Abstract
Dissecting complex biological systems requires target-specific control of protein function or abundance. Genetic perturbations are limited by off-target effects, multi-component complexity, and irreversibility. Most limiting is the requisite delay from modulation to experimental measurement. To enable the immediate and selective control of single protein abundance, we created a chemical biology system that leverages the potency of cell-permeable heterobifunctional degraders. The dTAG system pairs a novel degrader of FKBP12F36V with expression of FKBP12F36V in-frame with a protein of interest. By transgene expression or CRISPR-mediated locus-specific knock-in, we exemplify a generalizable strategy to study the immediate consequence of protein loss. Using dTAG, we observe an unexpected superior anti-proliferative effect of pan-BET bromodomain degradation over selective BRD4 degradation, characterize immediate effects of KRASG12V loss on proteomic signaling, and demonstrate rapid degradation in vivo. This technology platform will confer kinetic resolution to biological investigation and provide target validation in the context of drug discovery.
Graphic abstract
Genetic mechanisms for modulating abundance of gene products, such as RNA interference and CRISPR/Cas9 genome editing, are powerful means to determine the functional consequence of loss or gain of a target gene. However, these methodologies fall short in their ability to assess acute changes, particularly for proteins required for cell growth and survival. Small-molecule perturbations allow for the rapid assessment of biological responses, but are often limited by the time to creation of selective chemical matter, the selective disruption of discrete domain function, and off-target effects. We therefore undertook efforts to create a hybrid chemical biology system to leverage the temporal experimental benefits of small molecules with the precision of genetics.
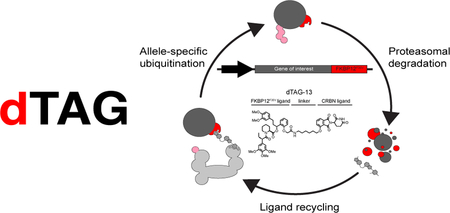
The degradation tag (dTAG) system is predicated upon our recent description of an all-chemical solution for target-specific protein degradation1. This prior research demonstrated selective degradation of BET bromodomain transcriptional co-activators (BRD2, BRD3, BRD4) using heterobifunctional degraders that bridge BET bromodomains to an E3 ubiquitin ligase (cereblon; CRBN). Catalytic-like biochemical activity of these small molecules allows rapid and target-specific turnover without degradation of the ligand. Use of E3-binding heterobifunctional degraders has now been validated for BRD41–4, and extended to FKBP121, ERRα5, RIPK25 and BRD96. Chemical probes established by this approach will powerfully enable the study of target biology, but each requires the up-front identification of a target-selective ligand. We therefore envisioned a single, generalizable approach to rapidly degrade allele-specific protein chimeras for biological investigation and early target validation.
Tag-based technologies to influence protein homeostasis have been pursued intently in the discipline of chemical biology. Exemplary technologies include induced displacement of cryptic degrons7, degradation of HaloTag fusion proteins via hydrophobic tagging8 or HaloPROTACs9, small molecule assisted shutoff (SMASh)10, and the auxin-inducible degron (AID)11. Selective stabilization of a target protein known as the Shield system has also been described. This system involves expression of a protein chimera involving a destabilized mutant allele of FKBP12, which is stabilized upon administration of a directed ligand, Shield-112. This system has been well utilized to study gain-of-function in complex biological systems13, 14. Inspired by this research, we endeavored to create a comparable loss-of-function solution to resolve target biology and dependencies in cells and in mice.
RESULTS
dTAG molecules engage FKBP12F36V and CRBN
To establish the more generalizable concept of all-chemical protein degradation, we previously reported the feasibility of degrading a cytosolic prolyl isomerase, FKBP12 (FKBP1A, also referred to herein as FKBP12WT), by heterobifunctional degraders (dFKBP-1 (1)) that recruit CRBN1. We initially considered expressing target proteins as an FKBP12WT chimera for biological investigation, but were concerned about confounding biological effects of endogenous FKBP12WT disruption. Engineered variants of FKBP12 were previously described by Ariad Pharmaceuticals (e.g. FKBP12F36V), that create a cavity (or “hole”) in FKBP12 allowing selective recognition by a “bumped” synthetic FKBP12F36V-directed ligand, AP186715. We hypothesized that an FKBP12F36V chimera might be selectively degraded by heterobifunctional FKBP12F36V and CRBN chemical inducers of dimerization (Fig. 1a).
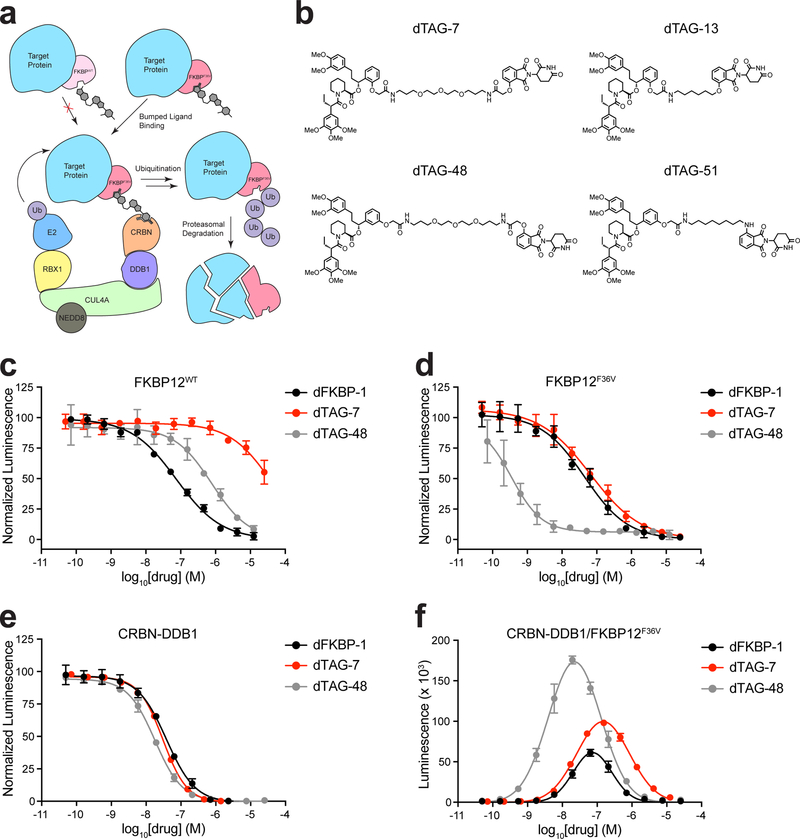
Figure 1 |. Heterobifunctional dTAG molecules engage and dimerize FKBP12F36V and CRBN in biochemical assays.
(a) Schematic depiction of the dTAG strategy. Bumped heterobifunctional dTAG molecules induce dimerization of FKBP12F36V fusion chimeras and the CRBN E3 ligase complex leading to CRBN-mediated degradation. (b) Chemical structures of heterobifunctional dTAG molecules. (c) DMSO normalized displacement of bio-SLF from GST-FKBP12WT by AlphaScreen with dFKBP-1, dTAG-7, or dTAG-48. (d) DMSO normalized displacement of bio-SLF from GST-FKBP12F36V by AlphaScreen with dFKBP-1, dTAG-7, or dTAG-48. (e) DMSO normalized displacement of bio-Thal from HIS-CRBN-DDB1 by AlphaScreen with dFKBP-1, dTAG-7, or dTAG-48. (f) dTAG molecule induced GST-FKBP12F36V and HIS-CRBN-DDB1 complex formation by AlphaScreen with dFKBP-1, dTAG-7, or dTAG-48. Data in c-f are presented as mean ± s.d. of n = 3 independent samples and are representative of n = 3 independent experiments.
To this end, we appended CRBN ligands to AP1867 as well as a novel regioisomer with varying linker length and composition to create a series of heterobifunctional degraders (dTAG-7 (2), dTAG-13 (3), dTAG-48 (4), dTAG-51 (5); Fig. 1b and Supplementary Fig. 1a). To biochemically evaluate the selectivity of these molecules, we developed FKBP12WT, FKBP12F36V, and CRBN-DDB1 ligand-displacement assays, using a nanomaterial-based proximity assay (AlphaScreen)16. These experiments test the ability of dTAG molecules to compete for binding to FKBP12WT and FKBP12F36V with a biotin-conjugated FKBP12WT-directed ligand (bio-SLF (6)) or to CRBN with a biotin-conjugated thalidomide (bio-Thal (7)) (Supplementary Fig. 1b–e). As expected, we observed a dose-dependent decrease in luminescence upon incubation of dFKBP-1 with FKBP12WT, FKBP12F36V, and CRBN, validating competition in each context (Fig. 1c–e and Supplementary Table 1). Comparative evaluation of dTAG molecules found that ortho-substituted molecules (dTAG-7 and dTAG-13) favorably engaged with FKBP12F36V and CRBN, with limited activity towards FKBP12WT (Fig. 1c-e, Supplementary Fig. 2a–c and Supplementary Table 1). In contrast, meta-substituted molecules (dTAG-48 and dTAG-51), despite being remarkably potent in their ability to engage FKBP12F36V and CRBN, had significant affinity for FKBP12WT.
To assess the adaptor function of the putative chemical degraders, we developed a CRBN-DDB1/FKBP12F36V dimerization assay, again using a luminescence proximity format as we previously reported for CRBN-DDB1/BRD41 (Supplementary Fig. 2d). dFKBP-1 and all dTAG molecules exhibited a dose-dependent increase in luminescence, supporting effective heterodimerization of CRBN-DDB1 and FKBP12F36V (Fig. 1f and Supplementary Fig. 2e). As expected, loss of luminescence with excess concentrations of dTAG molecules was observed, obeying the characteristic “hook effect” behavior17 upon saturation of CRBN and FKBP12F36V binding sites. Collectively, these biochemical data identify dTAG-7 and dTAG-13 as FKBP12F36V-selective heterodimerizers that effectively engage FKBP12F36V and CRBN.
Cellular target degradation by dTAG-7 and dTAG-13
To assay dose-dependent effects on FKBP12WT and FKBP12F36V degradation in cells, we employed a modified version of a dual luciferase assay18. FKBP12WT and FKBP12F36V were subcloned as in-frame fusions to nano-luciferase (Nluc) and expressed on a multicistronic mRNA with control firefly luciferase (Fluc) (Supplementary Fig. 3a). Measurement of Nluc:Fluc ratios allows for determination of FKBP12 fusion protein degradation relative to the internal Fluc control, independent of effects on transcription. We first transduced 293FT CRBN wild-type (293FTWT) cells with FKBP12WT-Nluc or FKBP12F36V-Nluc and observed that dTAG-7 and dTAG-13 potently reduced the bioluminescent signal of FKBP12F36V-Nluc, with limited effect on FKBP12WT-Nluc, consistent with the observed selectivity in biochemical experiments (Fig. 2a and Supplementary Table 2). By contrast, dFKBP-1 and dTAG-48 indiscriminately diminished bioluminescent signal of both FKBP12WT-Nluc and FKBP12F36V-Nluc, corroborating our biochemical experiments that dTAG-48 exhibits off-target activity (Fig. 2a, Supplementary Fig. 3b and Supplementary Table 2). We further evaluated the selectivity of our dTAG molecules by monitoring degradation of IKZF1, a previously described neo-substrate of CRBN upon immunomodulatory drug treatment18, 19, using a dual luciferase IKZF1 reporter system in 293T cells18. Compared to lenalidomide and pomalidomide, dTAG-7, dTAG-13 and dTAG-48 treatment had limited effect on bioluminescent signal, indicating little to no off-target IKZF1 activity (Supplementary Fig. 3c–d and Supplementary Table 3). However, dTAG-51 diminished bioluminescent signal to levels comparable to lenalidomide and pomalidomide, indicating off-target activity towards IKZF1. Due to the limited off-target activity towards FKBP12WT and IKZF1, we selected dTAG-7 and dTAG-13 as lead heterobifunctional degraders for further characterization.
