a typical conserved GSH‐binding site (G‐site) and a hydrophobic co‐substrate‐binding site (H‐site). At elevated H2O2 concentrations, methionine sulfur oxidation decreases its transferase activity. This oxidation increases the flexibility of the H‐site loop, which is reflected in lower activities for hydrophobic substrates. Determination of the transition state thermodynamic parameters shows that upon oxidation an increased enthalpic penalty is counterbalanced by a more favorable entropic contribution. All in all, to guarantee functionality under oxidative stress conditions, GSTF9 employs a thermodynamic and structural compensatory mechanism and becomes substrate of methionine sulfoxide reductases, making it a redox‐regulated enzyme.
Keywords: redox, methionine sulfoxide, X‐ray structure, methionine sulfoxide reductase, steady‐state kinetics, thermodynamics
Short abstract
Abbreviations
- AU
asymmetric unit
- CDNB
2,4‐Dinitrochlorobenzene
- CuOOH
cumene hydroperoxide
- FOX assay
ferrous oxidation‐xylenol orange assay
- GST
glutathione transferase
- GSTU23
glutathione transferase Tau23
- GSTF9
glutathione transferase Phi9
- GSH
glutathione
- GSOH
glutathione sulfenic acid
- GSSG
glutathione disulfide
- GSO3
glutathione sulfonate
- Met‐SO
methionine sulfoxide
- ROS
reactive oxygen species
Introduction
To understand the global effect of methionine oxidation in plants, we recently identified the sulfoxome (proteome of oxidized methionines) in Arabidopsis thaliana leaves exposed to high light stress and we mapped the modifications back to single amino acids, which is key for understanding possible consequences of this post‐translational modification on protein function.1 We found members of the Phi and Tau glutathione transferase (GST) families affected by methionine oxidation.
GST enzymes detoxify electrophilic substrates by glutathione (GSH) conjugation and by peroxide scavenging.2, 3 Particularly, they are involved in the detoxification of heavy metals4 and herbicides.5 Some GST members deglutathionylate their substrates,6 and others are involved in cell signaling by interacting with substrates that have an important signaling role.7 The A. thaliana genome encodes for 55 GST enzymes, which are differently expressed and regulated depending on the type of biotic and abiotic stress conditions.4 GST enzymes are subdivided into different classes depending on their amino acid sequence, catalytic residues (Ser, Tyr, or Cys), and their tertiary structure.8, 9 Although members of the plant GST classes have low primary sequence identity, they share an overall common structural fold.5, 10, 11
Most cytosolic GST enzymes are homodimeric and have an N‐terminal thioredoxin‐like domain, which contains the glutathione (GSH) binding site (G‐site) and a C‐terminal alpha‐helical domain, which contains the hydrophobic substrate‐binding site (H‐site).8 Upon GSH binding to the G‐site, the catalytic residue (Ser, Tyr, or Cys), which is Ser for Phi and Tau class GSTs, activates GSH (thiol) to GS−(thiolate), which is then conjugated to an electrophilic substrate located within the H‐site via a thio‐ether bond.
Some GST enzymes contain cysteines, located close to the active site, which are highly sensitive to oxidation. For example, we recently showed that A. thaliana GSTU23 forms a reversible disulfide that protects against oxidative damage.12 Other GST enzymes have no cysteines, like some of the Phi class members, where methionine residues play a role in redox regulating their transferase activity.13
Here, we present the impact of oxidation on the structure and function of GSTF9, an enzyme with no cysteines. GSTF9 is constitutively expressed together with GSTF10, GSTU5, and GSTU13, indicating its importance as housekeeping enzyme.14 Further, Horvath et al. showed that an A. thaliana GSTF9 loss‐of‐function mutant has a decreased overall glutathione peroxidase activity.15 We found that the oxidation of methionines located near or within the G‐ and H‐sites of GSTF9 lead to increased flexibility of the H‐site loop, which results in a decrease in the affinity for the substrates, GSH and CDNB. Upon oxidation, GSTF9 glutathione peroxidase activity decreases for the more hydrophobic peroxides.
Results
GSTF9 has a universal GST structural fold
This study focuses on the impact of oxidation on the structure and function of A. thaliana glutathione transferase Phi 9 (GSTF9), which was identified within the list of the proteins with an oxidized methionine, the A. thaliana sulfoxome.1 Reduced GSTF9 (GSTF9red) was purified to homogeneity and crystallized using the hanging drop vapor diffusion method. We solved the structure to a resolution of 2.34 Å (PDB code 6EZY) (Table 1). As observed for other GSTs, GSTF9red is a homodimer and forms trigonal crystals, with one GSTF9red dimer in the asymmetric unit. Each monomer contains a thioredoxin domain followed by an α‐helical domain (Fig. 1). The thioredoxin domain contains the glutathione (GSH) binding site (G‐site), while the hydrophobic substrate binding site (H‐site) is embedded within the C‐terminal helical bundle domain. The G‐site shows a high degree of structural conservation, while the conservation of the H‐site is rather limited.16 The catalytic Ser12 within the G‐site is responsible for GSH activation (Fig. 1). We observed a GSH molecule residing the G‐site of the GSTF9red structure, while the H‐site is occupied by glutathione sulfenic acid (GSOH) (Fig.1). The presence of two GSH molecules, or even a glutathione disulfide (GSSG) has been shown previously for the E. coli Nu class of GST enzymes.17, 18 Overlay of these structures with the GSTF9red structure shows that the GSH molecules in YghU and YfcG are differently oriented.
Table 1.
