Abstract
A small number of high-burden countries account for the majority of tuberculosis cases worldwide. Detailed data are lacking from these regions. To explore the evolutionary history of M. tuberculosis in China — the third highest TB burden country — we analyzed a countrywide collection of 4,578 isolates. Little genetic diversity was detected within the large M. tuberculosis population in China, with 99.4% of the bacterial population belonging to lineage 2 and three sublineages of lineage 4. The deeply rooted phylogenetic positions and geographic restriction of these four genotypes indicate that their populations expanded in situ following a small number of introductions to China. Coalescent analyses suggest that these bacterial sub-populations emerged in China around 1,000 years ago, expanded in parallel from the 12th century onward, and the whole population peaked in the late 18th century. More recently, sublineage L2.3, which is indigenous to China and exhibited relatively high transmissibility and extensive global dissemination, came to dominate the population dynamics of M. tuberculosis in China. Our results indicate that historical expansion of four M. tuberculosis strains shaped the current TB epidemic in China, and highlight the long-term genetic continuity of the indigenous M. tuberculosis population.
INTRODUCTION
Mycobacterium tuberculosis complex (MTBC), which causes tuberculosis (TB), has circulated among human populations for thousands of years1. With more than ten million cases and 1.7 million deaths each year, TB remains the leading cause of death due to an infectious disease. The burden of TB is unevenly distributed, with 30 TB high-burden countries accounting for 87% of all TB cases in the world2. TB is a typical disease of poverty and all the high-burden countries are from the developing world3,4.
China ranks as the third highest TB burden country in the world with about one million incident cases each year2. Numerous literary sources describe a disease resembling TB in ancient China, suggesting that TB has affected Chinese populations for thousands of years5–8. The oldest literary description suggestive of TB is from ~5,700 years ago, predating the first dynasty Xia in China6. MTBC DNA was detected in human skeletons from Xinjiang province dating back ~2,000 years before present5. Based on these observations, the current epidemic of TB in China may have very deep historical roots. However, unlike Western Europe, which was devastated by the so-called White Plague of TB during the 18th and 19th centuries9,10, the historical record does not contain any similar descriptions of severe epidemics of TB in China11,12. It thus remains unclear when epidemic forms of TB first arose in East Asia, and what course these epidemics may have followed throughout Chinese history11,13. Starting in the second half of the last century, China underwent major social changes, including strong population growth, massive internal migrations of rural workers to urban areas, increases in household crowding, and later the incursion of the HIV pandemic14,15. It is not clear what role these factors have played in enabling epidemic forms of TB to flourish in the more recent past.
The global spread of MTBC strains is mainly driven by human activities. Prior research suggests it was globally disseminated as a result of human movements driven by exploration, migration, trade and conquest16–19. Although more recent human movements linked to globalization might disturb phylogeographic patterns, MTBC lineages still vary in their distribution between countries and continents20,21. These geographic patterns can shed light on historical phenomena that contributed to the spread of TB22,23. Whole-genome sequencing studies of MTBC populations in Greenland and Nunavik have demonstrated a single recent origin for the regional population of pathogens24,25, while the studies from Ethiopia and Vietnam point to more ancient and complex origins for the endemic population of MTBC26,27. In this study, we aimed to investigate historical migration events and bacterial population history underlying the current TB epidemic in China. To this end, we integrated analyses of genotyping and whole genome sequence data from 4,578 MTBC isolates collected throughout China. Our findings demonstrate that, although the extant population of MTBC in China is large, almost all strains currently circulating in the country descend from only four ancestors introduced into China around the turn of the second millennium.