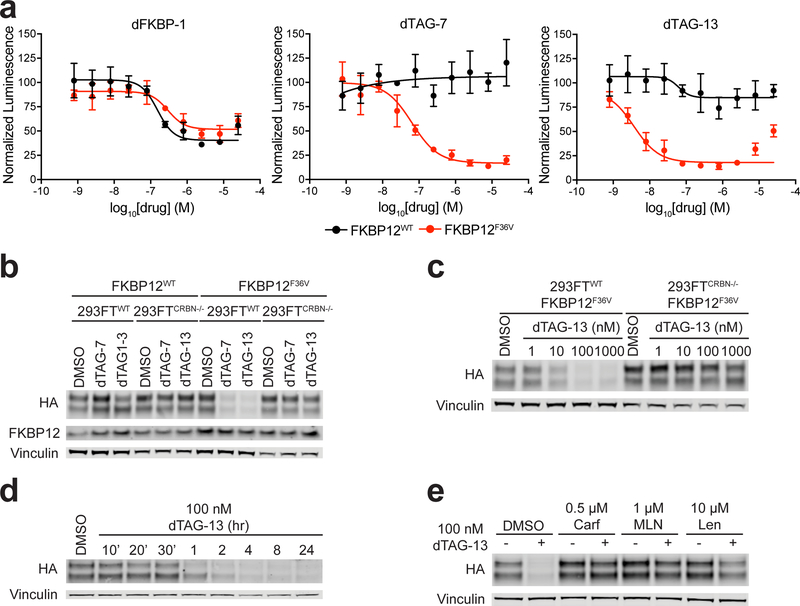
Figure 2 |. dTAG-7 and dTAG-13 selectively degrade FKBP12F36V in a CRBN-dependent manner in cells.
(a) DMSO normalized ratio of Nluc/Fluc signal of 293FTWT cells expressing FKBP12WT-Nluc or FKBP12F36V-Nluc treated with dFKBP-1, dTAG-7, or dTAG-13 for 24 hours. Data are presented as mean ± s.d. of n = 4 biologically independent samples and are representative of n = 3 independent experiments. (b) Immunoblot analysis of 293FTWT and 293FTCRBN−/− cells expressing FKBP12WT-Nluc or FKBP12F36V-Nluc treated with DMSO, 100 nM dTAG-7, or 100 nM dTAG-13 for four hours. (c) Immunoblot analysis of 293FTWT and 293FTCRBN−/− cells expressing FKBP12F36V-Nluc treated with DMSO or dTAG-13 at the indicated doses for four hours. (d) Immunoblot analysis of 293FTWT cells expressing FKBP12F36V-Nluc treated with DMSO or dTAG-13 for the indicated time-course. (e) Immunoblot analysis of 293FTWT cells expressing FKBP12F36V-Nluc pretreated with DMSO, Carfilzomib (Carf), MLN4924 (MLN), or Lenalidomide (Len) for two hours prior to DMSO or dTAG-13 treatment for four hours. Data in b-e are representative of n = 2 independent experiments. Uncropped immunoblots are displayed in Supplemental Fig. 11.
To evaluate the potency, selectivity, and CRBN-dependency of dTAG-7 and dTAG-13 in cells, we transduced isogenic 293FT CRBN deficient (293FTCRBN−/−) cells18 with FKBP12WT-Nluc or FKBP12F36V-Nluc and used immunodetection of the hemagglutinin (HA)-tag to monitor changes in Nluc. dTAG-7 and dTAG-13 treatment, at doses as low as 100 nM, potently reduced FKBP12F36V-Nluc levels in 293FTWT cells and had no activity in 293FTCRBN−/− cells, indicating the requirement of CRBN for the observed effects (Fig. 2b-c). dTAG-7 and dTAG-13 treatment did not alter the levels of endogenous FKBP12WT or exogenous FKBP12WT-Nluc (Fig. 2b). Temporal analysis revealed that dTAG-13 treatment led to rapid degradation of FKBP12F36V-Nluc in 293FTWT cells, as early as one hour after treatment (Fig. 2d). Notably, these data compare very favorably to a peptide-based proteolysis targeting chimeric molecules (PROTAC) approach that targeted EGFP-FKBP12F36V for degradation with HIF1α-peptides20, where apparent activity was observed at 25 μM ligand concentration.
To corroborate the cellular requirement of CRBN binding, E3 complex activation, and proteasome function for effective target degradation, 293FTWT cells stably expressing FKBP12F36V-Nluc were pre-treated separately with carfilzomib (proteasome inhibitor), MLN4924 (Nedd8 activating enzyme inhibitor), or lenalidomide (CRBN binder) prior to application of dTAG-13. Each co-incubation attenuated target degradation, consistent with a requirement for functional engagement of the ubiquitin-proteasome system, cullin-RING E3 ligases, and CRBN (Fig. 2e). Collectively, these data indicate that dTAG-7 and dTAG-13 treatment leads to rapid and selective CRBN-mediated degradation of FKBP12F36V in cells.
BRD4 degradation by CRISPR-mediated FKBP12F36V knock-in
Having established allele-specific degradation by our dTAG molecules, we next endeavored to knock-in FKBP12F36V at a genomic locus to provide control over a discrete, endogenously expressed target. BET bromodomains (BRD2, BRD3, and BRD4) are co-activator proteins that influence elongation of RNA polymerase in the context of gene expression21–25. The study of BET bromodomains has been powerfully facilitated by inhibitors and degraders that we and others have innovated26 (e.g. JQ127, MT128, dBET11, and dBET64), but these compounds do not discriminate between BET isoforms. We therefore sought to knock-in FKBP12F36V at the BRD4 genomic locus toward pharmacological control of a single BET bromodomain protein.
First, we tested the feasibility of degrading exogenously expressed FKBP12F36V tagged BRD4 fusion chimeras. Pairwise modification of the pLEX_305 lentiviral plasmid allowed expression of FKBP12F36V as an in-frame amino-terminal (N-terminal, FKBP12F36V-gene of interest) or carboxy-terminal (C-terminal, gene of interest-FKBP12F36V) chimera with any target gene using a Gateway recombination cloning strategy (Fig. 3a). Each plasmid includes two tandem HA-tags for immunodetection of gene products and a puromycin selectable marker. We first subcloned in the short isoform of BRD4 and stably expressed BRD4(short)-FKBP12F36V in MV4;11 cells. Treatment of MV4;11 cells expressing BRD4(short)-FKBP12F36V with dTAG-7 and dTAG-13 led to robust degradation of BRD4 (Supplementary Fig. 4a). dTAG-13 treatment led to rapid degradation of BRD4 within one hour, with no effect on endogenous FKBP12WT (Fig. 3a).
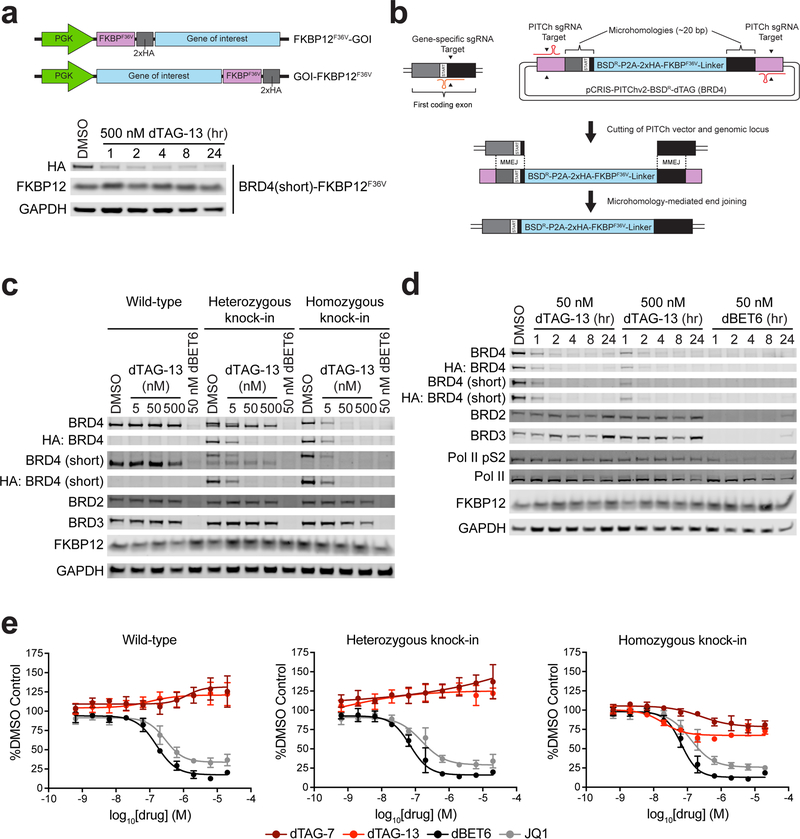
Figure 3 |. Selective pharmacological degradation of endogenously tagged BRD4.
(a) Schematic depiction of lentiviral expression strategy (Top). Immunoblot analysis of MV4;11 cells expressing BRD4 (short isoform)-FKBP12F36V treated with DMSO or dTAG-13 for the indicated time-course (Bottom). (b) Schematic depiction of knock-in strategy. Gene-specific and PITCh-specific sgRNAs expressed from pX330A-nBRD4/PITCh (not visualized, see methods) target the genomic locus and the pCRIS-PITChv2-BSDR-dTAG construct, respectively. This allows for both cutting of the genomic locus and liberation of the FKBP12F36V-containing cassette. MMEJ leads to repair of the double strand break through insertion of the FKBP12F36V-containing cassette, yielding an endogenously tagged product. (c) Immunoblot analysis of 293T wild-type, heterozygous BRD4 knock-in, or homozygous BRD4 knock-in cells treated with DMSO, dTAG-13, or dBET6 for four hours. (d) Immunoblot analysis of 293T homozygous BRD4 knock-in cells treated with DMSO, dTAG-13, or dBET6 for the indicated time-course. Data in a, c-d are representative of n = 2 independent experiments. Uncropped immunoblots are displayed in Supplemental Fig. 11–12. (e) DMSO normalized anti-proliferation of 293T wild-type, heterozygous BRD4 knock-in, or homozygous BRD4 knock-in cells treated with the indicated compounds for 72 hours. Data are presented as mean ± s.d. of n = 4 biologically independent samples and are representative of n = 3 independent experiments.