GSTF9 X‐Ray Data Collection and Refinement Statistics
| Dataset | GSTF9red | GSTF9H2O2+NaOCl | GSTF9ox |
|---|---|---|---|
| PDB code | 6EZY | 6F01 | 6F05 |
| Data collection | |||
| Beamline | IO4@DLS | IO4@DLS | IO4@DLS |
| Wavelength | 0.9795 | 0.9795 | 0.9795 |
| Processing | |||
| Space group | P31 2 1 | P31 2 1 | P1 |
| Cell parameters (Å/°) | 61.43 93.97 107.71 93.19 101.57 101.97 | 114.76 114.76 90.31 90 90 120 | 61.43 93.97 107.71 93.19 101.57 101.97 |
| Resolution, Å (outer shell)a | 48.13–2.34 (2.48–2.34) | 66.84–2.5 (2.57–2.5) | 62.7–2.2 (2.26–2.2) |
| Total reflections | 282,989 (43,703) | 158,033 (11,796) | 268,584 (19,908) |
| Unique reflections | 2880 (4594) | 24179 (1781) | 116579 (8561) |
| Completeness (%) | 98.7 (96.8) | 99.8 (99.9) | 97.8 (97.1) |
| Multiplicity | 9.9 (9.5) | 6.5 (6.6) | 2.3 (2.3) |
| CC1/2 (%) | 99.9 (55.9) | 99.5 (71.1) | 99.7 (73.3) |
| Rmeas (%) | 11.2 (137.9) | 17.7 (124.8) | 7.4 (60.4) |
| <I/σ(I)> | 18.3 (1.64) | 11.56 (2.09) | 10.67 (2.08) |
| Refinement | |||
| Resolution range (Å) | 49.41–2.35 | 66.84–2.3 | 65.73–2.06 |
| Percentage observed (%) | 98.79 | 99.84 | 97.66 |
| R cryst (%)b | 15.63 | 16.43 | 16.15 |
| R free (%)c | 19.66 | 20.89 | 20.93 |
| RMS | |||
| Bonds (Å) | 0.012 | 0.010 | 0.008 |
| Angles (°) | 1.267 | 1.268 | 1.18 |
| Ramachandran plot | |||
| Most favored (%) | 97.83 | 97.31 | 97.76 |
| Additionally allowed (%) | 1.69 | 2.2 | 1.68 |
| Disallowed (%) | 0.48 | 0.49 | 0.56 |
| Bound ligand | GSH (G‐site) and GSOH (H‐site) | GSO3 (G‐site) and GSOH (H‐site) | GSO3 (G‐site) |
Values between brackets are for the highest resolution shell.
R cryst = ΣǁF obs| − |F calcǁ/Σ|F obs|, F obs and F calc are observed and calculated structure factor amplitudes.
R free is the same as R cryst, but using a random subset of 5% of the data excluded from the refinement.
DLS, diamond light source.
Figure 1.
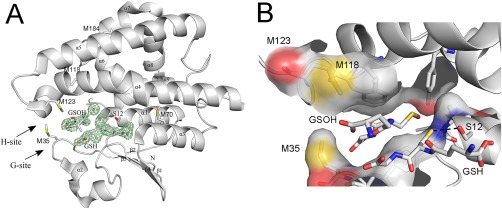
Crystal structure of reduced GSTF9. (A) The X‐ray crystal structure of reduced GSTF9red shows a typical GST‐fold. The crystal structure of GSTF9red is shown in a gray cartoon representation with the alpha helices and beta sheets labeled. The glutathione (GSH) binding site (G‐site) and the hydrophobic substrate‐binding site (H‐site) are indicated with arrows. A glutathione molecule is observed in the G‐site, and sulfenylated GSH (GSOH) in the H‐site. The catalytic serine (S12), the Met residues (M35, M70, M118, M123 and M184), and the N‐ and C‐terminus are indicated. An omit map contouring the GSH and GSOH at 3σ is shown in green. (B) A closer view of the three methionines (M35, M118 and M123) located close to the G‐ and H‐site. Surface representation is shown for atoms within 4 Å of H‐GSOH. The H‐bond between the catalytic S12 and the sulfur of GSH is 3.3 Å (black dotted line).
Oxidation impacts transferase activity but not H2O2 peroxidase activity
GSTF9 belongs to the Phi class of glutathione transferases, for which both glutathione transferase and peroxidase activities have been described.14, 19 To study the impact of oxidation on transferase and peroxidase activities (Fig. 2), GSTF9red was oxidized with H2O2 (1:100 molar ratio) to obtain GSTF9ox, followed by oxidant removal. The transferase activity of GSTF9 was measured by monitoring the increase of absorbance at 340 nm for the conjugation of the thiol group of GSH to the CDNB substrate in function of time. The initial velocities were plotted for several conditions [Fig. 2(A)]. As previously reported by Jacques et al.,1 we observed a 30% decrease in initial velocity for GSTF9ox [Fig. 2(A)]. To study the influence of oxidation on the glutathione peroxidase activity of GSTF9, we used a FOX assay [Fig. 2(B)]. Progress curves [Fig. 2(B)] show that GSTF9red and GSTF9ox remove H2O2 with similar rates.
Figure 2.
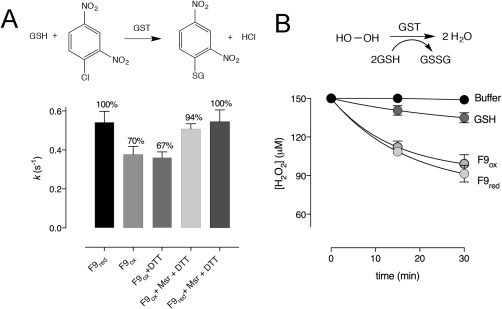
GSTF9 oxidation has a different impact on the transferase and glutathione peroxidase activity. (A) Bar graphs show the percentage of transferase activity measured in rate constants (k) of GSTF9red and GSTF9ox (0.2 µM), where the GST catalysed conjugation of GSH (0.5 mM) onto CDNB (3.8 mM) is followed at 340 nm. When oxidized, GSTF9ox activity significantly (P < 0.001) decreases from 100% (0.54 s−1) to 70% (0.38 s−1). MsrA and MsrB treatment causes activity increase from 70% to 94% (0.51 s−1), indicating that Msr enzymes are capable of recovering GSTF9ox activity. Two control samples were used: GSTF9ox incubated with DTT and GSTF9red incubated with MsrA, MsrB and DTT. The data from at least 3 independent experiments are presented as a mean ± SD. (B) FOX assay was used to compare the peroxidase activity of GSTF9red and GSTF9ox in the presence of 150 µM H2O2. Remained H2O2 concentrations were determined for samples taken after 15 and 30 min. Upon treatment of GSTF9 with hydrogen peroxide (GSTF9ox), glutathione peroxidase activity is not influenced. As a control, the influence of GSH on H2O2 reduction was also recorded. The data from 3 independent experiments were normalized and presented as a mean ± SD.
G‐ and H‐site methionines form a sulfoxide
In our previous study on GSTF9,1 we reported sulfur oxidation of Met35, which is located close to the G‐site (Fig. 1). We decided to determine whether the methionines (Met35, Met118, and Met123) located within or near the G‐ and H‐ sites of GSTF9 could influence its activity upon hydrogen peroxide treatment (Fig. 1). Therefore, both GSTF9red and GSTF9ox samples were trypsin or chymotrypsin digested, and the formation of methionine sulfoxide and sulfone was analyzed by mass spectrometry. We found sulfoxide and sulfone formation on Met35 (Fig. 3), which is located close to the G‐site, and sulfoxide on Met118 and Met123, which are located close to the H‐site (Fig. 1). Aside from the active site methionines, also Met184, which is surface exposed and located at the opposite side of the active site, was oxidized to methionine sulfoxide. Taken its location at distance of more than 18 Å away from the active site Ser12 (Fig. 1), it is highly unlikely that Met184 oxidation would directly influence the activity of GSTF9.