RESULTS
A large MTBC population with low genetic diversity
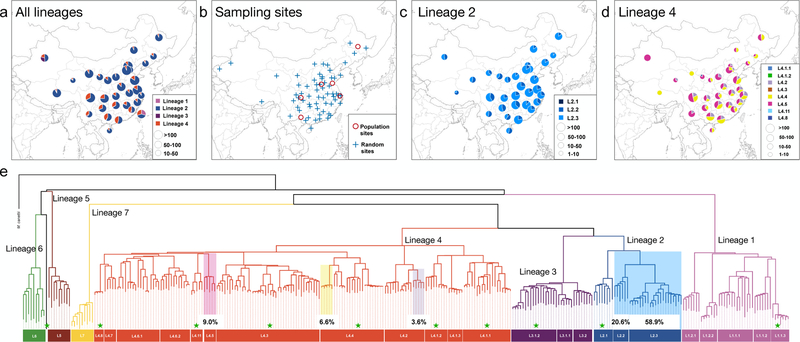
We first collected spoligotyping data from MTBC isolates sampled throughout China to obtain an initial picture of the MTBC population structure in the country. A total of 16,621 isolates’ spoligotyping records were obtained from 26 studies covering all 32 provinces in the country, and 15,217 of them can be successfully assigned to known lineages (Supplementary Table 1). Among these, 12,302 (80.8%) were classified as MTBC lineage 2 (L2), 2,570 (16.9%) were assigned to lineage 4 (L4), while 227 (1.5%) and 118 (0.8%) were assigned to lineage 1 (L1) and lineage 3 (L3), respectively (Fig. 1a). These data are in line with the previous observations that the Beijing family strains (belonging to L2) are most prevalent in China28,29. Our results indicate that L2 and L4 are distributed throughout the country, whereas L1 is most prevalent in Taiwan (Southeast) and L3 in Xinjiang (Northwest) (Fig. 1a). The overwhelming majority of TB cases in China were caused by L2 and L4 strains.
Fig. 1. Genotyping results of countrywide collected MTBC strains in China.
(a) The prevalence of different MTBC lineages in 32 provinces based on spoligotyping data from 16,221 isolates collected throughout China. (b) The 76 county sites from which MTBC isolates were sampled for this study: “population sites” are counties where MTBC isolates were collected through exhaustive sampling from 2009–2010, and “random sites” are counties where MTBC isolates were randomly sampled in 2007. SNP typing results of L2 strains (c) and L4 strains (d) show the relative proportion of each sublineage in each province. (e) Phylogeny of 301 MTBC isolates reflecting diversity found worldwide. Branches are colored according to the convention described in Comas et al 2010. Sublineages found commonly in China are highlighted, with a notation of their prevalence. Sublineages that were rarely encountered in China are marked with green pentacles; the remaining unmarked sublineages were not identified in China.
To characterize the genetic diversity within L2 and L4, we further genotyped a countrywide collection of 4,578 MTBC isolates using previously validated phylogenetic SNPs21,30 (Fig. 1b, Supplementary Table 2). Ninety-nine percent of the 4,578 MTBC isolates were from L2 (79.8%) and L4 (19.6%). Within L2, sublineage L2.3 (“modern” Beijing) was the most prevalent throughout the country and accounted for 73.9% of all L2 isolates (Fig. 1c); L2.2 (“ancient” Beijing) accounted for 25.9% and was widely distributed with a greater concentration in the south relative to north; L2.1 (proto-Beijing) showed a low frequency (0.2%) and was restricted to provinces in far southwest China (Fig. 1c). A total of eight L4 sublineages were found, however, 96.7% of all L4 strains in China belonged to only three of these sublineages (L4.2, L4.4 and L4.5), which were widely distributed throughout the country. The other sublineages of L4 were identified sporadically across the country (Fig. 1d, Supplementary Table 2). Taken together, these results indicate that the TB epidemic in China largely traces its origin to a handful of MTBC sublineages (Fig. 1e).
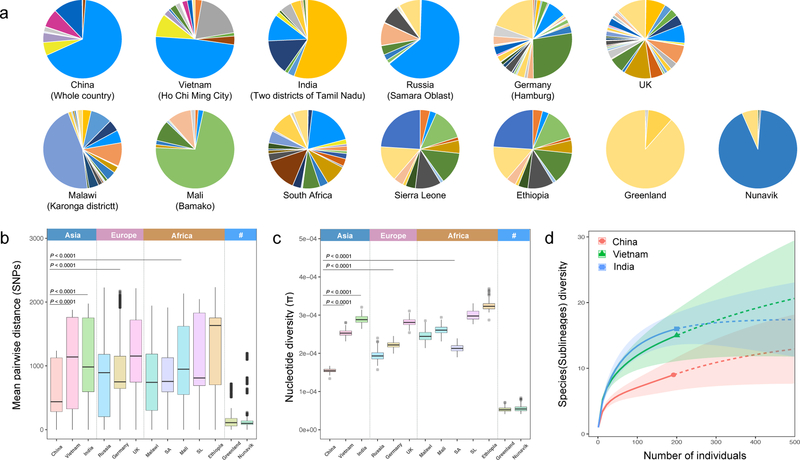
To put the MTBC strains from China into the wider context of the global population of the MTBC, we selected 306 representative isolates out of 4,578 genotyped strains for whole genome sequence analysis (Supplementary Table 3). In addition, we analyzed 15,591 previously published MTBC genomes to represent the global diversity of the MTBC (Supplementary Table 4). These data illustrate the homogeneous composition of the MTBC population in China, in contrast to most other countries harboring a greater diversity of MTBC (Fig. 2a). A formal comparison of diversity, pairwise SNP genetic distances, nucleotide diversity (π) and rarefaction analyses consistently demonstrate that the genetic diversity of MTBC in China as a whole was lower even than that of regional samples from individual cities in other countries (Fig. 2b-2d). It is striking that the world’s third-largest MTBC population exhibits so little genetic diversity.