Having established the feasibility of degrading a BRD4-FKBP12F36V chimera, we next developed a means to knock-in FKBP12F36V into the N- or C-terminus of BRD4 using a modified version of the Precise Integration into Target Chromosome (PITCh) system29. This strategy employs microhomology-mediated end-joining (MMEJ) to achieve selective knock-in, and does not require large homology arms. We cloned an N-terminal or C-terminal targeting BRD4 sgRNA into the pX330A PITCh vector, which expresses a PITCh-sgRNA and Cas9. We also modified the pCRIS-PITChv2 donor vector by replacing the original EGFP-2A-Puro cassette with a cassette that includes a blasticidin selectable marker (BSD), P2A sequence, two tandem HA-tags, and FKBP12F36V, which is flanked by 5’ and 3’ BRD4 microhomology sequences (Fig. 3b). To evaluate the efficiency of this approach, we co-transfected the N- or C-terminal targeting constructs into 293T cells, which are notably hypotriploid. While we were unable to recover homozygous C-terminal FKBP12F36V knock-in clones, we isolated clonal cell lines with heterozygous and homozygous N-terminal FKBP12F36V knock-in at the BRD4 locus, prompting further characterization of these cells (Supplementary Fig. 4b–d).
We first examined the consequences of dTAG treatment on BRD4 function and stability, compared to a pan-BET bromodomain inhibitor (JQ1) or degrader (dBET6). dTAG-13 treatment led to rapid and potent degradation of the BRD4 fusion chimera in the heterozygous and homozygous knock-in clones, with no effect on endogenous FKBP12WT (Fig. 3c-d and Supplementary Fig. 5a). Importantly, we did not detect changes in BRD2 or BRD3 protein levels in FKBP12F36V integrant clones and did not detect changes in BRD2, BRD3, or BRD4 levels in wild-type cells, establishing a powerful chemical-genetic system to study BRD4 function in isolation from BRD2 and BRD3. Upon dBET6 treatment, we confirmed the degradability of BRD2, BRD3, and BRD4 in wild-type cells and FKBP12F36V integrant clones (Fig. 3c-d and Supplementary Fig. 5a). BRD4 degradation was also observed with dTAG-7, while BRD4 and FKBP12WT degradation was observed with dTAG-48, further confirming the selectivity of dTAG-7 and the affinity of dTAG-48 towards both FKBP12WT and FKBP12F36V (Supplementary Fig. 5a).
We recently demonstrated that pan-BET bromodomain degradation results in a collapse in global transcriptional elongation4. Leveraging the selective BRD4 degradation afforded by the dTAG system, we tested whether BRD4 alone was required for productive transcriptional elongation by monitoring RNA polymerase II (Pol II) phosphorylation on the C-terminal repeat domain (CTD) at serine 2 (pS2). While pan-BET bromodomain degradation upon dBET6 treatment led to a rapid decrease in Pol II pS2, selective BRD4 degradation upon dTAG-13 treatment had no observable effect on Pol II pS2 levels in 293T cells (Figure 3d). This suggests that BRD4 alone is not required for Pol II CTD-phosphorylation in 293T cells, although we anticipate that the contributions of individual BET bromodomains will be context-dependent. We next explored the differential effects of BRD4 loss versus pan-BET bromodomain inhibition or degradation on cellular proliferation. In the homozygous BRD4 knock-in clone, treatment with dTAG-7 and dTAG-13 led to a modest anti-proliferative effect, while JQ1 and dBET6 treatment had a significantly greater impact (lower Emax) (Fig. 3e). Treatment of 293TWT cells and the heterozygous BRD4 knock-in clone were similarly sensitive to JQ1 and dBET6 treatment, and insensitive to dTAG-7 and dTAG-13 treatment, further confirming selectivity of the dTAG molecules. These data suggest that BRD2 and BRD3 may partially compensate for loss of BRD4 in 293T cells, and that pan-BET bromodomain activity of inhibitors or degraders is important for achieving maximal anti-proliferative effects in this cellular context.
Generalizability of dTAG to study divergent proteins
With the successful degradation of the BRD4 fusion chimera, we used our facile N- and C-terminal lentiviral expression system to assess pharmacological CRBN-mediated degradation of diverse target proteins. In parallel, we cloned a panel of wild-type and mutant genes, generating N-terminal fusions including FKBP12F36V-KRASG12V and FKBP12F36V-EZH2, and C-terminal fusions including HDAC1-FKBP12F36V, MYC-FKBP12F36V, and PLK1-FKBP12F36V. Upon stable expression of these fusion chimeras in MV4;11 cells, we evaluated degradation upon dTAG-7 or dTAG-13 treatment. Each fusion chimera was potently degraded with as little as 50 nM of dTAG-7 or dTAG-13, supporting the generalizable nature of this approach (Supplementary Fig. 5b). We also compared the kinetics of degradation of each fusion chimera upon dTAG-13 treatment (Fig. 4a). While we observed pronounced degradation of HDAC1-FKBP12F36V, MYC-FKBP12F36V, FKBP12F36V-EZH2, and PLK1-FKBP12F36V within one hour treatment, near complete degradation of FKBP12F36V-KRASG12V was observed after four to eight hours of treatment, indicating that different fusion chimeras may have slightly varying kinetics of degradation. These data further validate rapid and potent degradation of nuclear and cytoplasmic tagged proteins using the dTAG system.
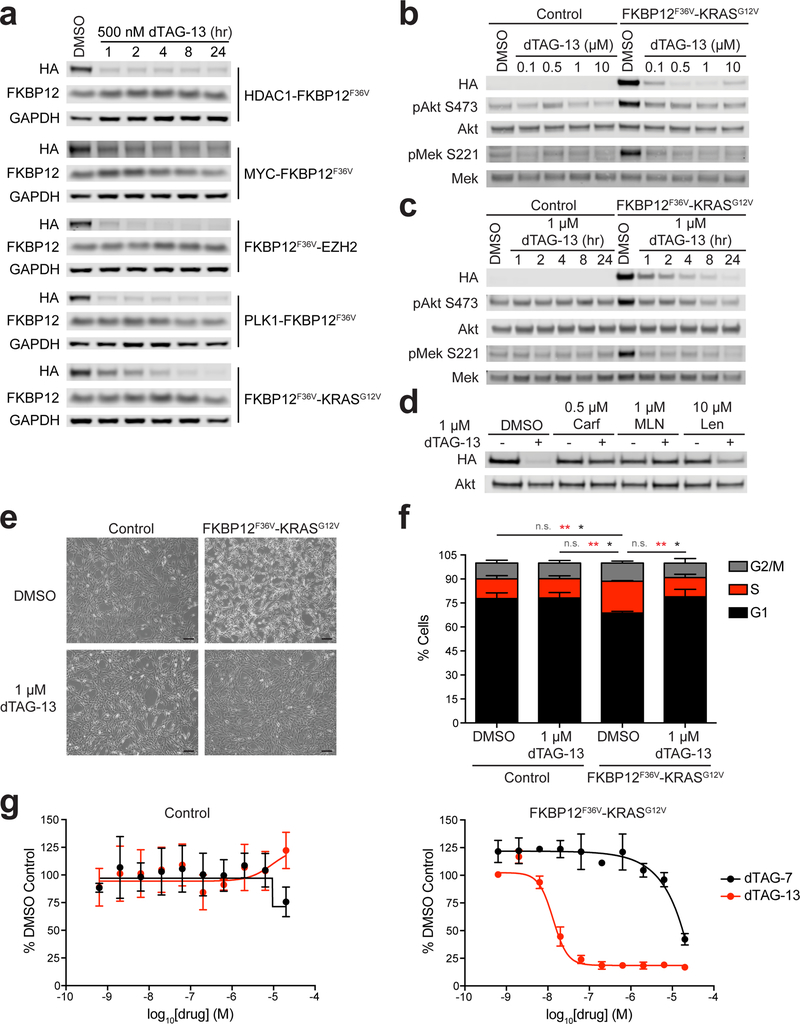
Figure 4 |. Rapid degradation of nuclear and cytoplasmic FKBP12F36V fusion chimeras.
(a) Immunoblot analysis of MV4;11 cells expressing the indicated FKBP12F36V fusions treated with DMSO or dTAG-13 for the indicated time-course. (b) Immunoblot analysis of mock transduced (Control) NIH/3T3 cells or NIH/3T3 cells expressing FKBP12F36V-KRASG12V treated with DMSO or dTAG-13 for 24 hours. (c) Immunoblot analysis of mock transduced (Control) NIH/3T3 cells or NIH/3T3 cells expressing FKBP12F36V-KRASG12V treated with DMSO or dTAG-13 for the indicated time-course. (d) Immunoblot analysis of NIH/3T3 cells expressing FKBP12F36V-KRASG12V pretreated with DMSO, Carfilzomib (Carf), MLN4924 (MLN), or Lenalidomide (Len) for two hours prior to DMSO or dTAG-13 treatment for four hours. (e) Phase contrast images of mock transduced (Control) NIH/3T3 cells or NIH/3T3 cells expressing FKBP12F36V-KRASG12V treated with DMSO or dTAG-13 for 24 hours. Scale bar represents 100 μm. Data in a-e are representative of n = 2 independent experiments. Uncropped immunoblots are displayed in Supplemental Fig. 12–13. (f) Propidium iodide analysis of the percentage of mock transduced (Control) NIH/3T3 cells or NIH/3T3 cells expressing FKBP12F36V-KRASG12V in different phases of the cell cycle treated with DMSO or dTAG-13 for 48 hours. Data are presented as mean ± s.d. of n = 3 independent experiments. P values derived from a two-tailed Student’s t-test are color coded for each phase of the cell cycle. * P < 0.05, ** P < 0.01, not significant (n.s.) (G1, P values for indicated comparisons: P = 0.0140 for Control DMSO versus FKBP12F36V-KRASG12V DMSO, P = 0.0105 for Control 1 μM dTAG-13 versus FKBP12F36V-KRASG12V DMSO, P = 0.0205 for FKBP12F36V-KRASG12V DMSO versus FKBP12F36V-KRASG12V 1 μM dTAG-13; S, P values for indicated comparisons: P = 0.0030 for Control DMSO versus FKBP12F36V-KRASG12V DMSO, P = 0.0020 for Control 1 μM dTAG-13 versus FKBP12F36V-KRASG12V DMSO, P = 0.0028 for FKBP12F36V-KRASG12V DMSO versus FKBP12F36V-KRASG12V 1 μM dTAG-13; G2/M, P values for indicated comparisons were n.s.). (g) DMSO normalized anti-proliferation of mock transduced (Control) NIH/3T3 cells or NIH/3T3 cells expressing FKBP12F36V-KRASG12V treated with the indicated compounds for 72 hours. Data are presented as mean ± s.d. of n = 4 biologically independent samples and are representative of n = 3 independent experiments.
Immediate effects of KRASG12V loss on cellular signaling
To establish the utility of the dTAG system in the setting of fast biology, we selected FKBP12F36V-KRASG12V for further study. RAS proteins interact with downstream signaling effectors to promote cellular growth, survival, migration, and differentiation30. In cancer, RAS family members (KRAS, NRAS, and HRAS) are together among the most prevalent oncogenes, with KRAS lesions predominating in aggressive epithelial malignancies31. Although KRAS has been largely impervious to coordinated efforts in drug discovery, chemical degradation comprises a potentially new path to RAS-targeted therapeutics, as heterobifunctional degraders could disassociate the need for active site directed ligands.