Figure 3.
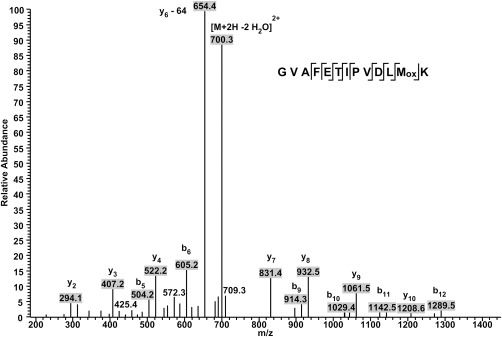
Identification of Met35 of GSTF9 as a methionine sulfoxide. Identification of oxidation on Met35 of GSTF9 under H2O2 exposure. The LC‐MS spectrum shows MS2 data obtained from a +2 parent ion with m/z 718.3. The y‐ and b‐ series of ions allow exact localization of the methionine sulfoxide. A neutral loss of 64 Da is observed from the y6 daughter ion and corresponds to the release of methane sulfenic acid (CH3SOH) from the side chain of methionine sulfoxide.
Oxidized GSTF9 is a methionine sulfoxide substrate
As methionine oxidation causes a decrease in the transferase activity of GSTF9 [Fig. 2(A)], we wondered whether the transferase activity could be recovered by methionine sulfoxide reductase. Therefore, we incubated 10 µM GSTF9ox with a 5 µM mixture of methionine sulfoxide reductase A (MsrA) and B (MsrB) for 1 h at 25°C, and measured the initial velocity in the CNDB‐GSH conjugation assay. The drop in transferase activity seen for GSTF9ox was almost completely recovered [Fig. 2(A)], which indicates that Msr enzymes regulate the activity of GSTF9 under oxidative stress conditions.
Oxidation has only a marginal effect on the specificity constants
Next, we wanted to understand the impact of oxidation on the kinetic parameters of GSTF9, and this for both the G‐site binding GSH substrate and the H‐site binding CDNB substrate (Table 2 and Supporting Information, Fig. S1). From the Michaelis–Menten curves (Supporting Information, Fig. S1) under respective substrate saturating conditions, we determined K M and k cat values. GSTF9 oxidation increased the K M for CDNB from 1.3 to 1.8 mM, but the k cat only slightly varies from 0.98 to 0.72 s−1. For GSH, the K M decreases twofold from 131 µM to 71 µM, while the k cat only slightly varies from 0.85 to 0.5 s−1. The overall effect of oxidation on k cat/K M is only marginal as the specificity constants are within the same range, ∼102 M−1 s−1 for CDNB and ∼103 M−1 s−1 for GSH (Table 2).
Table 2.
Steady‐State Kinetics Parameters of GSTF9 Under Reducing and Oxidizing Conditions
| [GSH] | [CDNB] | |||
|---|---|---|---|---|
| F9red | F9ox | F9red | F9ox | |
| K M | 131 ± 9 μM | 71 ± 7 μM | 1.3 ± 0.1 mM | 1.8 ± 0.2 mM |
| k cat (s−1) | 0.85 ± 0.02 | 0.50 ± 0.01 | 0.98 ± 0.02 | 0.72 ± 0.04 |
| k cat/K M (M−1s−1) | 6.5 × 103 | 7.0 × 103 | 7.5 × 102 | 4.0 × 102 |
Kinetic parameters were determined using varying concentrations of GSH or CDNB and the values were plotted using the Michaelis–Menten equation.
GSTF9 oxidation induces H‐site flexibility
To understand the impact of oxidation at the molecular level, we decided to solve the structure of oxidized GSTF9 crystals. First, crystals were grown using GSTF9ox (GSTF9 oxidized using a 1:100 GSTF9:H2O2 ratio). The resulting GSTF9ox structure was solved to 2.2 Å resolution (Table 1, PDB code 6F05). Second, we exposed the GSTF9red crystals to the extreme oxidation conditions of 0.5 M H2O2 and 1 mM NaOCl for 1 h (GSTF9H2O2+NaOCl, PDB code 6F01). While GSTF9red (Fig. 1) formed trigonal crystals, the GSTF9ox formed triclinic crystals (Table 1), with 10 copies of GSTF9 in the asymmetric unit (AU). Overlay of all the chains of GSTF9ox and GSTF9H2O2+NaOCl clearly shows mobility of the Met35, Met118 and M123 side chains, and also the electron density surrounding amino acids 120–127 is absent in all the 10 copies in the AU [Fig. 4(A)]. Unfortunately, within the crystal structures of the oxidized GSTF9, GSTF9ox and GSTF9H2O2+NaOCl, the side chain of Met35 was not always well defined, as it is localized on a highly mobile loop between the β2‐strand and the α2‐helix (amino acids 34–43). In the structures of both oxidized GSTF9, GSTF9ox and GSTF9H2O2+NaOCl, the Met35, Met118, and Met123 sulfurs do not show any sulfoxide or sulfone formation under the applied oxidation conditions [Fig. 4(A)], which is different from what we observed in our mass spectrometric results.
Figure 4.
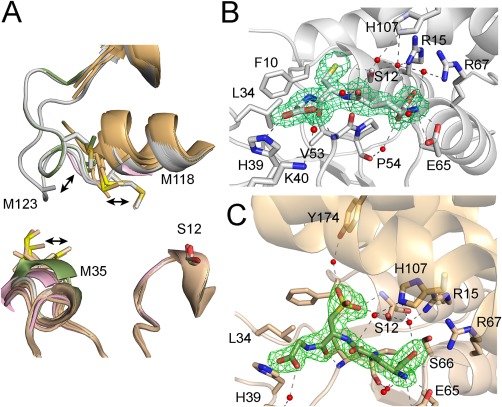
Oxidation leads to GSTF9 H‐site disorder and oxidized glutathione (GSO3) formation in the G‐site. (A) Overlay of the 10 copies of GSTF9 in the AU of GSTF9ox (shown in gold cartoon) and the two chains present in the AUs of both the GSTF9red (chain A and B in gray cartoon) and GSTF9_H2O2+NaOCl structures (chain A in pink, chain B in green) shows a high mobility for the α2‐β2 and especially the α4‐α5 loops. Residues 120–127 in the latter loop become entirely disordered in the GSTF9ox structure, as well as in chain A of the GSTF9_ H2O2+NaOCl structure. The side chains of Met35, Met118 and Met123 shown in sticks are highly mobile. (B‐C) The active site H‐bond network of GSFT9red (gray cartoon) has a GSH molecule (B), which is oxidized to GSO3 (C) within the GSTF9ox G‐site (gold cartoon). The residues (gold for GSTF9ox and gray for GSTF9red) interacting with GSH and GSO3 are shown in stick representation. Leu34 and Phe10 delineate the G‐site. Hydrogen bonds are in gray or black dashed lines. Water molecules H‐bonded to GSH or GSO3 are in red spheres. An omit map contouring the ligands, GSH and GSO3, at 3σ is shown in green.