Fig. 2. Low genetic diversity in China’s MTBC population.
(a) Pie charts showing the relative prevalences of MTBC sublineages in different countries, where each sublineage is assigned a color according to a recent defining scheme. Mean pairwise SNP distance between MTBC strains (b), nucleotide diversity (π) in MTBC population (c) from each country/region/population. “SA” refers to South Africa, “SL” refers to Sierra Leone, and 95% confidence intervals are shown. (d) Rarefaction analysis predicted the sublineage diversity of MTBC population in China, India, and Vietnam. Two hundred isolates were randomly sampled from each of the three countries; solid lines show the captured sublineages while the dashed lines show the predicted changes as the sample size is increased.
Phylogeographically restricted patterns indicate single origins
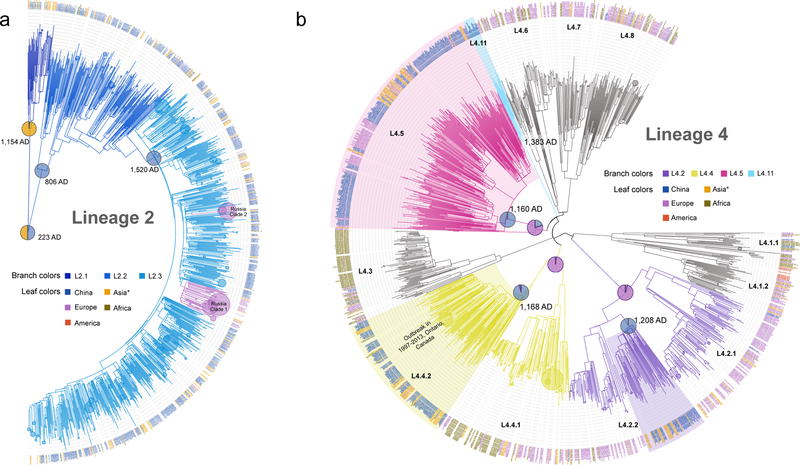
Published phylogeographic studies have inferred an African origin for the MTBC16,18,31, suggesting that MTBC strains in other continents were introduced via human migration. The diversity found within MTBC sublineages in China could have been generated before or after the introduction of the ancestral strains. To distinguish between these possibilities, we determined the phylogenetic positions of strains from China in global MTBC phylogenies. In the L2 phylogeny, although the most recent common ancestor of L2 has a mixed probability of China/Southern East (SE) Asian origin, the ancestors of L2.2 and L2.3 had a predicted origin in China with posterior probabilities of 99.6% and 98.6%, respectively (Fig. 3a and Supplementary Fig. 1, 2). This observation was robust to resampling analyses, in which we randomly reduced the number of isolates from China (Supplementary Fig. 3). These results indicate that L2 diversified locally following the migration event that established its most recent common ancestor (L2.2) in China. Moreover, the globally extant L2 appears to trace to more recent migration events out of China and SE Asia.
Fig. 3. Single origins of the four indigenous genotypes.
The phylogenetic trees of lineage 2 (a) and lineage 4 (b) were reconstructed with 1,242 isolates and 1,569 isolates respectively. To reduce the complexity in both trees, terminal branches with branch length < 0.008 (indicating clusters diversified very recently, e.g. the two Russian clades in L2.3) were automatically collapsed into circles. The circle sizes of those collapsed branches were proportional to the number of leaves that were collapsed. The estimated origin times of indigenous genotypes are shown at the relevant nodes, and their inferred geographic states are shown as pies with the colors indicating the isolates’ country origin. Asia* refers to Asian countries and regions excluding China. The three dominant clades of L4 in China were highlighted.