To model the mechanism and consequence of allosteric site mediated mutant KRAS degradation, in advance of possessing heretofore elusive chemical matter, we stably expressed FKBP12F36V-KRASG12V in murine NIH/3T3 cells. Stable chimeric oncoprotein expression led to characteristic changes in signaling, morphology and growth, akin to those observed in focused studies of mutant RAS32–34. FKBP12F36V-KRASG12V expression increased phosphorylation of MEK (pMEK) and AKT (pAKT), indicating that the chimeric oncoprotein is functional (Fig. 4b-c). dTAG-13 treatment led to rapid and pronounced degradation of FKBP12F36V-KRASG12V, while dTAG-7 treatment resulted in modest degradation at high doses, and therefore, we focused on dTAG-13 for further characterization (Fig. 4b-c and Supplementary Fig. 6a). Strikingly, FKBP12F36V-KRASG12V degradation decreased pMEK and pAKT to baseline levels within one hour of dTAG-13 treatment (Fig. 4c). Washout of dTAG-13 led to rapid recovery of FKBP12F36V-KRASG12V and particularly robust recovery of pMEK, pERK, and pAKT within one hour of washout, indicating the reversibility of the observed changes (Supplemental Fig. 6b). Pre-treatment with Carfilzomib, MLN4924 or lenalidomide prior to dTAG-13 treatment prevented FKBP12F36V-KRASG12V degradation, confirming the requirement of the proteasome and activated cullin-RING E3 ligases for the observed effects (Fig. 4d). Immediate biological changes were also observed upon oncoprotein degradation. Upon dTAG-13 treatment, FKBP12F36V-KRASG12V expressing cells reverted to control NIH/3T3 cell morphology in twenty-four hours (Fig. 4e and Supplemental Fig. 6c). These cellular changes corresponded with a significant reduction in the number of FKBP12F36V-KRASG12V expressing cells in S-phase, which returned to control NIH/3T3 levels in forty-eight hours, and a significant anti-proliferative effect in seventy-two hours (Fig. 4f-g). Collectively, these data demonstrate that FKBP12F36V-KRASG12V is a functional oncoprotein, and that selective CRBN-mediated degradation of FKBP12F36V-KRASG12V rapidly reverses the biology of KRASG12V-transformed cells to the basal cellular state.
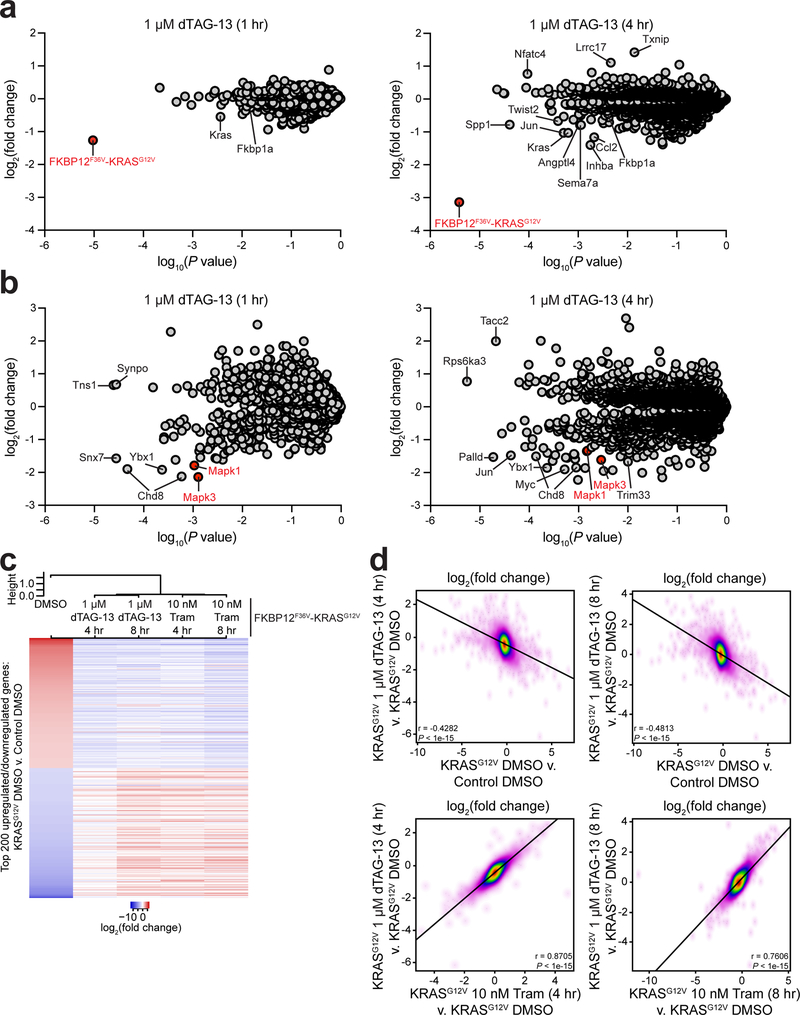
We next used multiplexed quantitative mass spectrometry-based proteomics35, 36 and phospho-proteomics37 to evaluate the specificity and consequences of FKBP12F36V-KRASG12V degradation upon dTAG-13 treatment. In the proteomics analysis, 8164 proteins qualified for quantification (Supplementary Dataset 1). Despite the high conservation between mouse Kras and human KRAS, as well as mouse Fkpb1a and human FKBP1a, we were able to monitor two peptides unique to FKBP12F36V-KRASG12V (KKVGTMTEYKL and KLEGGYPYDVPDYAGYPYDVPDYAGIDRS). Using these signature peptides, we observed highly significant and selective depletion of FKBP12F36V-KRASG12V upon dTAG-13 treatment (58.3% at one hour and 88.6% at four hours; Fig. 5a). Endogenous mouse Kras and mouse Fkbp1a peptides that were quantified are conserved between mouse and human, and therefore likely represent both human and murine peptides. Although we observe partial depletion of endogenous mouse Kras and Fkbp1a peptides, these differences are likely due to changes in the human fusion chimera upon degradation and not endogenous mouse protein levels. In the phospho-proteomics analysis, 8316 phospho-serine/threonine and 33 phospho-tyrosine sites qualified for quantification (Supplementary Dataset 2–3). Focusing on unique phospho-serine/threonine sites, we observed highly significant reductions in the doubly phosphorylated activation loop of Mapk3 (Erk1) and Mapk1 (Erk2) at one and four hours of dTAG-13 treatment, further confirming the functionality of the KRASG12V fusion chimera (Fig. 5b and Supplementary Fig. 7a). No effect on total Mapk1 or Mapk3 levels was observed (Supplementary Fig. 7b). We also observed highly significant reductions in previously described Jun38, Myc39, and Palld40 phosphosites, as well as chromatin regulators including Chd8 and Trim33. Together, these data support the high degree of potency and selectivity of KRASG12V degradation using the dTAG system, and highlight the immediate collapse of cellular signaling upon KRASG12V degradation.
Figure 5 |. KRASG12V degradation rapidly reverses the deregulated proteomic and transcriptional signaling program of transformed cells.
(a) Protein abundance after treatment of NIH/3T3 cells expressing FKBP12F36V-KRASG12V with dTAG-13 for one and four hours, compared to DMSO treatment. Volcano plot depicts fold change abundance of 8164 quantified proteins versus P value derived from a two-tailed Student’s t-test. Normalized percent relative abundance of quantified proteins are provided in Supplementary Dataset 1. (b) Phospho-serine/threonine abundance after treatment of NIH/3T3 cells expressing FKBP12F36V-KRASG12V with dTAG-13 for one and four hours, compared to DMSO treatment. Volcano plot depicts fold change abundance of 8316 quantified phosphosites versus P value derived from a two-tailed Student’s t-test. Normalized percent relative abundance of quantified phosphosites are provided in Supplementary Dataset 2. (c) Heatmap of fold change in expression of top 200 upregulated and top 200 downregulated genes upon comparison of NIH/3T3 cells expressing FKBP12F36V-KRASG12V treated with DMSO and mock transduced (Control) NIH/3T3 cells treated with DMSO. Fold change levels of NIH/3T3 cells expressing FKBP12F36V-KRASG12V cells treated with dTAG-13 or Trametinib (Tram) compared to NIH/3T3 cells expressing FKBP12F36V-KRASG12V treated with DMSO are depicted. ERCC spike-in normalized FPKM of genes included in the heatmap are depicted in Supplementary Dataset 4. (d) Scatterplots of fold change in gene expression of NIH/3T3 cells in the indicated comparisons. Pearson correlation coefficient and associated P values are indicated. Data in a-d are from n = 3 biologically independent samples.
KRASG12V loss rapidly alters transcriptional signaling
Temporal quantitative proteomics and phospho-proteomics revealed that FKBP12F36V-KRASG12V degradation rapidly diminished pERK1/2 within one hour and ERK pathway dependent proteins including Jun, Sema7a, and Angptl4 within four hours, suggesting KRASG12V degradation triggered acute transcriptional changes (Fig. 5a–b). Prior to evaluating the consequences of FKBP12F36V-KRASG12V degradation on the transcriptome, we treated parental NIH/3T3 cells with dTAG-13 and performed RNA-sequencing to evaluate off-target transcriptional changes. No significantly differentially expressed genes were identified upon dTAG-13 treatment, further validating the high degree of selectivity of dTAG-13 (Supplementary Fig. 7c). As expected, dBET6 treatment led to a collapse in the NIH/3T3 gene expression program. Having established the limited off-target activity of dTAG-13, we performed RNA-sequencing to quantify the acute transcriptional changes upon FKBP12F36V-KRASG12V degradation with dTAG-13 and MEK inhibition with trametinib (GSK112021), an allosteric MEK1/2 inhibitor approved for use in BRAF-mutant metastatic melanoma41, 42. A 10 nM dose of trametinib was used for these experiments, as the reduction in pERK levels and anti-proliferative effect at this dose were similar to levels achieved upon FKBP12F36V-KRASG12V degradation with dTAG-13 (Supplementary Fig. 8a–b).
Compared to control NIH/3T3 cells, FKBP12F36V-KRASG12V expression led to significant upregulation of transcripts including Sema7a, Itgb3, Fgf2, Dusp6, and Spry4 and downregulation of transcripts including Egfr and Cxcl1 (Supplementary Dataset 4). We previously identified these genes as dependent on oncogenic KRASG12V or HRASG12V,43 providing further support for the functionality of the FKBP12F36V-KRASG12V fusion chimera. Upon FKBP12F36V-KRASG12V degradation or MEK inhibition, we observed rapid reversal of the aberrant KRASG12V expression program to the baseline transcriptional state of NIH/3T3 cells (Fig. 5c, Supplementary Fig. 8c and 9a, and Supplementary Dataset 4). A striking correlation was observed between genes differentially expressed upon FKBP12F36V-KRASG12V expression or dTAG-13 treatment, as well as between genes differentially expressed upon dTAG-13 or trametinib treatment, suggesting that acute KRASG12V degradation predominantly alters ERK pathway genes (Fig. 5d). Furthermore, gene set enrichment analysis (GSEA) revealed significant on-pathway enrichment of ‘KRAS Up’ signatures in vehicle-treated FKBP12F36V-KRASG12V cells, and ‘KRAS Down’ signatures in dTAG-13 or trametinib treated cells (Supplementary Fig. 9b). Ingenuity Pathway Analysis identified similar deregulated signatures such as cellular growth and proliferation and cellular death and survival in each treatment comparison (Supplementary Fig. 9c). In sum, we find that KRASG12V degradation rapidly alters ERK dependent transcriptional signaling. In advance of discovery of selective mutant KRAS degraders, we present a chemical biology platform to evaluate the dynamic consequences of mutant KRAS degradation.