Glutathione sulfonate (GSO3) inhibits the transferase activity
In addition to the high loop mobility close to the active site in both oxidized GSTF9 structures, GSTF9H2O2+NaOCl and GSTF9ox, we observed GSH oxidized to glutathione sulfonate (GSO3) within the G‐site. GSO3 is bound in exactly the same orientation as GSH in the G‐site [Fig. 4(B,C)]. Both the GSH and GSO3 γ‐Glu moieties hydrogen bond to Glu65 and Ser66. The glycyl moieties hydrogen bond to His39 and via a water molecule to Lys40. Where the GSH Cys sulfur interacts with the Ser12 OH group [Figs. 1(B) and 4(B)], the SO3 group of GSO3 makes hydrogen bonds with the Ser12 OH group, as well as with the Ser12 backbone. Via a water molecule it also interacts with the side chain of Tyr174 [Fig. 4(C)]. The GSH and GSO3 seem to be strongly bound to the G‐site as all the size exclusion chromatography attempts to remove the GSH from GSTF9 were unsuccessful.
As a molecule of glutathione sulfonate (GSO3) resides in the active site of GSTF9ox and GSTF9H2O2+NaOCl, we decided to test the effect of GSO3 on GSTF9 transferase activity. GSTF9 was incubated with increasing concentrations of GSO3 (0–300 µM), and the transferase activity was measured. The increase in GSO3 concentration led to a decrease of the transferase activity with an inhibition constant (K i) of 34 µM [Fig. 5(A)]. Hence, formation of GSO3 during the oxidation of GSTF9 could be another reason for the decrease in transferase and peroxidase activity [Fig. 5(B)].
Figure 5.
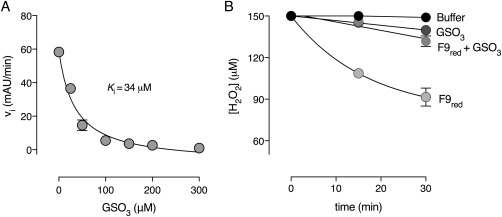
Glutathione sulfonate (GSO3) inhibits GSTF9 glutathione transferase and peroxidase activity. Increasing concentrations of GSO3 results in a decrease of the initial velocity of (A) the GSTF9 glutathione transferase and (B) the peroxidase activity. The inhibition constant (K i) for the transferase activity is 34 µM. The data represent the mean ± SD of 2 independent experiments.
GSTF9ox reduces hydrophobic peroxides less efficiently
Next, we decided to test whether increased H‐site flexibility observed within GSTF9ox might be the reason for the decreased transferase activity. Loss of a structured H‐site under oxidative stress [Fig. 4(A)] might affect the binding of hydrophobic substrates in the H‐site of GSTF9. The CDNB molecule used in the transferase assay is a hydrophobic molecule as it contains a benzene ring that needs to be correctly oriented in the H‐site for GSH transfer. Therefore, we decided to test the GSTF9red and GSTF9ox peroxidase activity with two hydrophobic peroxides: cumene hydroperoxide (with a benzene ring) and tert‐butyl hydroperoxide (with a more extended carbon chain) (Fig. 6, Table 3, and Supporting Information). The results show a similar glutathione peroxidase activity for GSTF9red and GSTF9ox towards H2O2 after 30 min incubation (Fig. 6 and Supporting Information, Table SI). On the other hand, when comparing the GSTF9red and GSTF9ox peroxide reduction after 30 min incubation with tert‐butyl peroxide and cumene hydroperoxide, it became apparent that less peroxide is being consumed for these more hydrophobic peroxides by GSTF9ox (Table 3 and Supporting Information). A clear trend was observed, peroxidase activity towards more hydrophobic peroxides decreases for GSTF9ox compared to GSTF9red (Fig. 6). The correlation between the concentration of peroxide consumed by GSTF9red versus the hydrophobicity index (ALogP) of the peroxides corroborates the hypothesis that GSTF9 oxidation impairs its activity towards hydrophobic substrates (Fig. 6).
Figure 6.
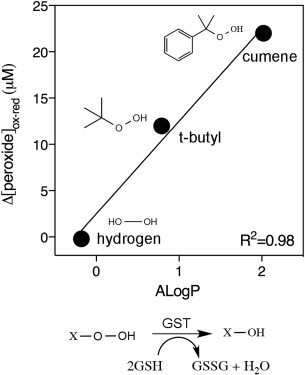
Oxidized GSTF9 reduces hydrophobic peroxides less efficiently. A difference plot of the peroxide concentration (µM) (H2O2, t‐butyl peroxide, CuOOH) reduced after 30 min with GSTF9ox and GSTF9red versus the hydrophobicity index (ALogP) of the peroxides is presented. The more hydrophobic the peroxide, the larger the difference between GSTF9red and GSTF9ox peroxide reduction (Δ[peroxide]). The data represent the mean ± SD of 3 independent experiments. The reduction scheme for peroxides (X) with the release of oxidized glutathione (GSSG) and a water molecule is shown. The structures of the three peroxides (H2O2, t‐butyl peroxide, CuOOH) are generated using ChemBioDraw 14.0.
Table 3.
Glutathione Peroxidase Activity of GSTF9red and GSTF9ox Using Different Peroxides
| Time (min) | [H2O2] (μM) | [t‐butyl peroxide] (μM) | [CuOOH] (μM) | |
|---|---|---|---|---|
| F9red | 0 | 200 ± 0 | 200 ± 0 | 200 ± 0 |
| 15 | 171 ± 1 | 180 ± 4 | 117 ± 4 | |
| 30 | 161 ± 2 | 162 ± 7 | 84 ± 9 | |
| F9ox | 0 | 200 ± 0 | 200 ± 0 | 200 ± 0 |
| 15 | 163 ± 0 | 191 ± 4 | 147 ± 7 | |
| 30 | 162 ± 1 | 174 ± 0 | 106 ± 7 |
The concentration of peroxide present within the sample containing GSTF9red and GSTF9ox with 1 mM GSH at time 0, 15, and 30 min is shown.
