Abstract
Siderophore A (SidA) from Aspergillus fumigatus is a flavin‐containing monooxygenase that hydroxylates ornithine (Orn) at the amino group of the side chain. Lysine (Lys) also binds to the active site of SidA; however, hydroxylation is not efficient and H2O2 is the main product. The effect of pH on steady‐state kinetic parameters was measured and the results were consistent with Orn binding with the side chain amino group in the neutral form. From the pH dependence on flavin oxidation in the absence of Orn, a pK a value >9 was determined and assigned to the FAD‐N5 atom. In the presence of Orn, the pH dependence displayed a pK a value of 6.7 ±0.1 and of 7.70 ±0.10 in the presence of Lys. Q102 interacts with NADPH and, upon mutation to alanine, leads to destabilization of the C4a‐hydroperoxyflavin (FADOOH). Flavin oxidation with Q102A showed a pK a value of ~8.0. The data are consistent with the pK a of the FAD N5‐atom being modulated to a value >9 in the absence of Orn, which aids in the stabilization of FADOOH. Changes in the FAD‐N5 environment lead to a decrease in the pK a value, which facilitates elimination of H2O2 or H2O. These findings are supported by solvent kinetic isotope effect experiments, which show that proton transfer from the FAD N5‐atom is rate limiting in the absence of a substrate, however, is significantly less rate limiting in the presence of Orn and or Lys.
Keywords: flavin‐dependent monooxygneases, solvent kinetic isotope effect, pH profile, oxidation, siderophore, ornithine hydroxylase, hydroperoxyflavin
ABBREVIATIONS
- FAD
flavin adenine dinucleotide
- Orn
ornithine
- PDB
Protein Data Bank
- SidA
ornithine N5‐monooxygenase siderophore A
- SKIE
solvent kinetic isotope effects
Introduction
Flavin‐dependent monooxygenases that catalyze the hydroxylation of nitrogen atoms (N‐monooxygenases, NMO) are important in the biosynthesis of isoxazolidinone rings and hydroxamate functional groups in siderophores.1, 2 Because siderophores are essential virulence factors in bacterial and fungal pathogens, NMOs have been identified as potential drug targets.3, 4 NMOs catalyze the NADPH‐ and oxygen‐dependent hydroxylation of ornithine, lysine, histamine, and propyl or butyl‐diamines.5, 6 Siderophore A (SidA) from Aspergillus fumigatus is the most biochemically and structurally characterized member of this family of enzymes. The reaction begins with binding of NADPH to SidA with the FAD in its oxidized form (FADOX) [Fig. 1(A)].9, 10 This is followed by sterospecific (pro‐R) transfer of a hydride equivalent, resulting in NADP+ and reduced FAD (FADred), which completes the reductive half‐reaction (Fig. 1).11 In the oxidative half‐reaction, the FADredNADP+ complex reacts with molecular oxygen forming the C4a‐hydroperoxyflavin (FADOOH) intermediate.7, 12 In the absence of ornithine (Orn), this intermediate is stable and decays to hydrogen peroxide very slowly (k H2O2 ~ 0.01 s−1). However, in the presence of Orn, the enzyme is “activated” and rapid turnover is observed (k cat ~1.0 s−1).7 Stabilization of the FADOOH in the absence of Orn ensures that oxidation of NADPH is coupled to the hydroxylation of Orn, thereby, preventing the production of H2O2 and the wasting of NADPH. The active site architecture provides high specificity for Orn, with almost 100% efficient hydroxylation by the FADOOH intermediate, with little or no H2O2 being released (~100% coupled). SidA, however, can also bind lysine (Lys). This amino acid is hydroxylated with lower efficiency than Orn, with most of the FADOOH releasing as H2O2 (~10% coupled).13
Figure 1.
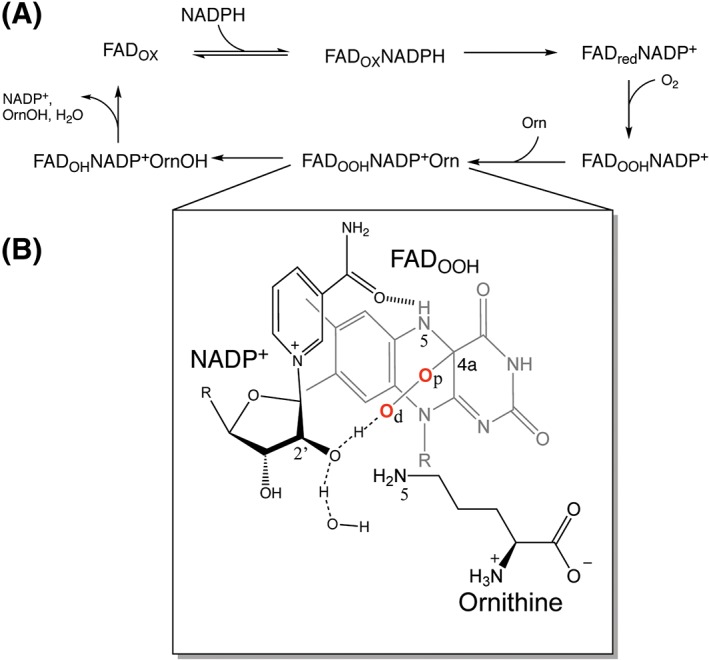
(A) Kinetic mechanism for the reaction catalyzed by SidA. The enzyme begins with the flavin in the oxidized form (FADOX). NADPH binds and reduces the flavin (FADred). The reduced FAD/NADP+ complex reacts with oxygen, forming the C4a‐hydroperoxyflavin (FADOOH). This is followed by Orn binding and hydroxylation at the Orn N5‐atom. The next step is release of H2O from the hydroxyflavin (FADOH). Release of products is shown to occur in one step. (B) Scheme of the FADOOH, NADP+, Orn complex obtained from previous molecular dynamics simulation and density functional theory studies.7, 8
Previously, we studied the mechanism of formation and stabilization of FADOOH using a combination of rapid reaction kinetics, isotope effects, and density functional theory. These studies showed that NADP+ plays several key roles in this process. First, NADP+ is involved in the protonation of the distal oxygen of FADOO‐ to form FADOOH via the 2’‐OH of the nicotinamide ribose [Fig. 1(B)]. Second, NADP+ provides binding interactions that are essential for stabilization of this intermediate, including hydrogen bonding of the NADP+ amide carbonyl with the FADOOH‐N5‐H and hydrogen bonding between the 2’‐OH of the NADP+ ribose and the distal oxygen of FADOOH. Third, NADP+ allows Orn to be in proper orientation for hydroxylation [Fig. 1(B)].7, 8, 13
Here, we present steady‐state, rapid reaction kinetics, pH studies, and solvent kinetic isotope effects, focusing mainly on decay of FADOOH in the absence and presence of Orn or Lys. The data are consistent with Orn binding with the side chain amino group in the deprotonated form. Favin oxidation in the absence of Orn substrate is very slow due to stabilizing interactions with the FAD‐N5‐H atom, which results in a pK a value >9. When Orn or Lys are present, the pK a value decreases to ~7 and flavin oxidation no longer limits the catalytic cycle. We present a complete description of the oxidative half‐reaction of SidA and briefly discuss previous studies that show conflicting results.