Rapid and potent degradation in vivo
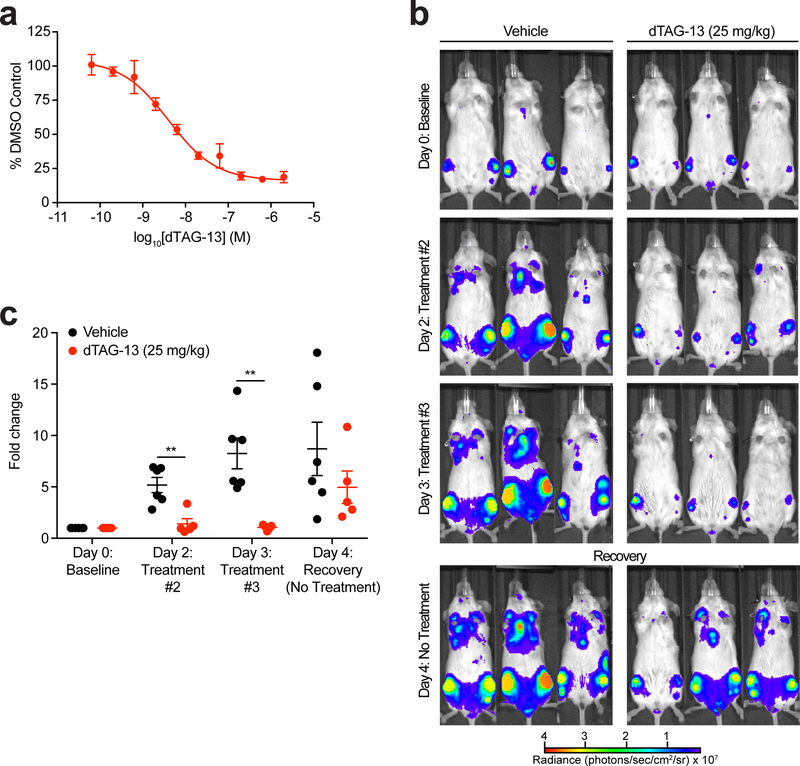
Having established extensible strategies to study target-specific degradation in cells, we endeavored to establish a system to evaluate degradation in living organisms. We aimed to use non-invasive bioluminescent measurements to report on the potency, kinetics, and recovery upon treatment with dTAG molecules. We stably expressed luciferase as a FKBP12F36V chimera (luc-FKBP12F36V) in a human leukemia cell line (MV4;11), to allow for non-invasive monitoring of ligand-induced target protein degradation in a highly aggressive disseminated leukemia model. Treatment of MV4;11 cells expressing luc-FKBP12F36V with dTAG-13 in vitro led to effective loss of bioluminescent signal (Fig. 6a). Based on this potent degradation, and the favorable pharmacokinetic (PK) parameters of dTAG-13, we proceeded to evaluate dTAG-13 in vivo (Supplementary Table 4). Following bone marrow engraftment of MV4;11 cells expressing luc-FKBP12F36V in mice, we monitored bioluminescent signal after vehicle or dTAG-13 administration (Supplemental Fig. 10a). A significant, rapid, and durable effect on bioluminescent signal was observed four hours after dTAG-13 administration, indicating effective degradation of luc-FKBP12F36V (Fig. 6b-c). Twenty-eight hours following the final treatment, we observed recovery of cellular bioluminescence to levels comparable between vehicle and dTAG-13 treatment groups. Together, these data support the use of the dTAG system to rapidly and reversibly evaluate the function of target proteins in living organisms.
Figure 6 |. Evaluation of rapid and reversible degradation in vivo.
(a) DMSO normalized ratio of Fluc signal of MV4;11 cells expressing luciferase-FKBP12F36V treated with dTAG-13 for 16 hours. Data are presented as mean ± s.d. of n = 4 biologically independent samples and are representative of n = 3 independent experiments. (b) Images of average radiance of vehicle (n = 6 biologically independent mice) or dTAG-13 (n = 5 biologically independent mice) treated mice. Three representative mice are shown and the same mouse is shown on the same scale at each time-point. (c) Fold change in average radiance of each mouse normalized to baseline signal at Day 0 of vehicle (n = 6 biologically independent mice) or dTAG-13 (n = 5 biologically independent mice) treated mice. P value derived from a two-tailed Welch’s t-test. ** P < 0.01 (P = 0.0025 for Day 2 and P = 0.0048 for Day 3).
DISCUSSION
Chemical genetic model systems capable of precise temporal control of target proteins can provide unique insights into cellular signaling and transcription. Here, we describe a series of dTAG molecules of varying linker composition, linker length, selectivity, and potency that recruit the CRBN E3 ligase complex to targets fused with FKBP12F36V. We present two generalizable approaches to engineer targeted protein degradation: a CRISPR/Cas9 system to achieve locus-specific FKBP12F36V knock-in, and a lentiviral system to exogenously express FKBP12F36V fused to a target of interest. We show that ortho-substituted degraders, dTAG-7 and dTAG-13, require CRBN for their effect, and are highly selective for FKBP12F36V, typically exhibiting activity at sub-micromolar concentrations in cells. We also develop a model system to non-invasively evaluate in vivo degradation, and demonstrate that dTAG-13 can induce potent degradation in living organisms. dTAG-13, which effectively induced rapid and potent target-specific degradation in all cell lines tested and in mice, will serve as a lead degrader for future studies.
Prior studies have achieved genetic knock-in of a destabilizing FKBP12 domain14 and an AID tag44, allowing Shield-1 mediated stabilization or auxin-inducible degradation of endogenously tagged protein targets, respectively. In the AID approach, high doses of auxin and exogenous expression or knock-in of the SCFTIR1 E3 ubiquitin ligase substrate recognition component, TIR1, are required to achieve degradation. By contrast, our dTAG system relies on hijacking the cell’s intrinsic degradation machinery, obviating the need to express additional ubiquitin proteasome system componentry. We recently achieved FKBP12F36V knock-in at the YY1 locus using homology-directed repair, demonstrating that targeted degradation of YY1 disrupts enhancer-promoter loop formation and gene expression in murine embryonic stem cells45. Here, we employed a second knock-in strategy, the PITCh approach29, which uses MMEJ and eliminates the need for long homology arms. The cassettes used for these two strategies can be modified using Gibson assembly for FKBP12F36V knock-in at any genomic locus. To demonstrate the feasibility of the modified PITCh approach, we generated clonal cell lines with homozygous N-terminal FKBP12F36V knock-in at the BRD4 locus. While we were not successful in isolating clones with homozygous C-terminal FKBP12F36V knock-in, it is possible that endogenous tagging altered the C-terminal P-TEFb interaction domain of BRD4, which is required for displacement of HEXIM1 and P-TEFb activation46, 47. When employing the dTAG system, we advise using our lentiviral plasmids to first evaluate exogenous expression of N- or C-terminus tagged targets to ensure protein functionality, prior to CRISPR-mediated knock-in. Our N-terminal FKBP12F36V knock-in BRD4 clones allowed comparisons between selective BRD4 degradation and pan-BET bromodomain inhibition or degradation. Selective BRD4 degradation had no effect on Pol II pS2 and led to a moderate anti-proliferative effect, while inhibition and degradation of BRD2, 3 and 4 was significantly more efficacious in 293T cells. In this cellular context, these data support the mechanism-based clinical development of pan-BET bromodomain inhibitors or degraders and indicates that isoform-selective BET bromodomain inhibitors and degraders may display unique pharmacology compared to pan-BET bromodomain agents. Further efforts leveraging the dTAG system to evaluate contributions of BRD2, 3, and 4 in disease-relevant and BRD4-dependent contexts will aid in the assessment of selective BRD4 degradation.
In addition to genetic knock-in, prior studies have induced target-specific degradation by fusing target genes to FKBP12F36V,20 FKBP12F36V appended with a 19-residue cryptic degron7, a HaloTag8, 9, a SMASh tag10, or an AID tag11. Here, we developed a lentiviral expression system, amenable to Gateway cloning, as a facile means to express a target gene fused to FKBP12F36V. In contrast to the mechanisms of degradation implemented in the described approaches, we co-opted the CRBN E3 ligase machinery to rapidly degrade a panel of fusion chimeras (BRD4, HDAC1, EZH2, MYC, PLK1, and KRASG12V). We selected KRASG12V to evaluate the functionality and efficacy of our chemical strategy, due to the lack of available chemical matter to directly inhibit mutant KRAS. While exciting progress has been made with covalent KRASG12C inhibitors, inhibitors targeting other common KRAS mutants are far from clinical translation30, 31. Prior studies examined degradation of HaloTag fusions including HRASG12V,8 MEK19, and ERK19, with limited assessment of the selectivity of these approaches, functionality of HaloTag fusions, or the downstream consequences of HRASG12V, MEK1, or ERK1 degradation. We leverage the dTAG system to perform unbiased global temporal measurements to confirm the specificity of our dTAG molecules, demonstrate the functionality of the KRASG12V fusion chimera, and identify the rapid proteomic and transcriptional signaling changes that occur upon acute KRASG12V degradation. Our data provides further validation that pharmacological degradation can relieve the oncogenic effects of mutant KRAS.
The extension of tag-based strategies to control target proteins in mouse models has been limited to studies using Shield-1 mediated stabilization of targets fused to a destabilizing FKBP12 domain48 and degradation of HaloTag-HRASG12V to reduce tumor burden in mice8. To our knowledge, other degron strategies, such as HaloPROTACs, SMASh, or AID have not been implemented in mouse studies. We employed a disseminated model of leukemia to non-invasively validate the use of the dTAG system for targeted degradation studies in mice. The absolute selectivity, defined mechanism of action, and potent and rapid performance of dTAG-13, supports the broad utility of the dTAG system for biological research in mice.
In addition to our studies described herein, we recently employed the dTAG system to evaluate the consequences of acute degradation of ENL, a transcriptional regulator and core component of the super elongation complex, and MELK, a kinase previously implicated in promoting proliferation of breast cancer cells. We demonstrated that targeted degradation of ENL leads to genome-wide transcriptional defects, providing mechanistic insight into the role of ENL in leukemia pathogenesis49. By contrast, we established that targeted degradation of MELK is not required for the proliferation of basal-like breast cancer cells, arguing against further optimization of MELK inhibitors or degraders50. Our studies establish the extensibility of a chemical biology system to study protein targets that we envision will broadly enable biological exploration and target validation in cells and in mice, notably in advance of direct-acting inhibitor or degrader drug molecules.
ONLINE METHODS
Molecule synthesis
Synthetic procedures and characterization of dFKBP-1, dTAG-7, dTAG-13, dTAG-48, dTAG-51, bio-SLF, and bio-Thal are detailed in the Supplementary Note.
Protein expression and purification
A construct containing residues 1–108 of FKBP12WT or FKBP12F36V in a gateway compatible pGEX-6P-1 vector (Amersham) was overexpressed in E. coli BL21 (DE3) in LB medium in the presence of 50 mg/ml of carbenicillin. Cells were grown at 37 ºC to an OD of 0.6, induced with 100 µM isopropyl-1-thio-D-galactopyranoside (IPTG), incubated for 4 hours at 37 ºC, collected by centrifugation, and stored at −80°C. Cell pellets were resuspended in buffer A (50mM HEPES pH 8.0, 250 mM NaCl, 0.1% Triton X-100, 1 mM TCEP), lysozyme was added to a concentration of 0.1 mg/mL, the pellet was incubated at 4 ºC for 15 minutes and subsequently sonicated. The resulting lysate was centrifuged at 15,000 xg for 20 minutes at 4 ºC. Supernatant was added to equilibrated Glutathione Sepharose 4B beads (GE Healthcare), incubated overnight at 4 ºC, and purified in batch mode with buffer A + 16 mM glutathione. Proteins were further purified on a HiLoad 16/600 Superdex 200 (GE Healthcare). Clean fractions were pooled and then frozen and stored at −80 ºC at a concentration of 0.44 mg/ml.