GSTF9ox employs a thermodynamic compensatory system
To further understand the source of the increased structural flexibility observed upon GSTF9 oxidation, we decided to compare the thermodynamic activation parameters (ΔH #, ΔS #, andΔG #) and the energy of activation (E a) of oxidized and reduced GSTF9. Therefore, we determined the initial velocities of the transferase activity with CDNB at different temperatures. We derived the thermodynamic parameters of the CDNB glutathione transferase reaction from the linear Eyring and Arrhenius plots and the respective equations (Table 4 and Fig. 7). We observed an enthalpy–entropy compensatory system where changes in enthalpy are balanced by changes in entropy, leading to similar ΔG # values for GSTF9red and GSTF9ox.20 The energy of activation (E a) and enthalpy (ΔH #) values indicate a larger energetic barrier to be overcome by oxidized GSTF9 compared to reduced GSTF9. The increased energy barrier is accompanied by a less pronounced conformational change of GSTF9ox to reach the activated complex, leading to this enthalpy–entropy compensatory mechanism.
Table 4.
Thermodynamic Activation Parameters for CDNB Glutathione Transferase Activity with Reduced and Oxidized GSTF9
| E a (kcal mol−1) | ΔH # (kcal mol−1) | TΔS # (kcal mol−1) | aΔG # (kcal mol−1) | Δ (ΔH #)ox‐red (kcal mol−1) | Δ(TΔS #)ox‐red (kcal mol−1) | Δ(ΔG #)ox‐red (kcal mol−1) | |
|---|---|---|---|---|---|---|---|
| GSTF9red | 9.4 (±0.5) | 8.8 (±0.5) | −9.0 (±0.9) | 17.9 (±1) | 2.3 | 2 | 0.3 |
| GSTF9ox | 11.7 (±0.6) | 11.1 (±0.6) | −7.0 (± 0.6) | 18.2 (±1) |
ΔG # was calculated at 25°C.
Δ(ΔH #)ox‐red and Δ(TΔS #)ox‐red represent the variation of the thermodynamic parameters between GSTF9ox and GSTF9red.
Figure 7.
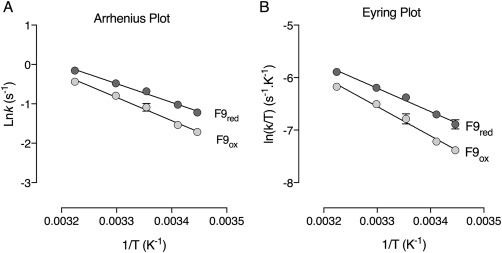
Arrhenius (A) and Eyring (B) plot of GSTF9red and GSTF9ox. Temperature dependence of lnk and ln (k/T) of the glutathione transferase catalyzed reaction by reduced and oxidized GSTF9 at pH 6.4 in a temperature range from 17°C to 37°C is shown. The linearity of the plots suggests that there is no change in the rate‐limiting step or protein inactivation within this temperature range.
Discussion
Glutathione transferase (GST) enzymes play an important role in the protection of plants against different stress conditions including xenobiotic and peroxide stress.3, 21 Understanding how these enzymes function and how their activities are influenced under stress conditions is important to understand how they survive extreme conditions. Prior to this study, two members of the GST family, GSTU23 and GSTF9, were found present in the sulfoxome of A. thaliana leafs exposed to high light stress,1 and as such sensitive to methionine oxidation. In a recent study, we have investigated how the activity of GSTU23 is influenced under hydrogen peroxide stress.12 We found that at lower H2O2 levels (100 µM), MetSO formation is the major post‐translational modification on GSTU23, which is recycled by methionine sulfoxide reductases (Msr). Here, we focus on the impact of oxidation on GSTF9, an enzyme with no cysteines.
GSTF9 has no cysteines, but six methionines, three of which (Met35, Met118, and Met123) are located within or close to the G‐ and H‐sites (Fig. 1), and which could interfere with GSH or hydrophobic substrate binding. We showed with mass spectrometry sulfoxidation of these three methionines and how they influence the GSTF9 glutathione transferase activity [Fig. 2(A)] and the glutathione peroxidase activity [Fig. 2(B), 6]. A similar case is observed in a study performed by Lee et al.,13 where oxidation of methionines in A. thaliana GSTF2 and GSTF3 causes a decrease in the GST transferase activity, and this activity could be recovered with methionine sulfoxide reductase B7 (MsrB7). Important to note is that similar to GSTF9, both GSTF2 and GSTF3 also contain no Cys residues.13 Similarly, we showed a decrease in glutathione transferase activity of A. thaliana GSTU23 upon methionine oxidation, which is restored when treated with MsrA and MsrB.12 Here, we show the importance of MsrA and MsrB in restoring GSTF9 transferase activity [Fig 2(A)]. Treatment with the Msr enzymes recovers the transferase activity of GSTF9ox up to 94%; however, 6% remains unrecovered. This might be due to the presence of a methionine sulfone, as observed in the MS analysis, or the presence of GSO3, which was observed in the active site of GSTF9ox structure [Fig. 4(C)].
The active site of GST enzymes consists of a highly conserved GSH binding site, also known as the G‐site, and a less conserved hydrophobic H‐site, which coordinates the binding of the hydrophobic co‐substrates to which GSH is transferred.22 Mechanistically, H‐bonding of GSH with a catalytic Ser residue of the G‐site results in a drop of the sulfur pKa,23, 24, 25 stabilizing it as a thiolate for a nucleophilic attack on an electrophilic co‐substrate in the H‐site.26 To find out how methionine sulfoxide modifications could interfere with the binding of the substrates to both active sites, we determined the steady‐state kinetic parameters by varying both substrates concentrations (GSH and CDNB) (Table 2 and Supporting Information, Fig. S1). The results show especially an increase in the K M for CDNB, and a decrease of the K M for GSH, with less variation in the k cat values (Table 2). GSTF9 does not use a K M–k cat compensatory mechanism to deal with increased H2O2 concentrations as observed for A. thaliana GSTU23,12 or for cytosolic malate dehydrogenase 1 (MDH1).27
We clearly observed increased flexibility of the loop carrying Met35 (between β2 and α2) [Fig. 4(A)], located close to the G‐site, and the loop carrying Met118 and Met123 (between α4 and α5), located at the H‐site [Fig. 4(A)]. Although methionine oxidation was observed with mass spectrometry and we showed activity recovery with methionine sulfoxide reductases, methionine sulfoxide formation was not observed in the structures, as electron density for the α4–α5 loop is missing in the oxidized structures. Oxidation of protein methionines, which lead to local unfolding and impact the protein activity, have been reported in several other studies.28, 29 For example, the chaperone‐like activity of A. thaliana chloroplast Hsp21 is influenced due to the oxidation of its N‐terminal methionines (Met49, Met52, Met55, and Met59). Here, methionine sulfoxide formation causes partial unfolding due to the local loss of the helical structure.28 For GSTF9, it looks that methionine oxidation induces local flexibility of the loops between β2–α2 and between α4–α5, which could make these loops more accessible to the A. thaliana MsrA or MsrB, and thus facilitating sulfoxide reduction.