By contrast, the tree topology of L4 revealed a deep separation between distinct L4 sublineages found in China, with other global clades interspersed between the Chinese clades (Fig. 3b). This indicates that the indigenous sublineages of L4 diverged prior to their introduction to China. Chinese strains of L4.2 and L4.4 formed sister clades to strains from the same sublineage found in other regions, while the L4.5 branch is almost entirely composed of strains from China, except for two early diverged strains that were isolated in Russia. Average pairwise genetic distances between strains from China versus other regions were 407 and 526 SNPs for L4.2 and L4.4, respectively, while the corresponding distance between strains from within China were 267 and 298 SNPs, respectively (P<0.0001, Permutation test) (Supplementary Fig. 4). The ancestors of the Chinese clades in L4.2, L4.4 and L4.5 consistently had a most likely origin in China with posterior probabilities of 99.4%, 96.1% and 99.6%, respectively (Supplementary Fig. 5). These results suggest that L4 strains in China diversified locally with minimal global exchanges following their original introduction. We identified a novel sublineage provisionally termed L4.11 that appears to be private to China (Fig. 3b), and principal component analysis showed a clear division from its sister clades L4.5 and L4.6 (Supplementary Fig. 6). These results suggest that the indigenous L4 sublineages arose from separate parallel migrations followed by local diversification.
Historical origins and expansions of indigenous M. tuberculosis population
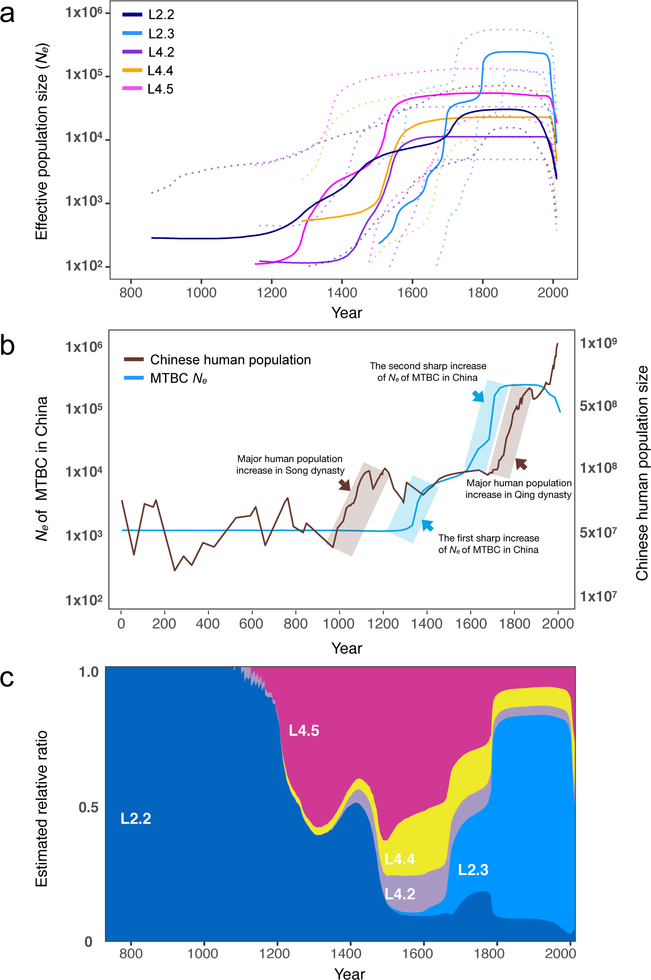
We estimate that L2 emerged around 223 AD, L2.2 in 806 AD and L2.3 in 1520 AD under the MTBC-6 substitution rate model, which are very similar to previous estimates using the same model1,16. The indigenous L4 sublineages originated over a similar timescale (Table 1, Fig. 3b). It is remarkable that new sublineages formed in situ and important external introduction events occurred during a short window (1150–1268 AD) (Supplementary Fig. 7). These numerous contemporaneous emergences indicate that epidemic expansion of TB was occurring during this period, likely as a result of environmental conditions that favored the pathogen. None of the currently prevalent sublineages were introduced after 1383 AD, indicating that the current TB epidemic in China has largely been shaped by introduction events in the early second millennium. We identified stepwise growths in effective population size (Ne) in all of the indigenous sublineages between 12th-18th centuries (Fig. 4a). Intriguingly, L2.2 and the three L4 sublineages appear to have followed parallel demographic trajectories: each started to expand around the 12th century, then experienced two or three waves of major expansions, plateauing, before going through a precipitous decline starting in the 1950s (Fig. 4a).