FKBP12WT and FKBP12F36V ligand-displacement AlphaScreen assay
In 384-well AlphaPlates (PerkinElmer), 90 nM GST-FKBP12 (WT or F36V) and 62.5 nM bio-SLF were diluted in 20 µL assay buffer (PBS containing 0.1% BSA and 0.01%Triton X-100) containing competitor compound or DMSO. Following a 30-minute incubation, 20 µL detection solution containing Streptavidin Donor Beads and Glutathione Acceptor Beads diluted to 20 ng/µL in assay buffer was added to each well. After a one hour incubation at room temperature, luminescence was measured on an Envision 2104 plate reader (PerkinElmer). Percent activity values were calculated by setting the average background (no protein wells) to 0% and the average DMSO wells to 100% activity. Data were analyzed and plotted using GraphPad PRISM v6 and IC50 values were determined using the ‘log(inhibitor) vs. response -- Variable slope (four parameters)’ analysis module.
CRBN-DDB1 ligand-displacement AlphaScreen assay
In 384-well AlphaPlates (PerkinElmer), 50 nM CRBN-DDB1 (6XHIS-tagged) and 125 nM bio-Thal were diluted in 20 µL assay buffer (50 mM HEPES pH 7.4, 200 mM NaCl, 1 mM TCEP, and 0.1% BSA) containing competitor compound or DMSO. Following a 30-minute incubation, 20 µL detection solution containing Streptavidin Donor Beads and Nickel Chelate AlphaLISA Acceptor Beads (PerkinElmer) diluted to 20 ng/µL in assay buffer was added to each well. After a one hour incubation at room temperature, luminescence was measured on the Envision 2104 plate reader (PerkinElmer). Percent activity values were calculated by setting the average background (no protein wells) to 0% and the average DMSO wells to 100% activity. Data were analyzed and plotted using GraphPad PRISM v6 and IC50 values were determined using the ‘log(inhibitor) vs. response -- Variable slope (four parameters)’ analysis module.
CRBN-DDB1/FKBP12F36V dimerization assay
Detection of chemical-induced dimerization was measured as previously described1. GST-FKBP12F36V and CRBN-DDB1 (6XHIS-tagged) were diluted to 125 nM and 250 nM, respectively, in assay buffer (50 mM HEPES pH 7.4, 200 mM NaCl, 1 mM TCEP, and 0.1% BSA), and 20 µL of protein mixture was added to each well of a 384-well AlphaPlate (PerkinElmer). Compounds were then added at 100 nL per well from DMSO stock plates using a Janus Workstation (PerkinElmer). Following a one hour incubation at room temperature, 20 µL Nickel Chelate AlphaLISA Acceptor and Glutathione AlphaLISA Donor beads (PerkinElmer) diluted to 20 ng/µl in assay buffer was added to each well. After a one hour incubation at room temperature, luminescence was measured on the Envision 2104 plate reader (PerkinElmer). Data were analyzed and plotted using GraphPad PRISM v6 and dimerization amplitude, mean, and standard deviation values were determined using the ‘Gaussian’ analysis module.
Cell lines
MV4;11 (ATCC, RPMI1640 supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine), NIH/3T3 (ATCC, DMEM supplemented with 10% bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin), 293FT and 293T (Thermo Fisher Scientific, DMEM supplemented with supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine) cells were cultured at 37 ºC and 5% CO2. Cells matched their expected cell type morphology and were not independently verified. Cells routinely tested negative for mycoplasma using the MycoAlert kit (Lonza). None of the cell lines used are among those that are commonly misidentified as listed by ICLAC.
Lentivirus production and transduction
Lentiviral production was performed using 293FT cells, which were co-transfected with pMD2.G (Addgene, #12259), psPAX2 (Addgene, #12260), and dTAG lentiviral plasmids using Lipofectamine 2000 (Thermo Fisher Scientific), according to manufacturer’s instructions. Viral particles were collected 48 and 72 hours after transfection, filtered through a 0.45 µM membrane, and concentrated using 1:3 Lenti-X Concentrator (Clontech), according to manufacturer’s instructions. Concentrated viral supernatants were applied to NIH/3T3 and 293FT cells in the presence of 4 µg/mL polybrene, and transduced cells were selected with 2 µg/mL puromycin. MV4;11 were transduced by spinoculation with concentrated virus for one hour at room temperature in the presence of 8 µg/mL polybrene, and transduced cells were selected with 2 µg/mL puromycin.
FKBP12WT and FKBP12F36V dual luciferase assay
293FT cells were transduced with a lentiviral construct containing FKBP12WT or FKBP12F36V fused to nanoluciferase (Nluc). Nluc/Fluc plasmids were a kind gift from W. Kaelin (Dana-Farber Cancer Institute). Cells were plated at 2000 cells per well in 40 µL of appropriate media in 384-well white culture plates (Thermo Fisher Scientific). Cells were allowed to adhere overnight before adding 100 nL of compound in DMSO from compound stock plates using a Janus Workstation pin tool (PerkinElmer). After addition of compound, plates were incubated for 24 hours at 37 ºC, prior to luminescence assessment. Plates were brought to room temperature prior to reagent addition. 20 µL Fluc buffer (200 mM Tris, 15 mM MgSO4, 100 µM EDTA, 1 mM DTT, 1 mM ATP, 200 µM Coenzyme A, 400 µM D-Luciferin, 0.1% Triton X-100) was added to each well for 15 minutes at room temperature, and luminescence was measured on the Envision 2104 plate reader (PerkinElmer). Subsequently, 20 µL of Nluc buffer (25 mM Na4PPi, 10 mM NaOAc, 15 mM EDTA, 500 mM NaSO4, 500 mM NaCl, 24 µM coelenterazine, 50 µM 4-(6-methyl-1,3-benzothiazol-2-yl)aniline) was added to each well for 15 minutes at room temperature, and luminescence was measured on the Envision 2104 plate reader (PerkinElmer). Nluc values were normalized by dividing by control Fluc values. Nluc/Fluc ratios were then normalized by setting DMSO only wells to 100%. Data were analyzed and plotted using GraphPad PRISM v6 and IC50 values were determined using the ‘log(inhibitor) vs. response -- Variable slope (four parameters)’ analysis module.
IKZF1 dual luciferase assay
A clonal 293T line stably transfected with pCMV-IRES-RenillaLUC-IRES-IKZF1-FireflyLUC reporter plasmid18 (a kind gift from W. Kaelin, Dana-Farber Cancer Institute) was used to evaluate compound activity on IKZF1-Fluc fusion protein levels. Cells were plated at 2000 cells per well in 50 µL of appropriate media in 384-well white culture plates (Thermo Fisher Scientific). Cells were allowed to adhere overnight before adding 100 nL of compound in DMSO from compound stock plates using a Janus Workstation pin tool (PerkinElmer). After addition of compound, plates were incubated for 24 hours at 37 ºC, prior to luminescence assessment. Fluc and Rluc detection was performed as described above for the FKBP12WT and FKBP12F36V dual luciferase assay (with Rluc activity being measured using Nluc buffer). Fluc values were normalized by dividing by control Rluc values. Fluc/Rluc ratios were then normalized by setting DMSO only wells to 100%. Data were analyzed and plotted using GraphPad PRISM v6 and IC50 values were determined using the ‘log(inhibitor) vs. response -- Variable slope (four parameters)’ analysis module.
FKBP12F36V luciferase assay
MV4;11 cells expressing luciferase-FKBP12F36V were plated at 2000 cells per well in 50 µL of appropriate media in 384-well white culture plates (Thermo Fisher Scientific). 100 nL of compound in DMSO from compound stock plates was added to each well using a Janus Workstation pin tool (PerkinElmer). After addition of compound, plates were incubated for 16 hours at 37 ºC, prior to luminescence assessment. Plates were brought to room temperature prior to reagent addition. 25 µL Fluc buffer (as described above for the FKBP12WT and FKBP12F36V dual luciferase assay) was added to each well for 10 minutes at room temperature, and luminescence was measured on the Envision 2104 plate reader (PerkinElmer). Luminescence values were then normalized by setting DMSO only wells to 100%. Data were analyzed and plotted using GraphPad PRISM v6 using the ‘log(inhibitor) vs. response -- Variable slope (four parameters)’ analysis module.
Immunoblotting
MV4;11 and 293T cells were lysed with RIPA buffer supplemented with halt protease inhibitor cocktail (Thermo Fisher Scientific) and 0.1% benzonase (Novagen) on ice for 60 minutes. Lysates were clarified by centrifugation at 20,000 xg for 10 minutes at 4 ºC. NIH/3T3 and 293FT cells were lysed with RIPA buffer supplemented with cOmplete protease inhibitors (Roche) and PhosSTOP phosphatase inhibitors (Roche) on ice for 60 minutes. Lysates were clarified by centrifugation at 20,000 xg for 10 minutes at 4 ºC. Protein concentration was determined by a BCA assay (Pierce), and all samples were run with equal total protein content. The following antibodies were used in this study: HA (Cell Signaling, #3724 and #2367), BRD4 long isoform (Bethyl labs, #A301-985A), BRD4 short isoform (Abcam, #ab128874), BRD3 (Abcam, #ab56342), BRD2 (Bethyl labs, #A302–582A), FKBP12 (Abcam, #ab24373), vinculin (Santa Cruz, #Sc-25336), GAPDH (Millipore, #MAB374), phospho-AKT S473 (Cell Signaling, #4060), AKT (Cell Signaling, #2920), phospho-MEK1/2 S221 (Cell Signaling, #2338), MEK1/2 (Cell Signaling, #4694), phospho-ERK1/2 T202/Y204 (Cell Signaling, #4370), ERK1/2 (Cell Signaling, #4696), phospho-Pol II S2 (Millipore, #04-1571-1), and Pol II (Santa Cruz, #Sc-899). Imaging was performed by detection of fluorescently labelled infrared secondary antibodies (IRDye) on the Odyssey CLx Imager (LI-COR).
Lentiviral plasmid construction
We deposited a set of plasmids, maps, and sequences for the lentiviral dTAG system to Addgene. To develop pLEX_305-N-dTAG (Addgene, #91797) and pLEX_305-C-dTAG (Addgene, #91798) empty vectors, the pLEX_305 (Addgene, #41390) vector was modified. FKBP-2xHA cassettes were designed to include NheI and ClaI (N-dTAG) or AgeI and MluI (C-dTAG) restriction sites. Cassettes were synthesized by GeneScript and cloned into the pLEX_305 vector using the appropriate restriction sites.