Numerous glutathione transferase enzymes can also act as hydroxyl peroxidases but with rate constants much lower30, 31 than those for established glutathione peroxidases and peroxiredoxins.32, 33 While oxidation has been shown to decrease the H2O2 peroxidase activity of GSTU23,12 our results show that the GSTF9 glutathione peroxidase activity is resistant to oxidation. However, the glutathione peroxidase activity of GSTF9ox is more impaired for hydrophobic peroxides (Table 3, Supporting Information, SI, and Fig. 6). This suggests that the loss of a structured hydrophobic H‐site upon oxidation of GSTF9 forms the basis for the loss in peroxidase activity toward more hydrophobic peroxides.
To further understand the observed changes in kinetic efficiency of GSTF9, we studied the transition state thermodynamic parameters of reduced and oxidized GSTF9 (Fig. 7 and Table 4). To study the transition state of GSTF9red and GSTF9ox, thermodynamic parameters were calculated and the data of the activated complex show a compensatory system with different enthalpy and entropy values for oxidized and reduced GSTF9 (Table 4). Generally, when comparing both forms, the overall spontaneity of the E:S# formation is similar (reflected by similar ΔG # values), but a different intermediate transition state along the reaction coordinates might get temporally populated. The E a and ΔH # are a result of the sum of interatomic interactions that undergo rearrangement during the E:S activation step. These interatomic interactions are hydrogen bonds and van der Waals interactions between enzyme and substrates.34 A positive difference of 2.3 kcal/mol was found when the absolute value of ΔH # of the oxidized GSTF9 was subtracted from the ΔH # value obtained for its reduced counterpart. The less pronounced energetic barrier for GSTF9red indicates that the number of interactions that needs to be rearranged in the reduced form is smaller than in the oxidized form (Table 4), leading to the observed energetic difference. Subsequently, the E:S complex formed by the reduced form seems to be less pronounced and more prone to rearrangement to achieve the transition state. Entropy is the measure of the number of microstates available for a specific species and is more widely described as the degree of disorder. The generation of disorder when E:S is converting to E:S# comes from bond rearrangement and changes in solvation.35 The value obtained for Δ(TΔS #)ox‐red indicates that GSTF9red undergoes a larger conformational change to reach the transition state when compared with GSTF9ox.36 The more positive TΔS # and positive ΔH # indicates that the increased flexibility of the loops between β2–α2 and between α4–α5 is a positive feature, and to achieve the transition state a larger number of interatomic interactions need to be rearranged. Accordingly, to maintain its enzymatic activity under oxidative stress, GSTF9 seems to follow a structural compensatory system. The best scenario is composed by an increase in flexibility in the regions directly involved in catalysis and an increase in rigidity in all the other areas; however, GSTF9 oxidation increased the flexibility globally, leading to a neglectable Δ(ΔG #)ox‐red value.36
In conclusion, we showed that methionine oxidation near the two active sites causes local loop‐flexibility. As a consequence, oxidized GSTF9 becomes substrate for methionine sulfoxide reductases. By thermodynamic and structural compensatory mechanisms, GSTF9 has evolved to become an enzyme that is able to catalyze detoxification reactions under oxidative conditions.
Materials and Methods
GSTF9 purification and oxidation
Protein expression, purification, and oxidation were performed as described by Jacques et al.4 Prior to oxidation, GSTF9 was reduced with 10 mM DTT at RT for 30 min followed by gel filtration using a Superdex75 HR 10/30 size exclusion chromatography (SEC) column equilibrated in 1× PBS. The collected sample was incubated with 1:100 molar ratio of protein to H2O2 for 135 min at 10°C, as performed by Jacques et al.4 Micro Bio‐Spin® Chromatography Column (BIO‐RAD) equilibrated with 1× PBS was used to remove excess of H2O2.
Glutathione transferase activity assay
The assay was performed as described by Habig et al.37 Briefly, a UV–visible Varian Cary 100 spectrophotometer was used to measure the GSTF9 catalyzed conjugation of GSH onto CDNB at A 340. The reaction mixture was composed of 0.5 mM GSH and 0.2 µM GSTF9 in 250 mM MOPS pH 6.5 and 150 mM NaCl buffer. The assay was initiated by the addition of 3.8 mM CDNB to a final reaction volume of 500 µL.
To determine the influence of Msr incubation on the GSTF9ox transferase activity, a 2:1 molar ratio of GSTF9ox (10 µM) was incubated with Corynebacterium diphtheriae MsrA38 or MsrB12 (5 µM) for 1 h at 25°C in a 1× PBS buffer containing 0.5 mM DTT. As a control, GSTF9red was also incubated with MsrA, MsrB and DTT. The samples were diluted and 10 µL was transferred to the assay mixture (500 µL), where the final concentrations were: 0.2 µM GSTF9, 0.1 µM Msr, and 10 µM DTT. A sample with only GSTF9ox and DTT was included as a control. Prior to the incubation of GSTF9 with Msr enzymes, both MsrA and MsrB were incubated with 20 mM DTT for 30 min at room temperature. Following reduction, DTT was removed on a Micro Bio‐Spin® Chromatography Column (BIO‐RAD) equilibrated with 1× PBS.
To determine the kinetic parameters of GSTF9, at varying concentrations of GSH (0–750 µM) or CDNB (0–4.5 mM) the initial velocities were measured and the Michaelis–Menten Eq. (1) was used to fit these data and obtain the kinetic parameters k cat and K M. For varying [GSH], the CDNB concentration used was 3.8 mM and for varying [CDNB], 0.5 mM GSH was used. Two independent replicates were measured for each substrate concentration. In Eq. (1), v represents the initial velocity, V max the maximum velocity, K M the Michaelis constant, and [S] the substrate concentration.
| (1) |
GSTF9 FOX assay with GSH and GSO3
Ferrous oxidation‐xylenol orange (FOX) assay was used to monitor the peroxidase activity of GSTF9.39 The assay was performed as described by Tossounian et al.12 Briefly, the reaction mixture included 50 µM GSTF9red or GSTF9ox, 1 mM GSH, and 150 µM H2O2 in 100 mM potassium phosphate buffer at pH 6.4, which was incubated at room temperature (RT). At three time points (0, 15, and 30 min), 10 µL of the sample was mixed with 490 µL of the FOX assay mixture and incubated in the dark. After 30 min, SpectraMax340PC spectrophotometer (Molecular Devices) was used to measure the absorption at 560 nm. To determine the effect of GSO3 on the glutathione peroxidase activity of GSTF9, the same procedure was followed with the addition of 0.5 mM GSO3. As background, the reaction mixture without enzyme was used. A reaction mixture containing 1 mM GSO3 was also used as a control. Glutathione peroxidase activity of GSTF9 was also tested by replacing H2O2 (200 µM) with two hydrophobic peroxides, cumene (200 µM) or tert‐butyl hydroperoxide (200 µM). Three independent measurements were performed and the data representing the Δ[peroxide]ox‐red versus ALogP value of H2O2 (−0.18), tert‐butyl hydroperoxide (0.79), and CuOOH (2.02) was plotted using GraphPad prism7. The AlogP values were obtained from ChEMBL.