Results
kcat and kcat/Km pH profiles
The pH profile of SidA activity under steady‐state conditions was determined using oxygen consumption assays at a fixed concentration of NADPH (1 mM) and varying Orn. The data show that as the pH increases, the k cat value of SidA slightly increases with a single pK a value of 7.0 ±0.2 [Fig. 2(A)]. The dependence of k cat/K m in Figure 2(B) shows a similar trend with increasing values as the pH increases with a single pK a value of 8.0 ±0.2. The amplitude of the k cat/K m pH profile is greater than the k cat pH profile because, as the pH increases, the apparent K m for Orn decreases ~39‐fold across the pH range studied [Fig. 2(C), Table S1].
Figure 2.
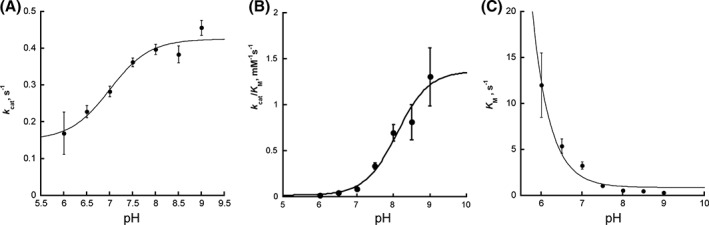
Effect of pH on the steady‐state oxygen consumption activity of SidA. (A) Effect of pH on the k cat value. (B) Effect of pH on the k cat/K m value. The pH profiles were analyzed using Eq. (3) and pK a values of 7.0 ±0.2 and 8.0 ±0.2 were obtained for k cat and k cat/K m, respectively. (C) Changes in the K m value as a function of pH. Values used in all plots are listed in Table S1. The line on panel C connects the points for viewing purposes.
pH dependence of FADOOH formation
The pH dependence of the rate of formation of the FADOOH intermediate was determined in both the presence and the absence of Orn to determine if any ionizable groups contributed to oxygen activation in SidA. The presence of Orn enhanced the rate constant for formation of the FADOOH intermediate. However, the rate constant for FADOOH formation in the presence or absence of Orn did not significantly change as a function of pH, resulting in an almost flat pH profile, as previously observed by our group and others (Supporting Information Fig. S1).12, 14
pL dependence and SKIE for flavin oxidation in the absence of substrate
The pL profile for flavin oxidation in the absence of substrate was performed to calculate the pK a value and SKIE of flavin oxidation. This experiment reports on the pL dependence of H2O2 elimination from FADOOH. It is essential to calculate SKIEs in the pL‐independent region as the pK a values of ionizable groups can change in D2O. The data in the absence of substrate in Figure 3(A) show a single pK a value for flavin oxidation that is >9 in H2O and a pK a value >10 in D2O, where the rate constant for flavin oxidation increases as the pL increases. The pK a values could not be accurately determined as SidA is unstable at pH >10 and the upper rate limit of flavin oxidation could not be defined. These data are consistent with previous experiments with SidA where a high pK a value was observed for flavin oxidation in the absence of a substrate.14, 15 The kinetic traces at 452 nm occurs mainly in a single phase regardless of pH (not shown). To properly measure the SKIE, it is essential to perform the reaction in the region where the kinetic constant is pL independent. This is necessary to prevent measuring an apparent SKIE that is due to the shift in the pK a value in D2O. For example, in Figure 3(A) at pL of 8.5, a significant different in the rate constant in D2O vs. H2O is observed. However, this is not a KIE, instead it is due to the shift in the pK a value. Therefore, the SKIE was calculated at pL 7.0 and a value of 2.30 ±0.05 was determined. This is consistent with our previous study, where we had a pL of 7.5 (Table 1).7
Figure 3.
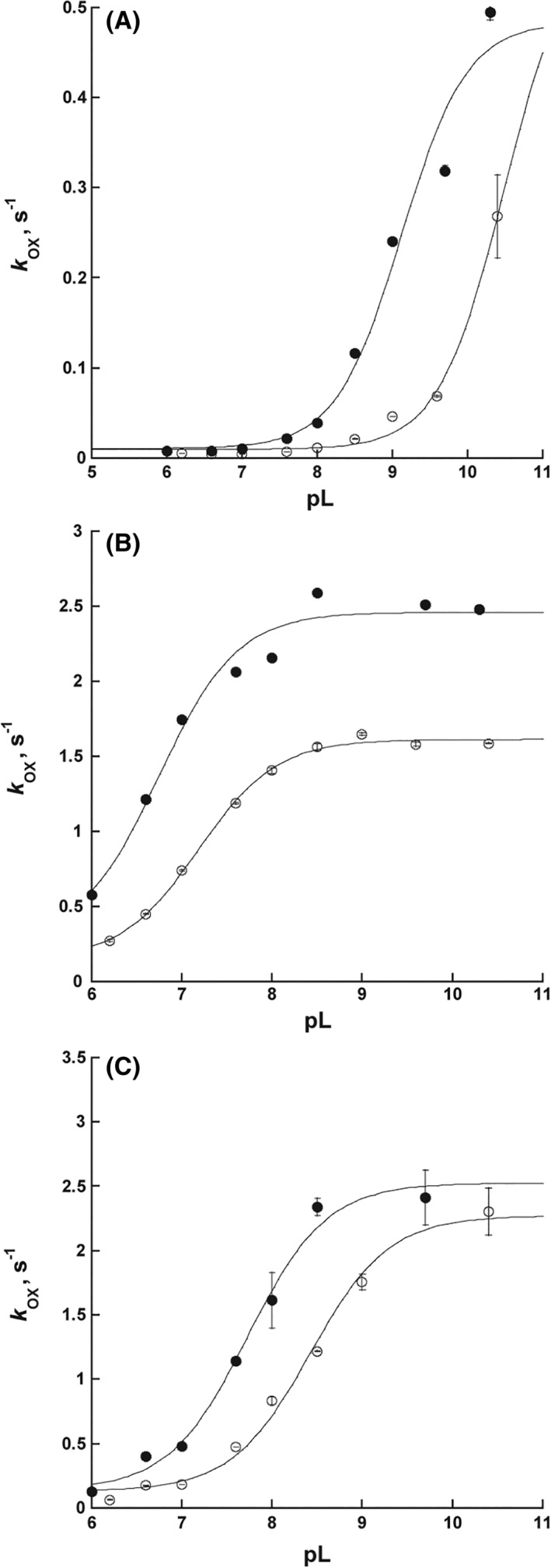
Rate constant for flavin oxidation (k OX) of SidA monitored at 452 nm as a function of pL in the absence or presence of amino acid substrates in H2O (●) or D2O (○). (A) Changes in the oxidation rate constant as a function of pH in the absence of amino acid substrate (H2O2 elimination). (B) In the presence of Orn (100 mM). (C) In the presence of 15 mM Lys.