Table 1.
TMRCA and population growth rates of the indigenous sublineages.
| Sublineage | TMRCA | Start of growth, AD | Fast Growth Interval, AD | Growth Rate by Interval, % | |
|---|---|---|---|---|---|
| MTBC-6, ADa | 95% HPD, AD | ||||
| L2.3 | 1520 | 1311 ~ 1726 | 1504 | 1504–1816 | 2.320 |
| L2.2 | 806 | 250 ~ 1272 | 858 | 1105–1750 | 0.669 |
| L4.2b | 1208 | 823 ~ 1528 | 1365 | 1365–1560 | 1.189 |
| L4.4b | 1268 | 946 ~ 1576 | 1285 | 1400–1610 | 1.556 |
| L4.5b | 1160 | 787 ~ 1510 | 1152 | 1240–1560 | 1.610 |
Timing estimates from MTBC-6 model are shown in Anno Domini (AD);
L4.2, L4.4 and L4.5 here only refer to the Chinese specific clades in the relative sublineages. Start of growth, fast growth interval, and growth rate by interval were given by the results from MTBC-6 model.
Fig. 4. Historical expansions of indigenous MTBC genotypes.
(a) Estimated effective population size changes of the major MTBC sublineages in China. L2.3 was separated from L2.2 in Bayesian Skyline Plot analysis, and the dashed lines represent the 95% HPD. (b) Comparison of Chinese human population growth curve and MTBC Ne curve (all indigenous genotypes). (c) The inferred past population dynamics of each sublineage in China estimated from the effective population growth.
To explore a potential correlation between MTBC population growth and human demography, we compared the human population growth curve32 with the MTBC Ne curve (Fig. 4b). We observed similar trends between human and MTBC population growth: the first sharp increase of MTBC Ne followed the major human population expansion during the Song dynasty (960–1279 AD); the second substantial increase of MTBC Ne was parallel to the population boom during the Qing dynasty (1616–1912 AD); there was a period (13th-16th century) when both human and MTBC populations tended to be stationary. More recently (second half of the twentieth century), we observe a dramatic decline in MTBC Ne that is coincident with the availability of anti-TB drugs and improved control of the TB epidemic: China has halved the prevalence of TB in the last 20 years and maintained a steady decrease in TB incidence each year2,33. The effects of drug therapy on MTBC diversity may extend beyond a decrease in case counts, in that positive selection imposed by antibiotics could promote displacement of diverse lineages via clonal expansion of drug-resistant strains34. Twentieth-century environments may also have introduced greater individual-to-individual variability in TB transmission as high-density, crowded locations allow extended chains of transmission to develop; this transmission variability is also expected to be reflected in reduced pathogen Ne35. These concordances between human historical phenomena and bacterial demography suggest that the current epidemic of TB in China was enabled by the historical growth of human populations, likely including non-linear effects such as crowding and urbanization. These results also highlight the continuity of the MTBC population in China over the past 1,000 years.