GSTF9‐GSO3 inhibition assays
To determine the GSTF9 inhibition by GSO3, the SpectraMax340PC spectrophotometer (Molecular Devices) was used, where increasing concentrations of GSO3 (0, 25, 50, 100, 200, 300, 400, and 500 µM) were added to the reaction mixture containing 0.4 µM reduced GSTF9, 0.5 mM GSH and 3.8 mM CDNB in 250 mM MOPS pH 6.5 and 150 mM NaCl buffer. The reaction started by the addition of CDNB. As background, the reaction mixture without enzyme was used. The inhibition constant for the inactivation of GSTF9 by GSO3 was determined from the plot of the initial velocities versus the GSO3 concentration [Eq. (2)]. For all data, two independent measurements were performed and GraphPad prism7 was used to plot the data. In Eq. (2), v i represents the initial velocity, K i the inhibition constant, V the maximum velocity, and [GSO3] the concentration of inhibitor.
| (2) |
GSTF9 thermodynamic parameter determination
GSTF9 thermodynamic parameters were assessed by measuring the initial velocity variation in function of temperature. The reactions were carried out at 17°C, 20°C, 25°C, 30°C, and 37°C in quartz cuvettes using a UV–visible Varian Cary 100 spectrophotometer and were monitored for 180 s with GSTF9 (reduced or oxidized) final concentration 0.2 µM, 0.5 mM GSH, and 3.8 mM CDNB in 250 mM MOPS pH 6.5 and 150 mM NaCl buffer. The values presented correspond to two replicates. Subsequently, the thermodynamic parameters were obtained fitting the data to Arrhenius [Eq. (3)] and Eyring [Eq. (4)] approaches. The Gibbs free energy of activation (ΔG #) was calculated using Eq. (5). In Eq. (3), E a represents the reaction activation energy, R, the gas constant (8.314 J mol−1 K−1) and A, the product of the collision frequency (Z). For Eq. (4), k B represents the Boltzmann constant (1.3805 × 10−23 J K−1), h, the Planck's constant (6.6256 × 10−34 J s). For Eq. (5), ΔH # and ΔS # represent the enthalpy and entropy variation of activation, respectively.
| (3) |
| (4) |
| (5) |
Mass spectrometric analysis of Met oxidation
For the identification of oxidized Met residues (methionine sulfoxide and methionine sulfone), GSTF9red and GSTF9ox were analyzed by mass spectrometry as described by Tossounian et al.12
Author Contributions
GSTF9 purification was performed by MAT, KW, and IVM. Biochemical, kinetic, and thermodynamic assays were performed by MAT. Crystallization was performed by KW and IVM, and X‐ray crystal structures were solved by IVM. Mass spectrometry and data analysis was performed by DV. MAT, IVM, LAR, and JM wrote the manuscript. JM supervised all aspects of the project.
Accession Codes
The structures of GSTF9red (PDB 6EZY), GSTF9H2O2+NaOCl (PDB 6F01), and GSTF9ox (PDB 6F05) have been deposited in the Protein Data Bank.
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Supporting information
Supporting Information
Acknowledgments
The authors thank the beamline scientist at the Diamond Light Source beamline IO4 for their technical support. Furthermore, they thank Kristof Moonens from VIB‐VUB Center for Structural Biology for X‐ray data collection on the GSTF9 crystals. They thank Frank Van Breusegem from Ghent University, Belgium for giving us At‐gstF9 plasmid.
Word statement: We report the impact of oxidation on the structure and on the active site methionine residues of GSTF9 under hydrogen peroxide stress. Formation of methionine sulfoxide causes an increased flexibility of the active site loop inducing a decrease in its transferase activity, and a decrease of its glutathione peroxidase activity toward hydrophobic substrates. We also show redox regulation of GSTF9 by methionine sulfoxide reductase enzymes, which restore the GSTF9 activity.
References
- 1. Jacques S, Ghesquiere B, De Bock PJ, Demol H, Wahni K, Willems P, Messens J, Van Breusegem F, Gevaert K (2015) Protein methionine sulfoxide dynamics in Arabidopsis thaliana under oxidative stress. Mol Cell Proteom 14:1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeRidder BP, Dixon DP, Beussman DJ, Edwards R, Goldsbrough PB (2002) Induction of glutathione S‐transferases in Arabidopsis by herbicide safeners. Plant Physiol 130:1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dixon DP, Hawkins T, Hussey PJ, Edwards R (2009) Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J Exp Bot 60:1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marrs KA, Walbot V (1997) Expression and RNA splicing of the maize glutathione S‐transferase Bronze2 gene is regulated by cadmium and other stresses. Plant Physiol 113:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prade L, Huber R, Bieseler B (1998) Structures of herbicides in complex with their detoxifying enzyme glutathione S‐transferase ‐ explanations for the selectivity of the enzyme in plants. Structure 6:1445–1452. [DOI] [PubMed] [Google Scholar]
- 6. Menon D, Board PG (2013) A role for glutathione transferase Omega 1 (GSTO1‐1) in the glutathionylation cycle. J Biol Chem 288:25769–25779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laborde E (2010) Glutathione transferases as mediators of signaling pathways involved in cell proliferation and cell death. Cell Death Differ 17:1373–1380. [DOI] [PubMed] [Google Scholar]
- 8. Sheehan D, Meade G, Foley VM, Dowd CA (2001) Structure, function and evolution of glutathione transferases: implications for classification of non‐mammalian members of an ancient enzyme superfamily. Biochem J 360:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deponte M (2013) Glutathione catalysis and the reaction mechanisms of glutathione‐dependent enzymes. Biochim Biophys Acta 1830:3217–3266. [DOI] [PubMed] [Google Scholar]
- 10. Thom R, Dixon DP, Edwards R, Cole DJ, Lapthorn AJ (2001) The structure of a zeta class glutathione S‐transferase from Arabidopsis thaliana: characterisation of a GST with novel active‐site architecture and a putative role in tyrosine catabolism. J Mol Biol 308:949–962. [DOI] [PubMed] [Google Scholar]
- 11. Thom R, Cummins I, Dixon DP, Edwards R, Cole DJ, Lapthorn AJ (2002) Structure of a tau class glutathione S‐transferase from wheat active in herbicide detoxification. Biochemistry 41:7008–7020. [DOI] [PubMed] [Google Scholar]
- 12. Tossounian MA, Van Molle I, Wahni K, Jacques S, Gevaert K, Van Breusegem F, Vertommen D, Young D, Rosado LA, Messens J (2018) Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage. Biochim Biophys Acta 1862:775–789. [DOI] [PubMed] [Google Scholar]