Table 1.
Observed pK a Values and Solvent Kinetic Isotope Effect Values for SidA Oxidation
| Parameter | No substrate | Orn | Lys |
|---|---|---|---|
| pK a, H2O | >9 | 6.7 ± 0.10 | 7.70 ±0.10 |
| pK a, D2O | >10 | 7.18 ±0.05 | 8.42 ±0.07 |
| k ox, H2O | 0.0100 ±0.00020 | 2.50 ±0.07 | 2.52 ±0.10 |
| k ox, D2O | 0.0043 ±0.00010 | 1.60 ±0.02 | 2.30 ±0.08 |
| SKIE | 2.30 ±0.05a | 1.56 ±0.14b | 1.10 ±0.06b |
All pL experiments were performed at 25°C.
The SKIE values for flavin oxidation with no substrate were calculated at pL 7.0.
The SKIE values for flavin oxidation with Orn and Lys were calculated using the upper limits obtained from the fits of the pL profiles.
pL profile and SKIE for flavin oxidation with Orn
The pL profile for flavin oxidation in the presence of Orn (H2O elimination from FADOH) was performed to determine the ionizable groups important for hydroxylation. The data in Figure 3(B) show a significant shift in the pL profile compared with that in the absence of substrate with pK a values of 6.7 ±0.10 in H2O and 7.18 ±0.05 in D2O. A SKIE value of 1.56 ±0.14 was calculated from the upper limit of pL profiles (Table 1).
pL profile and SKIE for flavin oxidation with Lys
SidA is selective for hydroxylation of Orn. Although Lys binds to the active site, hydroxylation is not efficient, and the main outcome is the stimulation of hydrogen peroxide release from FADOOH.9, 16 The pL profile of flavin oxidation with Lys was performed to determine if its presence alters the pK a value and the SKIE associated with flavin oxidation. Results show a pL profile with pK a values of 7.70 ±0.10 in H2O and 8.42 ±0.07 in D2O [Fig. 3(C), Table 1]. A SKIE value of 1.10 ±0.06 was also calculated from the upper limit of the fits at high pL (Table 1).
Characterization of Q102A
The reaction of Q102A with NADPH was characterized in a stopped flow spectrophotometer as previously described.9, 17 The same experiment was performed on the wild‐type enzyme for comparison. Reduction occurred in two phases. A fast phase corresponding to flavin reduction and a slow phase that is independent of NADPH concentration has been suggested to be product release or a subpopulation of slow enzyme.11, 16 The affinity for NADPH is high, which prevents accurate calculation of the K D value using the stopped‐flow (changes in flavin absorbance are very small at low concentrations of NADPH). Thus, the reaction and apparent binding affinity for NADPH are not negatively affected in the Q102A enzyme (Table S2). The steady‐state kinetic parameters for Q102A were determined by monitoring the rate of oxygen consumption and the formation of hydroxylated Orn, as previously described.9, 17 The oxygen consumption for Q102A was determined using Orn as the variable substrate at fixed saturated concentrations of NADPH. The k cat value was ~45% higher than the value for SidA, while the K m value was ~30‐fold lower. This is an apparent K m value as the enzyme was highly active in the absence of Orn, indicative of high oxidase activity. The steady‐state analysis of Orn hydroxylation as a function of Orn concentrations under saturating concentrations of NADPH resulted in a k cat value of 0.32 s−1, which is ~50% lower than for SidA. The lower k cat value for hydroxylation compared with the value obtained in the oxygen consumption assay indicates that the mutant protein is only ~37% coupled compared with ~100% for SidA (Table 2). The low coupling in Q102A indicated that this enzyme is not efficient in stabilizing the FADOOH.
Table 2.
Steady‐State Kinetic Characterization
| Parameter | SidA | Q102A |
|---|---|---|
| Oxygen consumption assay | ||
| k cat, s−1 | 0.59 ±0.01 | 0.86 ±0.03 |
| K m (Orn), mM | 1.1 ±0.3 | 0.035 ±0.02 |
| k cat/K m (Orn), M−1 s−1 | 500 ±100 | 24,700 ±1400 |
| Orn hydroxylation assay | ||
| k cat, s−1 | 0.62 ± 0.02 | 0.32 ±0.04 |
| K m (Orn), mM | 1.0 ±0.2 | 0.64 ±0.03 |
| k cat/K m (Orn), M−1 s−1 | 600 ±100 | 510 ±20 |
| Coupling, % | 105 ±4 | 37.2 ±0.4 |
Conditions: 100 mM sodium phosphate, pH 7.5, and 25°C.
pH profiles for flavin oxidation with Q102A
Having established that the main effect of Q102A is on coupling, we measured the pH profile of the elimination of H2O2 from the FADOOH (e.g., in the absence of Orn). The increase in absorbance at 452 nm showed a single exponential rise, which was analyzed as described above with SidA. The results are shown in Figure 4 along with the pH profile for SidA for comparison. In the absence of Orn, k OX increases as a function of pH with a pK a value of 8.55 ± 0.05, which is lower than for the wild‐type enzyme. In the presence of Orn, k OX also increases as a function of pH with a pK a value of 6.4 ± 0.2, which is very similar to the value obtained for SidA (Table 1).