Gateway recombination cloning technology (Invitrogen) was used to clone targets and controls of interest into pLEX_305-N-dTAG and pLEX_305-C-dTAG. pDONR plasmids for HDAC1, MYC, EZH2, PLK1, and BRD4 (short) were obtained from the Harvard PlasmID Repository, and KRASG12V was obtained from Addgene (#31200)51. Luciferase was cloned into a Gateway compatible donor vector (pDONR223) using BP clonase after PCR with primers containing BP overhangs GGGGACAAGTTTGTACAAAAAAGCAGGCTTAACCATGGAAGATGCCAAAAACAT TAAGAAGGG (forward) and GGGGACCACTTTGTACAAGAAAGCTGGGTTCACGGCGATCTT GCCGCCCTT (reverse) from the dual luciferase vector described above. In brief, 150 ng pDONR vectors containing genes of interest were mixed with 150 ng of destination vector in 10 µl. 2 µl of LR clonase II enzyme mix (Life Technologies) was added to the plasmid mix and incubated for one hour at room temperature. To terminate the reaction, 1 µl of Proteinase K was added to each sample and incubated at 37 ºC for 10 minutes. Samples were transformed using Stbl3 competent cells, plated on Ampicillin selective media, and grown at 30 ºC. Colonies were picked and grown up in 50 mL LB medium overnight at 30 ºC. Plasmid DNA was purified using midi-preps (Qiagen) and insertions were confirmed by BsrGI-HF digestion (NEB) and gel electrophoresis. Plasmids were sequenced using the primers listed in Supplementary Table 5. Note that when designing plasmids for Gateway recombination cloning into the N-dTAG vector, a stop codon must be included at the end of the gene to prevent read-through into the recombination sequences. Supplementary Table 6 lists plasmids associated with the dTAG system.
dTAG knock-in vector construction
We deposited a set of plasmids, maps, sequences, and protocols for the knock-in dTAG system to Addgene. To develop the pCRIS-PITChv2-BSDR-dTAG (BRD4) (Addgene, #91792) vector, the pCRIS-PITChv2-FBL (Addgene, #63672)29 plasmid was digested with MluI-HF (NEB) to excise the original EGFP-2A-Puro cassette. The BSDR-P2A-2xHA-FKBP12F36V-linker cassette was designed to include flanking BRD4 microhomology sequences and overlapping sequences to aid in Gibson assembly and synthesized at IDT as a gBlock gene fragment. The cassette was assembled into the pCRIS-PITChv2 backbone using the NEBuilder HiFi DNA Assembly Cloning Kit (NEB), according to the manufacturer’s instructions. This protocol was also used to generate an N-terminal BRD4 tagging cassette (PuroR-P2A-2xHA-FKBP12F36V-linker) with a puromycin selectable marker (Addgene, #91793), and C-terminal BRD4 tagging cassettes (linker-FKBP12F36V-2xHA-P2A-BSDR/PuroR) with blasticidin or puromycin selectable markers (Addgene, #91795 and #91796). Note that Gibson assembly can be used to modify the flanking microhomology sequences to those of any target gene of interest.
pX330A-nBRD4/PITCh (Addgene, #91794) was cloned as previously described29, 52. Briefly, N-terminal targeting guides for BRD4 were designed using http://crispr.mit.edu/. Oligonucleotides from IDT were annealed and inserted into the pX330A-1×2 (Addgene, #58766) plasmid after BbsI (NEB) digestion to generate pX330A-1×2-nBRD4. The PITCh sgRNA sequence from pX330S-2-PITCh (Addgene #63670) was inserted into pX330A-1×2-nBRD4 using Golden Gate assembly (NEB). The resulting plasmid pX330A-nBRD4/PITCh contained both the PITCh sgRNA, BRD4-specific sgRNA, and Cas9. Supplementary Table 6 lists plasmids associated with the dTAG system.
BRD4 knock-in cell line development
293T cells were plated in 10 cm plates and co-transfected with 1 µg of pCRIS-PITChv2-BSDR-dTAG (BRD4) and 2 µg of pX330A-nBRD4/PITCh using Lipofectamine 2000 (Thermo Fisher Scientific), according to manufacturer’s instructions. After 6 hours of transfection, appropriate growth media was added to the cells for 48 hours. Subsequently, cells were selected with 10 µg/mL blasticidin, and single cell clones were isolated in 96-well format. To isolate genomic DNA, cells were washed twice with cold PBS, mixed with 60 μL of directPCR (tail) (Viagen Biotech) and 0.4 mg/mL proteinase K (Life Technologies), transferred to a v-bottom PCR plate (VWR), and incubated overnight at 55 ºC. To inactivate proteinase K, cells were incubated at 85 ºC for 1.5 hours. Cell debris was spun down at 500 xg for 5 minutes. To genotype the cells and confirm knock-in, PCR reactions were performed using 1 μl of genomic DNA, 2x Q5 hot start master mix (NEB), and the following primers: TCCCTCTGGCCAACTTGGCTA (forward) and TGGTCTGCCTCTTGGGCTTGTTA (reverse). Amplified DNA was purified by gel extraction (Qiagen) and sequenced using the PCR primers. The same protocol was used for the C-terminal targeting plasmids, pCRIS-PITChv2-dTAG-BSDR (BRD4) and pX330A-cBRD4/PITCh using the primers: TACGGGGCTGACTGCTGGTA (forward) and AATCCTCAGCTGCCCGTCA (reverse). While we did not successfully isolate C-terminal targeted clones with homozygous knock-in at the BRD4 locus, the pCRIS-PITChv2-dTAG-BSDR (BRD4) vector can be modified for C-terminal targeting of any gene of interest (Supplementary Fig. 4 displays genotyping and validation of select clones). pX330A-cBRD4/PITCh was not deposited to Addgene as sgRNA cloning can be performed as described above.
Analysis of cell viability by ATPLite
293T and NIH/3T3 cells were plated at 1000 cells per well in 50 µL of appropriate media in 384-well white culture plates (Thermo Fisher Scientific). Cells were allowed to adhere overnight before adding 100 nL of compound in DMSO from compound stock plates using a Janus Workstation pin tool (PerkinElmer). After addition of compound, plates were incubated for 72 hours at 37 ºC, and ATPlite 1 Step Luminescence Assay System (PerkinElmer) was used to determine ATP-dependent luminescence as an approximation of cellular viability. Plates were brought to room temperature prior to reagent addition. Lyophilized powder was resuspended in lysis buffer and diluted 1:3 with deionized water. 20 µL of this solution was added to each well, plates were incubated for 15 minutes at room temperature, and luminescence was measured on an Envision 2104 plate reader (PerkinElmer). Cell viability values were calculated by normalizing to DMSO treated wells for each cell line. Data were analyzed and plotted using GraphPad PRISM v6 and the ‘log(inhibitor) vs. response -- Variable slope (four parameters)’ analysis module.
Cell cycle analysis
75,000 NIH/3T3 cells were plated in 6-well plates and allowed to adhere overnight. Cells were incubated with DMSO or 1 µM dTAG-13 for 48 hours, trypsinized, washed with PBS, and fixed in 70% ethanol at 4 ºC overnight. Fixed cells were washed with PBS, resuspended in PBS with 100 µg/mL RNase A (Thermo Fisher Scientific) and 50 µg/mL propidium iodide (Sigma), and incubated at room temperature in the dark for 10 minutes. Guava Easycyte flow HT (Millipore) was used to detect stained cells, and data was analyzed FlowJo software (version 10.0.8r1, TreeStar).
Quantitative proteomics and phospho-proteomics
Sample preparation for mass spectrometry
Samples for peptide analysis were prepared as previously described1. Samples for phosphopeptide analysis were prepared as previously described37 with the following modifications. Following tandem mass tag (TMT) labeling (1:4; peptide:TMT label, Pierce), phospho-tyrosine (pY) peptides were enriched from the total phosphopeptide pool using pY-specific antibodies as described by the manufacturer (Cell Signaling, P-Tyr-100). The pY enriched peptide data was collected as a single 3 hour run on the mass spectrometer. The flow through from the pY-enrichment was used to collect the phospho-serine/threonine data as previously described37.
Liquid chromatography-MS3 spectrometry (LC-MS3)
An Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) equipped with a Proxeon Easy nLC 1000 was used to analyze peptide fractions with an LC-MS3 data collection strategy, as previously described1.
The scan sequence for the 12 of the 24 peptide fractions (every other fraction from the basic reverse phase step) for the Fusion Orbitrap began with an MS1 spectrum (Orbitrap analysis, resolution 120,000, 400−1400 m/z scan range with quadrapole isolation, AGC target 1 × 106, maximum injection time 100 ms, dynamic exclusion of 60 seconds). For MS2 analysis, the top 10 precursors (‘Top N’) were selected, which consisted of CID ion trap analysis (AGC 1.5 × 104, NCE 35, maximum injection time 150 ms), and quadrapole isolation of 0.5 Da for the MS1 scan. For MS3 analysis, the top ten fragment ion precursors from each MS2 scan were selected (synchronous precursor selection), in which precursors were fragmented by HCD prior to Orbitrap analysis (NCE 55, max AGC 1.5 × 105, maximum injection time 150 ms, MS2 quadrapole isolation was set to 2.5 Da, resolution 50,000).
The scan sequence for the 12 phospho-serine/threonine fractions for the Fusion Orbitrap began with an MS1 spectrum (Orbitrap analysis, resolution 120,000, 400−1400 m/z scan range with quadrapole isolation, AGC target 2 × 105, maximum injection time 100 ms, dynamic exclusion of 75 seconds). For MS2 analysis, the top 10 precursors (‘Top N’) were selected, which consisted of CID ion trap analysis (AGC 8 × 103, NCE 35, maximum injection time 150 ms), and quadrapole isolation of 0.5 Da for the MS1 scan. For MS3 analysis, the top three fragment ion precursors from each MS2 scan were selected (synchronous precursor selection), in which precursors were fragmented by HCD prior to Orbitrap analysis (NCE 55, max AGC 1 × 105, maximum injection time 250 ms, MS2 quadrapole isolation was set to 2.5 Da, resolution 60,000).
The scan sequence for the single pY sample for the Lumos Orbitrap began with an MS1 spectrum (Orbitrap analysis, resolution 120,000, 400−1350 m/z scan range with quadrapole isolation, AGC target 1 × 106, maximum injection time 100 ms, dynamic exclusion of 60 seconds). For MS2 analysis, the top 10 precursors (‘Top N’) were selected, which consisted of CID ion trap analysis (AGC 2.5 × 104, NCE 35, maximum injection time 150 ms), and quadrapole isolation of 0.5 Da for the MS1 scan. For MS3 analysis, the top ten fragment ion precursors from each MS2 scan were selected (synchronous precursor selection), in which precursors were fragmented by HCD prior to Orbitrap analysis (NCE 55, max AGC 2.2 × 105, maximum injection time 150 ms, MS2 quadrapole isolation was set to 1.2 when z=2 or 1 when z=3 or 0.8 when z=4–6, resolution 60,000).
LC-MS3 data analysis
In-house software tools were used to process the data as previously described1 with the following modification. Database search criteria also included variable phosphorylation of serine, threonine, and tyrosine (79.96633 Da) for pST or pY searches.