- 13. Lee SH, Li CW, Koh KW, Chuang HY, Chen YR, Lin CS, Chan MT (2014) MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress. J Exp Bot 65:5049–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wagner U, Edwards R, Dixon DP, Mauch F (2002) Probing the diversity of the Arabidopsis glutathione S‐transferase gene family. Plant Mol Biol 49:515–532. [DOI] [PubMed] [Google Scholar]
- 15. Horvath E, Bela K, Papdi C, Galle A, Szabados L, Tari I, Csiszar J (2015) The role of Arabidopsis glutathione transferase F9 gene under oxidative stress in seedlings. Acta Biol Hung 66:406–418. [DOI] [PubMed] [Google Scholar]
- 16. Mashiyama ST, Malabanan MM, Akiva E, Bhosle R, Branch MC, Hillerich B, Jagessar K, Kim J, Patskovsky Y, Seidel RD, Stead M, Toro R, Vetting MW, Almo SC, Armstrong RN, Babbitt PC (2014) Large‐scale determination of sequence, structure, and function relationships in cytosolic glutathione transferases across the biosphere. PLoS Biol 12:e1001843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wadington MC, Ladner JE, Stourman NV, Harp JM, Armstrong RN (2009) Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase. Biochemistry 48:6559–6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stourman NV, Branch MC, Schaab MR, Harp JM, Ladner JE, Armstrong RN (2011) Structure and function of YghU, a nu‐class glutathione transferase related to YfcG from Escherichia coli. Biochemistry 50:1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nutricati E, Miceli A, Blando F, De Bellis L (2006) Characterization of two Arabidopsis thaliana glutathione S‐transferases. Plant Cell Rep 25:997–1005. [DOI] [PubMed] [Google Scholar]
- 20. Eftink MR, Anusiem AC, Biltonen RL (1983) Enthalpy‐entropy compensation and heat capacity changes for protein‐ligand interactions: general thermodynamic models and data for the binding of nucleotides to ribonuclease A. Biochemistry 22:3884–3896. [DOI] [PubMed] [Google Scholar]
- 21. Moons A (2005) Regulatory and functional interactions of plant growth regulators and plant glutathione S‐transferases (GSTs). Vitam Horm 72:155–202. [DOI] [PubMed] [Google Scholar]
- 22. Etienne F, Resnick L, Sagher D, Brot N, Weissbach H (2003) Reduction of Sulindac to its active metabolite, sulindac sulfide: assay and role of the methionine sulfoxide reductase system. Biochem Biophys Res Commun 312:1005–1010. [DOI] [PubMed] [Google Scholar]
- 23. Axarli I, Dhavala P, Papageorgiou AC, Labrou NE (2009) Crystal structure of Glycine max glutathione transferase in complex with glutathione: investigation of the mechanism operating by the Tau class glutathione transferases. Biochem J 422:247–256. [DOI] [PubMed] [Google Scholar]
- 24. Axarli I, Georgiadou C, Dhavala P, Papageorgiou AC, Labrou NE (2010) Investigation of the role of conserved residues Ser13, Asn48 and Pro49 in the catalytic mechanism of the tau class glutathione transferase from Glycine max . Biochim Biophys Acta 1804:662–667. [DOI] [PubMed] [Google Scholar]
- 25. Labrou NE, Papageorgiou AC, Pavli O, Flemetakis E (2015) Plant GSTome: structure and functional role in xenome network and plant stress response. Curr Opin Biotechnol 32:186–194. [DOI] [PubMed] [Google Scholar]
- 26. Dixon DP, Lapthorn A, Edwards R (2002) Plant glutathione transferases. Genome Biol 3:REVIEWS3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang J, Khan Niazi A, Young D, Rosado LA, Vertommen D, Bodra N, Abdelgawwad MR, Vignols F, Wei B, Wahni K, Bashandy T, Bariat L, Van Breusegem F, Messens J,JPR (2017) Self‐protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis . J Exp Bot 10.1093/jxb/erx396. [DOI] [PubMed] [Google Scholar]
- 28. Gustavsson N, Kokke BP, Anzelius B, Boelens WC, Sundby C (2001) Substitution of conserved methionines by leucines in chloroplast small heat shock protein results in loss of redox‐response but retained chaperone‐like activity. Protein Sci 10:1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balog EM, Lockamy EL, Thomas DD, Ferrington DA (2009) Site‐specific methionine oxidation initiates calmodulin degradation by the 20S proteasome. Biochemistry 48:3005–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Flury T, Wagner E, Kreuz K (1996) An inducible glutathione S‐transferase in soybean hypocotyl is localized in the apoplast. Plant Physiol 112:1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gronwald JW, Plaisance KL (1998) Isolation and characterization of glutathione S‐transferase isozymes from sorghum. Plant Physiol 117:877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manta B, Hugo M, Ortiz C, Ferrer‐Sueta G, Trujillo M, Denicola A (2009) The peroxidase and peroxynitrite reductase activity of human erythrocyte peroxiredoxin 2. Arch Biochem Biophys 484:146–154. [DOI] [PubMed] [Google Scholar]
- 33. Toppo S, Flohe L, Ursini F, Vanin S, Maiorino M (2009) Catalytic mechanisms and specificities of glutathione peroxidases: variations of a basic scheme. Biochim Biophys Acta 1790:1486–1500. [DOI] [PubMed] [Google Scholar]
- 34. Du X, Li Y, Xia YL, Ai SM, Liang J, Sang P, Ji XL, Liu SQ (2016) Insights into protein‐ligand Interactions: mechanisms, models, and methods. Int J Mol Sci 17:E144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loftfield RB, Eigner EA, Pastuszyn A, Lovgren TNE, Jakubowski H (1980) Conformational‐changes during enzyme catalysis ‐ Role of water in the transition‐state. Proc Natl Acad Sci USA 77:3374–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lonhienne T, Gerday C, Feller G (2000) Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim Biophys Acta 1543:1–10. [DOI] [PubMed] [Google Scholar]
- 37. Habig WH, Pabst MJ, Jakoby WB (1974) Glutathione S‐transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem 249:7130–7139. [PubMed] [Google Scholar]
- 38. Tossounian MA, Pedre B, Wahni K, Erdogan H, Vertommen D, Van Molle I, Messens J (2015) Corynebacterium diphtheriae methionine sulfoxide reductase a exploits a unique mycothiol redox relay mechanism. J Biol Chem 290:11365–11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jaeger J, Sorensen K, Wolff SP (1994) Peroxide accumulation in detergents. J Biochem Biophys Methods 29:77–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