Figure 4.
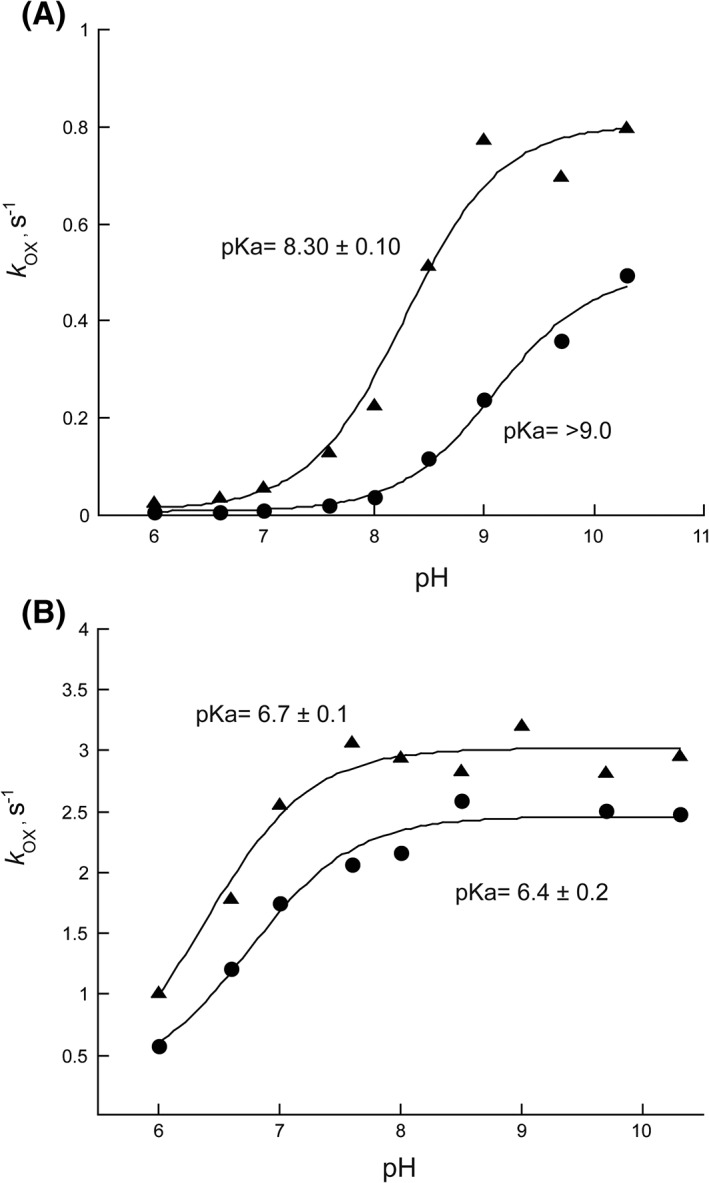
Effect of pH on the oxidation of Q102A (●) in the absence of Orn (A) or in the presence of 100 mM Orn (B). In both panels, SidA (π) is shown for comparison.
Discussion
Flavin‐dependent monooxygenases are important for adding chemical diversity to natural products as they are responsible for hydroxylating aromatic rings, aliphatic chains, sulfur, and nitrogen atoms.6, 18 In addition, the formation of epoxides, esters, or lactones are also outcomes of flavin‐dependent monooxygenation reactions.19, 20, 21 Our research group has been interested in the function of flavin‐dependent monooxygenases involved in the biosynthesis of hydroxamate‐containing siderophores.7, 8, 9, 11, 13, 22, 23, 24, 25 In this work, we were interested in determining whether functional groups with specific protonation states were required for the reaction catalyzed by SidA. The pH profile of k cat/K M showed that an ionizable group needs to be deprotonated for activity with a pK a value of ~8.0. We focused on the k cat/K M pH profile, since it reports on the protonation states of the free enzyme and substrate.26 The main effect observed was a decrease in the K M value for Orn as the pH of the solution was increased [Fig. 2(C)]. Thus, as an ionizable group becomes deprotonated, Orn binding becomes favorable. The structure of SidA in complex with Orn revealed that Lys107, Asn105, Ser469, and Asn323 are in hydrogen bonding distance to Orn (Fig. 5).13 Site‐directed mutagenesis showed that these residues are important for Orn binding.17 Asn105, Ser469, and Asn323 do not contain a side chain with a pK a value close to ~8.0. The pK a value of ~10.5 for the side chain of Lys107 could be lowered in the active site; however, the K m value for Orn would increase as Lys107 becomes deprotonated. Therefore, the ionizable group that needs to be deprotonated for Orn binding does not appear to belong to SidA.
Figure 5.
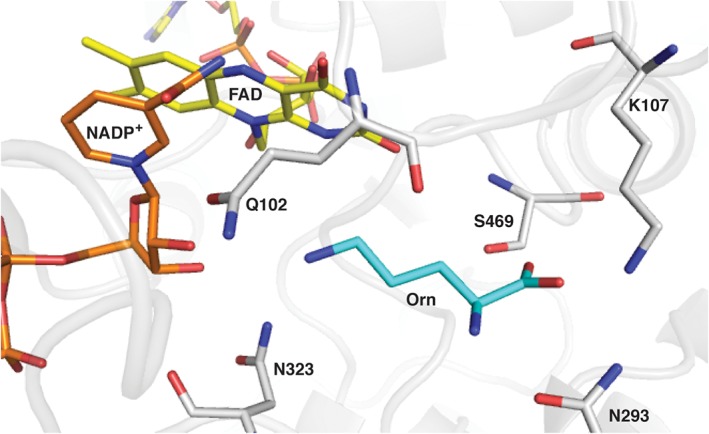
Active site of SidA showing the FAD (yellow carbons), NADP+ (orange carbons), and Orn (cyan carbons) (PDB code 4B63). The figure was made using PyMol.27
Alternatively, the ionizable group could be assigned to one of the amino groups of Orn. Although the pK a value of both groups is ~9, higher than the values observed in the k cat/K M pH profile, it has been well established that flavoproteins’ active sites modulate the protonation states of substrates to facilitate the chemical steps. For instance, p‐hydroxybenzoate hydroxylase (PHBH) modulates the protonation state of the p‐hydroxyl group such that it becomes deprotonated upon binding. This deprotonation is important to facilitating the nucleophilic aromatic hydroxylation.28, 29 Another well‐established modulation of the protonation state of the substrate is the case of amino acid oxidases. It has been shown that the amino acid binds with the α‐amino group in the deprotonated form, where deprotonation facilitates hydride transfer to the flavin.30 Thus, it is not unreasonable that SidA is capable of decreasing the pK a of the N δ‐Orn atom so that it binds in the deprotonated form. Previous computational studies have suggested that binding of Orn with the side chain in the deprotonated neutral form is more favorable.7, 8 Another ionizable group present in Orn is the α‐amino group. However, this group hydrogen bonds with Asn293 and deprotonation is not required for this interaction to occur (Fig. 5).13, 17 Our conclusions are different from that of Fredrick et al., who concluded that Orn binds in the protonated form and then becomes deprotonated in the ES complex. Their conclusion was mainly based on the pH profile of flavin oxidation that displayed a pK a value of ~7, which they assigned to Orn. However, as we will discuss below, we believe that this pK a value reports on the protonation state of the FAD‐N5 atom instead.