L2.3 dominates recent population dynamics
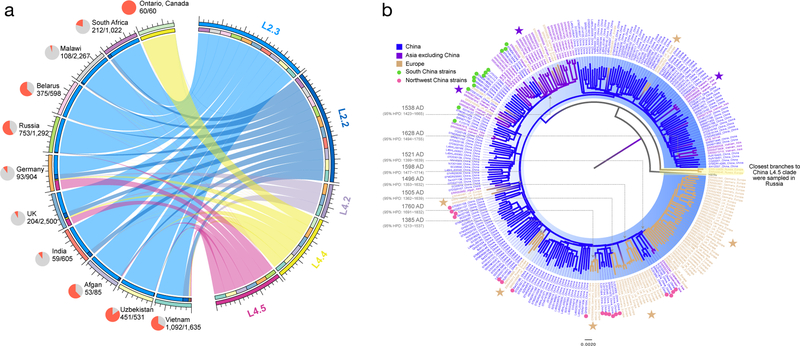
It is notable that L2.3 emerged relatively recently, i.e., ~450 years after the first expansion of L2.2 and ~300 years later than the three indigenous L4 sublineages. However, L2.3 is likely to have swept rapidly through the population (Fig. 4c). Our estimates of the population growth rate per year (based on changes in median Ne) imply that L2.3 may have expanded 1.4~3.5 times faster than that of the other sublineages (Table 1). It is also interesting that the growth of the other sublineages slowed or ceased as L2.3 started to expand (Fig. 4a, c). We further compared 15-locus MIRU-VNTR cluster rates (an indicator for recent transmission) of sublineages in six geographically distinct populations (population sites, Fig. 1b). Our data showed that isolates belonging to L2.3 were more likely to be clustered compared to the other sublineages (OR=3.7, 95%CI 2.9–4.8), consistent with higher rates of transmission (Table 2). This finding remained statistically significant when we repeated the comparison using different sets of VNTR loci to define clusters (Supplementary Table 5). These analyses suggest that relatively high transmission rates have contributed to L2.3’s dominance of modern MTBC populations. We also observed bias in the frequencies of L2.2 and L2.3 outside of China (Fig. 5a). Among L2 strains sampled in other non-SE Asian countries, 94.3% were caused by L2.3 while L2.2 accounted for only 5.7% (Chi-square test, P < 0.0001). This result suggests that the emergence of L2 strains across the globe has been driven primarily by L2.3.
Table 2.
VNTR cluster rates of the indigenous sublineages in six distinct populations.
| Sublineages | Individual County Sitea | All Sites | Odds Ratio | P value | |||||
|---|---|---|---|---|---|---|---|---|---|
| GX | HLJ | HN | SC | SD | SH | ||||
| L2.3 | 41% (24/58) |
66% (95/145) |
57% (70/123) |
23% (15/65) |
61% (76/125) |
58% (132/226) |
56% (412/742) |
1 | - |
| L2.2 | 31% (15/49) |
42% (5/12) |
40% (17/42) |
16% (7/45) |
25% (6/24) |
36% (29/80) |
31% (79/252) |
0.366 [0.267–0.500] |
<0.0001 |
| L4.2 | 13% (2/16) |
0% (0/0) |
22% (2/9) |
24% (4/17) |
0% (0/5) |
36% (8/22) |
23% (16/69) |
0.242 [0.127–0.440] |
<0.0001 |
| L4.4 | 20% (4/20) |
60% (6/10) |
0% (0/3) |
0% (0/23) |
27% (4/15) |
0% (0/17) |
16% (14/88) |
0.152 [0.078–0.277] |
<0.0001 |
| L4.5 | 40% (6/15) |
13% (2/15) |
0% (0/8) |
23% (10/44) |
26% (5/19) |
8% (2/25) |
20% (25/126) |
0.198 [0.120–0.319] |
<0.0001 |
The six county sites refer to the population sites, from which the MTBC isolates were collected through population-based approach from 2009 to 2010. GX refers to Guangxi, HLJ refers to Heilongjiang, HN refers to Henan, SC refers to Sichuan, SD refers to Shandong and SH refers to Shanghai.
Fig. 5. Global dispersal of Chinese indigenous genotypes.
(a) A circle plot with ribbons depicting the dispersal flows that led to the global emergence of Chinese indigenous sublineages. All the flows refer to “one-way” outflows and indicate direct or indirect exportation events with the ribbon width at each end proportional to the number of strains sampled in each country. The pie charts next to the country names show the proportion (red sector) of strains in the relative dataset that was found to be descendants of Chinese indigenous genotypes. (b) Global dispersal of Chinese L4.5 strains. Different leaf colors indicate the diverse geographic origins of those isolates. The Chinese L4.5 clade is highlighted in blue, and the branches are colored according to their geographic attributions. The major country transition events are marked with stars, and transition time of each event was estimated under MTBC-6 model. The European specific clades are nested within the Chinese L4.5 clade with the closest branches sampled from Northwest China. The closest branches to the strains sampled from Vietnam were mostly collected in South China.