RNA-sequencing with ERCC RNA spike-in normalization
350,000 NIH/3T3 cells were plated in 6-well plates and allowed to adhere overnight. NIH/3T3 cells were treated as indicated, lysed, homogenized using Qiashredder columns (Qiagen), and RNA was extracted using the RNeasy Plus Mini Kit (Qiagen), according to manufacturer’s instructions. Cell count normalized RNA samples were mixed with the ERCC RNA Spike-In Mix #1 (Life Technologies) prior to library preparation. Libraries for Illumina sequencing were prepared using TruSeq Stranded mRNA Library Prep Kit (Illumina) and equimolar multiplexed libraries were sequenced on an Illumina NextSeq 500 (single end 75 bp reads) by the Molecular Biology Core Facility at the Dana-Farber Cancer Institute. Fastq files were aligned to mouse genome build mm9 using HiSat with default parameters. Transcripts were assembled and FPKM values were generated using cuffquant and cuffnorm from the cufflinks pipeline53. FPKM values were then normalized to synthetic ERCC spike-in RNAs as described previously54. A transcript was considered expressed in each dataset, if in one experimental condition the average FPKM > 2.0 and in one experimental condition 2 out of 3 replicates had an FPKM > 2.0. snoRNAs, miRNAs, histone genes, ribosomal genes, and ERCC spike-in transcripts were removed from the analyses. Gene Set Enrichment Analysis (GSEA)55 analysis was performed using curated gene sets (C2) between experimental conditions. Significantly differentially expressed genes between experimental conditions were uploaded to the Ingenuity Pathway Analysis tool for functional and pathway enrichment analysis as previously described43.
Animal studies
1×106 viable MV4;11 cells expressing luciferase-FKBP12F36V were transplanted into 8-week-old immunocompromised female mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, NSG; Jackson Laboratory #005557) by tail-vein injection. Prior to transplantation, cells were tested at Charles River Laboratories to ensure lack of contaminating mycoplasma and rodent-infectious agents (Mouse Comprehensive Panel). Bioluminescence was used to monitor engraftment for two weeks, at which point mice were assigned to vehicle (n = 6 biologically independent mice) or dTAG-13 (25 mg/kg, n = 5 biologically independent mice) cohorts such that cohorts had similar variability in bioluminescent signal. Mice with no engraftment were excluded from the study. For treatments, dTAG-13 was formulated in a 20% solutol (Sigma), 5% DMSO solution in 0.9% sterile saline (Moltox) and intraperitoneal injections were performed. Each cohort was dosed and bioluminescence was monitored at the time-points indicated in Supplementary Fig. 10a. This animal experiment (protocol #13–053) was approved by the Dana-Farber Cancer Institute Animal Care and Use Committee (IACUC) and all experiments were adherent to institutional standards.
Pharmacokinetic assessment was performed at Shanghai ChemPartners Co., LTD. In brief, 1 mg/kg dTAG-13 and two other dTAG compounds (3 in 1 cassette dose) were administered via intraperitoneal injection to CD1 male mice (Shanghai SLAC Laboratory Animal Co. LTD, Certificate No.: SCXK (SH), 2012–0002 2015000512058, n = 3 biologically independent mice). Approximately 25 µL of blood from the facial vein of a mouse was collected into a K2EDTA tube at the following time-points: pre-dose, 0, 0.083, 0.25, 0.5, 1, 2, 4, 8 and 24 hours. 20 µL of blood was diluted with 60 µL sterile water, put on dry ice immediately after collection, and stored at approximately −70 °C until analysis. 30 µL of diluted blood sample was added to 150 µL acetonitrile containing 100 ng/mL internal standard (Diclofenac). The mixture was vortexed for 10 minutes, centrifuged at 5228 xg for 10 minutes, and 5 µL of the supernatant was used for LC-MS/MS analysis. MS analysis was performed using an AB SCIEX API4000 triple-quadrupole instrument with an electrospray ionization interface. The data acquisition and control system were created by using Analyst 1.5.2 software from AB SCIEX. The lower limit of quantification was 2.00 ng/mL in mouse blood samples. This animal experiment (protocol #A998HL0002) was approved by the Shanghai ChemPartners Co., LTD IACUC and all experiments were adherent to institutional standards.
Statistical analyses
Statistical tests are identified in the corresponding figure legends. Center values, error bars, P value cutoffs, number of replicates or samples, and number of independent experiments are detailed in the corresponding figure legends. Error bars are shown for all data points with replicates as a measure of variation within each data group. Data met the assumptions for all tests used and sample sizes were not predetermined using any statistical analyses. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. Kwiatkowski for critical reading of the manuscript, W. Kaelin for sharing referenced cell lines and the dual luciferase plasmids (Dana-Farber Cancer Institute, pLL3.7-EF1a-IRES-Gateway-nluc-2xHA-IRES2-fluc-hCL1-P2A-Puro), R. Kunz and the Thermo Fisher Scientific Center for Multiplexed Proteomics at the Harvard Medical School for the quantitative proteomics and phospho-proteomics assessment, and S. Nabet, A. Aguirre, W. Hahn, and members of the Bradner and Gray laboratories for helpful discussions. This work was supported by an American Cancer Society Postdoctoral Fellowship PF-17-010-01-CDD (B.N.), the Claudia Adams Barr Program in Innovative Basic Cancer Research (D.L.B.), Damon Runyon Cancer Research Foundation DRG-2196-14 (D.L.B.), and generous philanthropic gifts from the Hale Center for Pancreatic Cancer Research and the Katherine L. and Steven C. Pinard Research Fund.
COMPETING FINANCIAL INTERESTS
The authors claim the following competing financial interests: International Patent Application Nos. PCT/US2016/039048, PCT/US2016/046087, PCT/US2016/046088, PCT/US2016/046089, each filed in the name of Dana-Farber Cancer Institute, Inc. D.L.B., J.P., and A.S. are now employees of Novartis. G.E.W. is a consultant for C4 Therapeutics. N.S.G. is a Scientific Founder and member of the Scientific Advisory Board of Syros Pharmaceuticals, C4 Therapeutics, and Petra Pharmaceuticals and is the inventor on IP licensed to these entities. J.E.B. is a Scientific Founder of Syros Pharmaceuticals, SHAPE Pharmaceuticals, Acetylon Pharmaceuticals, Tensha Therapeutics (now Roche), and C4 Therapeutics and is the inventor on IP licensed to these entities. J.E.B. is now an executive and shareholder in Novartis AG.
Footnotes
Data availability statement
RNA-sequencing data reported in this paper has been deposited to the NCBI GEO and are available under the accession number: GSE92775. Normalized and scaled quantitative mass spectrometry-based proteomics and phospho-proteomics (phospho-serine/threonine and phospho-tyrosine) data are provided in Supplementary Datasets 1–3.
REFERENCES
- 1.Winter GE et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu J et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol 22, 755–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zengerle M, Chan KH & Ciulli A Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem. Biol 10, 1770–1777 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winter GE et al. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol. Cell 67, 5–18 e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondeson DP et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol 11, 611–617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remillard D et al. Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands. Angew. Chem. Int. Ed. Engl 56, 5738–5743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonger KM, Chen LC, Liu CW & Wandless TJ Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat. Chem. Biol 7, 531–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neklesa TK et al. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat. Chem. Biol 7, 538–543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckley DL et al. HaloPROTACS: Use of Small Molecule PROTACs to Induce Degradation of HaloTag Fusion Proteins. ACS Chem. Biol 10, 1831–1837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung HK et al. Tunable and reversible drug control of protein production via a self-excising degron. Nat. Chem. Biol 11, 713–720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T & Kanemaki M An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature methods 6, 917–922 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG & Wandless TJ A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoulders MD, Ryno LM, Cooley CB, Kelly JW & Wiseman RL Broadly applicable methodology for the rapid and dosable small molecule-mediated regulation of transcription factors in human cells. J. Am. Chem. Soc 135, 8129–8132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Q et al. A chemical genetics approach for the functional assessment of novel cancer genes. Cancer Res 75, 1949–1958 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Clackson T et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. U. S. A 95, 10437–10442 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts JM & Bradner JE A Bead-Based Proximity Assay for BRD4 Ligand Discovery. Curr. Protoc. Chem. Biol 7, 263–278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglass EF Jr., Miller CJ, Sparer G, Shapiro H & Spiegel DA A comprehensive mathematical model for three-body binding equilibria. J. Am. Chem. Soc 135, 6092–6099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu G et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kronke J et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneekloth JS Jr. et al. Chemical genetic control of protein levels: selective in vivo targeted degradation. J. Am. Chem. Soc 126, 3748–3754 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Anand P et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell 154, 569–582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradner JE, Hnisz D & Young RA Transcriptional Addiction in Cancer. Cell 168, 629–643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delmore JE et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deshpande AJ, Bradner J & Armstrong SA Chromatin modifications as therapeutic targets in MLL-rearranged leukemia. Trends Immunol 33, 563–570 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loven J et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shortt J, Ott CJ, Johnstone RW & Bradner JE A chemical probe toolbox for dissecting the cancer epigenome. Nat. Rev. Cancer 17, 160–183 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Filippakopoulos P et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M et al. Design and characterization of bivalent BET inhibitors. Nat. Chem. Biol 12, 1089–1096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakuma T, Nakade S, Sakane Y, Suzuki KT & Yamamoto T MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc 11, 118–133 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Stephen AG, Esposito D, Bagni RK & McCormick F Dragging ras back in the ring. Cancer Cell 25, 272–281 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Cox AD, Fesik SW, Kimmelman AC, Luo J & Der CJ Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feramisco JR, Gross M, Kamata T, Rosenberg M & Sweet RW Microinjection of the oncogene form of the human H-ras (T-24) protein results in rapid proliferation of quiescent cells. Cell 38, 109–117 (1984). [DOI] [PubMed] [Google Scholar]
- 33.Shih C, Padhy LC, Murray M & Weinberg RA Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature 290, 261–264 (1981). [DOI] [PubMed] [Google Scholar]
- 34.Stacey DW & Kung HF Transformation of NIH 3T3 cells by microinjection of Ha-ras p21 protein. Nature 310, 508–511 (1984). [DOI] [PubMed] [Google Scholar]
- 35.McAlister GC et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem 84, 7469–7478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAlister GC et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erickson BK et al. Evaluating multiplexed quantitative phosphopeptide analysis on a hybrid quadrupole mass filter/linear ion trap/orbitrap mass spectrometer. Anal. Chem 87, 1241–1249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smeal T, Binetruy B, Mercola DA, Birrer M & Karin M Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature 354, 494–496 (1991). [DOI] [PubMed] [Google Scholar]
- 39.Sears R et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14, 2501–2514 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin YR & Toker A The actin-bundling protein palladin is an Akt1-specific substrate that regulates breast cancer cell migration. Mol. Cell 38, 333–344 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilmartin AG et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res 17, 989–1000 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Robert C et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med 372, 30–39 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Nabet B et al. Deregulation of the Ras-Erk Signaling Axis Modulates the Enhancer Landscape. Cell reports 12, 1300–1313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natsume T, Kiyomitsu T, Saga Y & Kanemaki MT Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell reports 15, 210–218 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Weintraub AS et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171, 1573–1588 e1528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bisgrove DA, Mahmoudi T, Henklein P & Verdin E Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U. S. A 104, 13690–13695 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroder S et al. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J. Biol. Chem 287, 1090–1099 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banaszynski LA, Sellmyer MA, Contag CH, Wandless TJ & Thorne SH Chemical control of protein stability and function in living mice. Nat. Med 14, 1123–1127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erb MA et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang HT et al. MELK is not necessary for the proliferation of basal-like breast cancer cells. Elife 6, e26693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X et al. A public genome-scale lentiviral expression library of human ORFs. Nature methods 8, 659–661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ran FA et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loven J et al. Revisiting global gene expression analysis. Cell 151, 476–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.