The effect of pH on flavin oxidation in SidA was also studied. In this case, the FADred:NADP+ complex reacts with molecular oxygen to form the FADOOH intermediate, which decays to FADox and H2O2 if Orn is absent. Hydroxylation occurs in the presence of Orn and H2O is eliminated to form FADOX. The effect of pH on the formation and decay of the FADOOH intermediate was determined first. We observed that formation of the FADOOH is pH independent in the absence of Orn, whereas the presence of Orn enhanced the rate constant for formation of the FADOOH intermediate. However, this effect is also pH independent, as previously shown (Supporting Information Fig. S1).14, 15 This is also consistent with our previous work that showed that this enhancement is independent of the amino group of the side chain of Orn, since norvaline (which lack an amino group in the side chain) can also enhance the formation of FADOOH, albeit to a lesser extent.7
In contrast, decay of FADOOH in the absence of Orn was very slow at low pH values and accelerated as the pH of the solution was increased. A pK a value above 9 could only be approximated from our data (Fig. 3 and Table 1). It has been proposed that stabilization of the FADOOH intermediate is mediated by hydrogen bonding interactions with the FAD‐N5‐H atom, as this atom is release with H2O2 or H2O during oxidation.31, 32 We have shown that the carbonyl oxygen of the nicotinamide moiety of NADP+ in SidA plays a key role in the stabilization of this intermediate (Fig. 1).13 Thus, the pK a value might be assigned to the FAD‐N5‐H atom, which is tuned to prevent elimination of H2O2 in the absence of Orn. pH studies in other monooxygenase systems have shown similar high pK a values, which have also been assigned to the FAD‐N5‐H atom.32 We then measured the effect of pH on decay of FADOOH in the presence of Orn or Lys. Here, the amino acid binds either to the FADred:NADP+ or the FADOOH:NADP+ complex in a rapid equilibrium process.7, 16 Hydroxylation of Orn takes place and the FADOH intermediate is formed. This is followed by elimination of H2O, which yields the oxidized FADox. A pK a value of 6.7 was obtained from the k OX pH profile, indicating that a group in the ES complex needs to be deprotonated for optimal activity (Fig. 3 and Table 1). This pK a value is very similar to the pK a value of 7.0 obtained from the k cat pH profile, which reports ionizable groups in the enzyme substrate complex.26 In the presence of Lys, where hydroxylation occurs less than 10% of the time, a pK a value of ~7.7 was measured. We believe that this pK a value is for the FADOH‐N5‐H, which is tuned to a lower value to accelerate turnover. SKIE was used as an indirect measure of the relative strength of the FAD‐N5‐H bond. In the absence of Orn, the SKIE value is 2.3, suggesting that elimination of the step that involves the hydrogen atom equivalent from the FAD‐N5 is significantly rate limiting. The SKIE decreases to 1.6 in the presence of Orn (Table 1) and further decreases to a value of 1.1 when Lys is present, suggesting that breaking of the FAD‐N5‐H bond is facilitated in these cases. It is worth noticing that a previous work reported a SKIE value >6 with Lys. This high value most likely originated from performing the assay in the pH sensitive region of the curve and not in the pH independent region.14
Evidence of modulation of the FAD‐N5‐H atom comes from studies on flavin‐dependent oxidases, where flavin oxidation appears to be pH independent or more likely that the pK a value is much lower than what can be tested in vitro (possibly lower than 5). The decrease in the pK a value of FAD‐N5‐H allows for the fast elimination of H2O2 in oxidases.33, 34 To provide further evidence that the pK a value of the FADOOH‐N5‐H intermediate is decreased from >9 to facilitate elimination of H2O or H2O2, we performed the pH profile of Q102A. In this mutant, interactions with FAD‐N5 have been changed, leading to destabilization of the FADOOH intermediate and, thus, the elimination of H2O2, as suggested by only 37% coupling (Table 2). The pH profile of flavin oxidation in the absence of Orn shows a clear shift, with a lower pK a value (~8.5). These results suggest that in the absence of Orn, the hydrogen bonding network that tunes the pK a value of the FADOOH‐N5‐H atom to >9 in SidA is changed in the Q102A mutant, presumably by changing the position of NADP+. These changes result in a lower FADOOH‐N5‐H pK a value and, thus, a more facile elimination of H2O2. Binding of Orn and formation of product leads to further changes in the hydrogen bonding of the FADOH‐N5‐H, which further decreases the pK a value to ~6.4 (compared with 6.7 for the wild‐type). Computational studies of elimination of H2O2 or H2O from FADOOH have shown the requirement of a proton transfer network.35 Analysis of the structure of SidA:NADP+ in the oxidized form and SidA:NADP+ that had been reduced by soaking the crystal in dithionite only show changes in the position of the side chains of R144 and M101. These residues are located above the FAD‐N5‐H locus and might be involved in the hydrogen binding network that tunes the pK a atom, presumably mediating with water molecules (Fig. 6).
Figure 6.
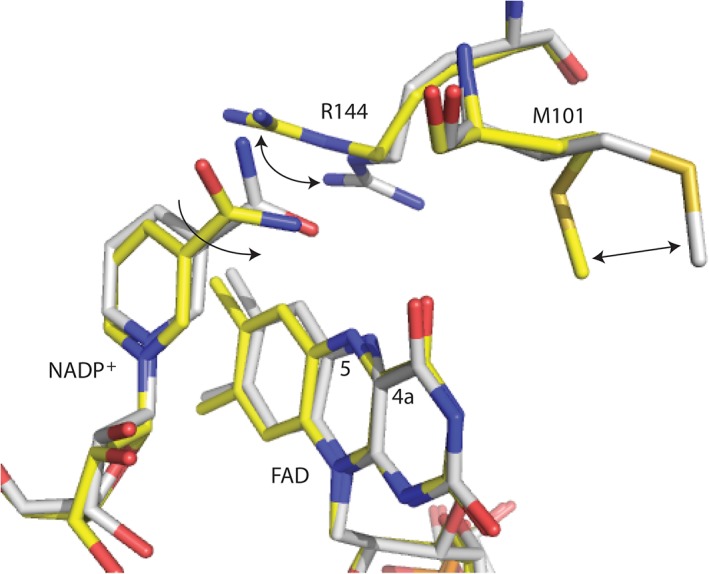
Observed conformational changes in SidA upon reduction. The oxidized form is shown with yellow carbons (PDB code 4B63) and the reduced form with white carbons (PDB code 4B65). The only observed changes are the rotation of residues R144 and M101, which are near the N5‐C4a. Typical bending of the FAD in the reduced form is also observed. Rotation of the amide bond is predicted based on the hydrogen bonding interactions with the oxidized flavin (where the amino group acts as a hydrogen bond donor) or reduced flavin (where the carbonyl oxygen acts as a hydrogen bond acceptor).
The results presented here show that Orn binds with the N5‐atom in the deprotonated form, which is required for the activity. The active site of SidA tunes the FAD‐N5‐H for optimal catalysis. The pK a value >9 in the absence of Orn stabilizes the FADOOH. Binding and hydroxylation of Orn leads to changes in the active site that decrease the FAD‐N5‐H pK a value to ~7.
Materials and Methods
Materials
Buffers and media were obtained from Fisher Scientific (Pittsburgh, PA). BL21(DE3)‐T1R chemically competent cells were obtained from Sigma‐Aldrich (St. Louis, MO). NADPH was obtained from EMD4 Biosciences (Billerica, MA). D2O was obtained from Cambridge Isotope Laboratories (Tewksbury, MA). Chromatography columns were obtained from GE Healthcare. All reagents were used without further purification.
Protein expression and purification
Wild‐type SidA (SidA) and Q102A were expressed in Escherichia coli BL21(DE3)‐T1R cells and purified as previously described.13 In general, ~25 mg of protein was obtained per liter of media. The purified proteins were stored in 30 μL aliquots in 100 mM sodium phosphate and 50 mM NaCl, pH 7.5, at −80°C at a concentration of ~200 μM. Protein concentration was calculated using the extinction coefficient at 450 nm of 13700 M−1 cm−1 for the FAD bound.9
pH effects on steady‐state kinetic parameters
The rate of oxygen consumption was measured using a Hansatech Oxygraph system (Norfolk, England). Assay solutions consisted of 1 mL of 100 mM sodium phosphate (pH 6 −8) and 100 mM Tris‐SO4 (pH 8.5−9.0) at 25°C. NADPH was kept constant at a concentration of 1 mM while Orn was varied between 0.1 and 10 mM. Reactions were initiated by the addition of 2 μM SidA and monitored for 5 min with constant stirring.
pL (pH or pD) effects on flavin oxidation
All rapid reaction experiments were carried out at 25°C using an SX‐20 stopped‐flow spectrophotometer (Applied Photophysics, Leatherhead, UK) in an anaerobic glove box (Coy, Grass Lake, MI). The preparation of anaerobic buffer was carried out with five cycles of vacuum (5 min) and flushing with O2‐free argon (0.5 min) with continuous stirring. This was repeated five times. The enzyme was made anaerobic with 10 cycles of vacuum (2 min) and flushing with O2‐free argon (10 s). The stopped‐flow was made anaerobic by flushing with 1 mL of anaerobic 100 mM sodium acetate, pH 5.0, containing 100 mM D‐glucose and 100 μg/mL glucose oxidase Type‐X. Substrates were made anaerobic by dissolving the appropriate mass in anaerobic buffer inside the glove box.
The rate constant of flavin oxidation (k OX, monitored at 452 nm) as a function of pH was measured in double mixing mode. Anaerobic SidA (60 μM before mixing) was first mixed with an equal volume of NADPH (60 μM before mixing) in 20 mM Tris‐Cl, 200 mM NaCl, pL 8.0 [pL refers to either the pH (H2O) or the pD (D2O)]. This mixture was incubated in an ageing loop for 60 s until the bound flavin was fully reduced. The reduced SidA−NADP+ complex was then allowed to react with air saturated buffer (130 μM oxygen after mixing at 1 atm and 25°C [O2] = 260 μM). This air‐saturated solution contained 200 mM of buffer, which rapidly increased the pL to the desired value in the flow cell. Between pL values of 6 and 8, sodium phosphate was used, between pL values of 8.5−9.0, Tris‐SO4 was used, and between pL values of 9.5−10.5, sodium carbonate/bicarbonate was used. In pL experiments with different amino acids present, 100 mM Orn and 15 mM Lys were used. For the reactions in D2O, SidA was concentrated to ~400 μM (based on flavin content) and diluted to 60 μM in a 100% D2O buffer of 20 mM Tris‐Cl, 200 mM NaCl, pD 8.0. This gave a concentration of ~85% D2O and, after two mixes in the stopped‐flow with 100% D2O buffer, a final D2O concentration of ~96% was achieved. The proper pD of all D2O buffers was calculated by adding 0.4 to the value on the pH meter, which is the variation from the change in the equilibrium on a hydrogen selective glass electrode.36 All solutions were checked for their proper pL values with a Fisher Scientific Accumet AB15+ Basic pH meter. Spectra were taken on a logarithmic time scale until complete flavin oxidation was observed. The rate constants were calculated from the average of at least three experiments.
Site‐directed mutagenesis
Replacement of Gln at Position 102 to Ala was performed using the QuikChange (Agilent Technologies, Santa Clara, CA) method following manufacturer's instructions. The wild‐type SidA gene, subcloned into the pET15b plasmid, was used as the template.11 The reaction was done using the forward primer (5′‐CCGGGCTCGAAGATGGCTATCAGCTTCATCAAG‐3′) and reverse primer (5′‐CTTGATGAAGCTGATAGCCATCTTCGAGCCCGG‐3′). Mutations were confirmed by DNA sequencing at the Virginia Biocomplexity Institute Core Sequencing Facility.
Characterization of Q102A
Steady‐state kinetic parameters were obtained by monitoring the reaction with oxygen using a Hansatech Oxygraph System (Norfolk, England). Reactions consisted of a 1 mL volume of 100 mM sodium phosphate, pH 7.5, at 25°C. NADPH was kept constant at a concentration of 1 mM while Orn was varied between 0.1 and 15 mM. Reactions were initiated by addition of 2 μM of Q102A. Reactions were monitored for 5 minutes with constant stirring. Hydroxylated Orn was monitored using a variation of the Csaky iodine oxidation assay.22, 37 The standard assay buffer contained 104 μL of 100 mM sodium phosphate (pH 7.5) with varying concentrations of Orn and NADPH held constant at 1 mM. Reactions were initiated by addition of 2 μM Q102A. Reactions were incubated for 10 min at 25°C with constant stirring at 750 rpm. The rates of flavin reduction with NADPH measured in the stopped‐flow spectrophotometer were done as previously described.11, 38, 39 The effect of pH on flavin oxidation in the absence or presence of Orn were performed as described above for wild‐type enzyme.
Data analysis
All data were fit using KaleidaGraph (Synergy Software, Reading, PA). For flavin oxidation studies, the increase in absorbance at 372 nm corresponding to formation of the FADOOH intermediate, was fit to Eq. (1) and describes a single exponential rise. Flavin oxidation in the absence of substrate, measured at 450 nm, was also fit to Eq. (1). In the presence of Orn or Lys the increase in absorbance at 452 nm, was fit to Eq. (2), which describes a double exponential rise. In both of these equations, c is the absorbance value at the beginning of the experiment, a is the total absorbance change, and k x is the rate constant for the formation of FADOOH (Eq. (1)) or flavin oxidation (Eq. (2)) at a particular pH value:
| (1) |
| (2) |
The pL dependence of flavin oxidation (monitored at 452 nm) and the pH dependences of k cat and k cat/K m were fit to Eq. (3), which describes a curve with a single pK a value with increasing activity as the pL increases and with plateau regions at high and low pL. The upper limits and lower limits for the pL profiles are denoted by C and A, respectively. The solvent kinetic isotope effects were calculated by dividing the value obtained for C in H2O by the value obtained in D2O:
| (3) |
Supporting information
Figure 1S. Effect of pH on the rate constant for formation of the FADOOH intermediate (kOOH) in the absence (A) or presence of Orn (B)
Table S1. Steady state kinetic parameters as a function of pH.
Table S2. Pre‐steady‐state kinetic parameters for flavin reduction by NADPH
Acknowledgments
This work was supported by a grant from the National Science Foundation MCB‐1021384 and CHE‐1506206. We thank the anonymous reviewers for their suggestions and comments.
Short statement for broader audience: Iron‐binding molecules, known as siderophores, are required for the pathogen Aspergillus fumigatus to scavenge iron during infection in humans. Siderophore A (SidA) is an enzyme that plays an essential role in the biosynthesis of siderophores. Oxidation of the flavin cofactor of SidA was probed using pH studies, site‐directed mutagenesis, and kinetic solvent isotope effects. Results indicate that the N5‐atom of the flavin cofactor is responsible for regulating the stability of the hydroxylating flavin intermediate and for oxidation. This information that is important for drug design.
References
- 1. Walsh CT, Wencewicz TA (2013) Flavoenzymes: versatile catalysts in biosynthetic pathways. Nat Prod Rep 30:175–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Olucha J, Lamb AL (2011) Mechanistic and structural studies of the N‐hydroxylating flavoprotein monooxygenases. Bioorg Chem 39:171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fischbach MA, Lin H, Liu DR, Walsh CT (2006) How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol 2:132–138. [DOI] [PubMed] [Google Scholar]
- 4. Frederick RE, Mayfield JA, DuBois JL (2009) Iron trafficking as an antimicrobial target. Biometals 22:583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esuola CO, Babaloa OO, Heine T, Schwabe R, Schlomann M, Tischler D (2016) Identification and characterization of a FAD‐dependent putrescine N‐hydroxylase (GorA) from Gordonia rubripertincta CWB2. J Mol Cataly B 134:378–389. [Google Scholar]
- 6. Huijbers MM, Montersino S, Westphal AH, Tischler D, van Berkel WJ (2014) Flavin dependent monooxygenases. Arch Biochem Biophys 544:2–17. [DOI] [PubMed] [Google Scholar]
- 7. Robinson R, Badieyan S, Sobrado P (2013) C4a‐hydroperoxyflavin formation in N‐hydroxylating flavin monooxygenases is mediated by the 2'‐OH of the nicotinamide ribose of NADP(+). Biochemistry 52:9089–9091. [DOI] [PubMed] [Google Scholar]
- 8. Badieyan S, Bach RD, Sobrado P (2015) Mechanism of N‐hydroxylation catalyzed by flavin‐dependent monooxygenases. J Org Chem 80:2139–2147. [DOI] [PubMed] [Google Scholar]
- 9. Chocklett SW, Sobrado P (2010) Aspergillus fumigatus SidA is a highly specific ornithine hydroxylase with bound flavin cofactor. Biochemistry 49:6777–6783. [DOI] [PubMed] [Google Scholar]
- 10. Robinson R, Franceschini S, Fedkenheuer M, Rodriguez PJ, Ellerbrock J, Romero E, Echandi MP, Martin Del Campo JS, Sobrado P (2014) Arg279 is the key regulator of coenzyme selectivity in the flavin‐dependent ornithine monooxygenase SidA. Biochim Biophys Acta 1844:778–784. [DOI] [PubMed] [Google Scholar]
- 11. Romero E, Fedkenheuer M, Chocklett SW, Qi J, Oppenheimer M, Sobrado P (2012) Dual role of NADP(H) in the reaction of a flavin dependent N‐hydroxylating monooxygenase. Biochim Biophys Acta 1824:850–857. [DOI] [PubMed] [Google Scholar]
- 12. Romero E, Robinson R, Sobrado P (2012) Monitoring the reductive and oxidative half‐reactions of a flavin‐dependent monooxygenase using stopped‐flow spectrophotometry. J Vis Exp 61:3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franceschini S, Fedkenheuer M, Vogelaar NJ, Robinson HH, Sobrado P, Mattevi A (2012) Structural insight into the mechanism of oxygen activation and substrate selectivity of flavin‐dependent N‐hydroxylating monooxygenases. Biochemistry 51:7043–7045. [DOI] [PubMed] [Google Scholar]
- 14. Frederick RE, Ojha S, Lamb A, Dubois JL (2014) How pH modulates the reactivity and selectivity of a siderophore‐associated flavin monooxygenase. Biochemistry 53:2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Romero E, Avila D, Sobrado P. Effect of pH on the reductive and oxidative half‐reactions of Aspergillus fumigatus Siderophore A In: MIller S, Hille R, Palfey B, Eds , 2013. Flavins and Flavoproteins. Raleigh, NC: Lulu. [Google Scholar]
- 16. Mayfield JA, Frederick RE, Streit BR, Wencewicz TA, Ballou DP, DuBois JL (2010) Comprehensive spectroscopic, steady state, and transient kinetic studies of a representative siderophore‐associated flavin monooxygenase. J Biol Chem 285:30375–30388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robinson R, Qureshi IA, Klancher CA, Rodriguez PJ, Tanner JJ, Sobrado P (2015) Contribution to catalysis of ornithine binding residues in ornithine N5‐monooxygenase. Arch Biochem Biophys 585:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teufel R (2017) Flavin‐catalyzed redox tailoring reactions in natural product biosynthesis. Arch Biochem Biophys 632:20–27. [DOI] [PubMed] [Google Scholar]
- 19. Leisch H, Morley K, Lau PC (2011) Baeyer‐Villiger monooxygenases: more than just green chemistry. Chem Rev 111:4165–4222. [DOI] [PubMed] [Google Scholar]
- 20. Torres Pazmino DE, Dudek HM, Fraaije MW (2010) Baeyer–Villiger monooxygenases: recent advances and future challenges. Curr Opin Chem Biol 14:138–144. [DOI] [PubMed] [Google Scholar]
- 21. Holtmann D, Fraaije MW, Arends IW, Opperman DJ, Hollmann F (2014) The taming of oxygen: biocatalytic oxyfunctionalisations. Chem Commun 50:13180–13200. [DOI] [PubMed] [Google Scholar]
- 22. Abdelwahab H, Robinson R, Rodriguez P, Adly C, El‐Sohaimy S, Sobrado P (2016) Identification of structural determinants of NAD(P)H selectivity and lysine binding in lysine N(6)‐monooxygenase. Arch Biochem Biophys 606:180–188. [DOI] [PubMed] [Google Scholar]
- 23. Qi J, Kizjakina K, Robinson R, Tolani K, Sobrado P (2012) A fluorescence polarization binding assay to identify inhibitors of flavin‐dependent monooxygenases. Anal Biochem 425:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shirey C, Badieyan S, Sobrado P (2013) Role of Ser‐257 in the sliding mechanism of NADP(H) in the reaction catalyzed by the Aspergillus fumigatus flavin‐dependent ornithine N5‐monooxygenase SidA. J Biol Chem 288:32440–32448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Binda C, Robinson RM, Martin Del Campo JS, Keul ND, Rodriguez PJ, Robinson HH, Mattevi A, Sobrado P (2015) An unprecedented NADPH domain conformation in lysine monooxygenase NbtG provides insights into uncoupling of oxygen consumption from substrate hydroxylation. J Biol Chem 290:12676–12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cook PF, Cleland WW. Enzyme kinetics and mechanism. Taylor & Francis Inc., 2007. New York, NY. [Google Scholar]
- 27. DeLano WL (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger.
- 28. Palfey BA, Moran GR, Entsch B, Ballou DP, Massey V (1999) Substrate recognition by "password" in p‐hydroxybenzoate hydroxylase. Biochemistry 38:1153–1158. [DOI] [PubMed] [Google Scholar]
- 29. Crozier‐Reabe K, Moran GR (2012) Form follows function: structural and catalytic variation in the class a flavoprotein monooxygenases. Int J Mol Sci 13:15601–15639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fitzpatrick PF (2010) Oxidation of amines by flavoproteins. Arch Biochem Biophys 493:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Orru R, Pazmino DE, Fraaije MW, Mattevi A (2010) Joint functions of protein residues and NADP(H) in oxygen activation by flavin‐containing monooxygenase. J Biol Chem 285:35021–35028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thotsaporn K, Chenprakhon P, Sucharitakul J, Mattevi A, Chaiyen P (2011) Stabilization of C4a‐hydroperoxyflavin in a two‐component flavin‐dependent monooxygenase is achieved through interactions at flavin N5 and C4a atoms. J Biol Chem 286:28170–28180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gannavaram S, Gadda G (2013) Relative timing of hydrogen and proton transfers in the reaction of flavin oxidation catalyzed by choline oxidase. Biochemistry 52:1221–1226. [DOI] [PubMed] [Google Scholar]
- 34. Carro J, Ferreira P, Martinez AT, Gadda G (2018) Stepwise hydrogen atom and proton transfers in dioxygen reduction by aryl‐alcohol oxidase. Biochemistry 57:1790–1797. [DOI] [PubMed] [Google Scholar]
- 35. Bach RD, Mattevi A (2013) Mechanistic aspects regarding the elimination of H2O2 from C(4a)‐hydroperoxyflavin. The role of a proton shuttle required for H2O2 elimination. J Org Chem 78:8585–8593. [DOI] [PubMed] [Google Scholar]
- 36. Schowen KB, Schowen RL (1982) Solvent isotope effects of enzyme systems. Methods Enzymol 87:551–606. [PubMed] [Google Scholar]
- 37. Csaky T (1948) On the estimation of bound hydroxylamine in biological materials. Acta Chem Scand 2:450–454. [Google Scholar]
- 38. Dhatwalia R, Singh H, Solano LM, Oppenheimer M, Robinson RM, Ellerbrock JF, Sobrado P, Tanner JJ (2012) Identification of the NAD(P)H binding site of eukaryotic UDP‐galactopyranose mutase. J Am Chem Soc 134:18132–18138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robinson RM, Rodriguez PJ, Sobrado P (2014) Mechanistic studies on the flavin‐dependent N(6)‐lysine monooxygenase MbsG reveal an unusual control for catalysis. Arch Biochem Biophys 550–551:58–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1S. Effect of pH on the rate constant for formation of the FADOOH intermediate (kOOH) in the absence (A) or presence of Orn (B)
Table S1. Steady state kinetic parameters as a function of pH.
Table S2. Pre‐steady‐state kinetic parameters for flavin reduction by NADPH