Internal and global dispersal routes of indigenous genotypes
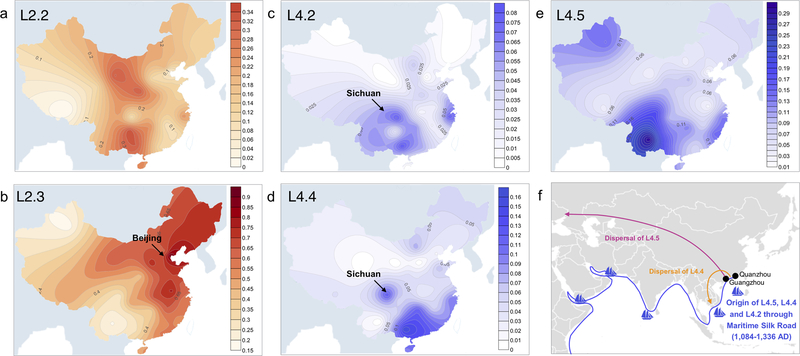
To explore possible dispersal routes of the indigenous MTBC genotypes in China, we created contour maps showing the relative concentrations of each sublineage across the country (Fig. 6). We identified hotspots for L2.2 in the southwest and northwest, suggesting this sublineage expanded out of two distinct geographic foci (Fig. 6a). L2.3 was concentrated in the northeast, with a single hotspot around Beijing and a gradual decrease in prevalence as a function of distance from the city (Fig. 6b). Beijing has been the capital city of China since the Yuan dynasty (1271–1368 AD). Since then, the human population in Beijing has doubled and has continued to grow rapidly36. As the political and trade center in China, Beijing was the central hub of population flow and interchange of resources between the 13th and 20th centuries37. L2.3’s emergence in the context of Beijing’s expanding and mobile human population may have contributed to its success.
Fig. 6. Contour maps show the countrywide prevalence of indigenous sublineages.
(a)-(e) The color ranges showing the prevalence of each sublineage in percentage based on the SNP typing data. (f) A scenario of Maritime Silk Road origins for L4 sublineages in China. The ship symbols mark the major ports on historical sea trade routes. The directions of dispersal of L4.5 and L4.4 are shown.
All three indigenous sublineages of L4 are concentrated in southern China (Fig. 6c-6e), suggesting a role of migrations to this region in the original establishment of these sublineages. In central China, L4.2 and L4.4 are concentrated in Sichuan province, which is consistent with the local history in that the inhabitants of Sichuan are mostly immigrants from Guangxi, Guangdong and other southern provinces where L4.2 and L4.4 are most prevalent (almost one million migrants moved to Sichuan following the great massacre in late Ming dynasty that almost emptied that province)38. Taken together, these results highlight the importance of human historical phenomena in shaping the genetic diversity of MTBC within China.
We also observed historical and recent exports of Chinese L4 strains. L4.5 strains were sampled in UK and Germany at relatively high frequencies (Fig. 5a), and those strains formed European specific clades nested within the Chinese L4.5 clade (Fig. 5b). Their deeply rooted positions suggest that they were exported in ancient times. Intriguingly, we noticed a consistent pattern in that the early diverging branches closest to European L4.5 clades were sampled exclusively in Northwest China (especially in Xinjiang province). This pattern could result from the dispersal of L4.5 to Europe through the ancient land Silk Road, on which Xinjiang acted as one of the crossroads of Eurasia39. A more recent example is a strain that caused an outbreak in Ontario, Canada in the 1990s, which turned out to be a descendant of the Chinese L4.4 clade (Supplementary Fig. 8). The origin of the relative ancestor was estimated to be around 1973 AD and the closest branches to that strain were sampled in Guangdong province (Southern China), suggesting this strain hitchhiked the Chinese migrant waves from Southern China to North America in the second half of last century40–42.
A scenario of Maritime Silk Road Origin
We inferred a European origin for the ancestors of L4.2, L4.4 and L4.5 (Supplementary Fig. 5), consistent with the designation of L4 as a “Euro-American” lineage43. In our reconstructions of the individual ancestral states for L4.2, L4.4 and L4.5, we found the three China-specific clades within those sublineages to have the highest probability of an origin in southern China (Supplementary Fig. 10, 11). The Chinese clades of L4 could have been introduced from Europe to South China via two major routes. The overland route traverses the Middle East, South Asia and Southeast Asia44. Alternatively, MTBC could have migrated by sea, for example along the well-known Maritime Silk Road45,46. In the former scenario, we would expect the L4 sublineages to be prevalent in the countries along the migration route to China, and the corresponding L4 sublineages there should form early-diverged branches to China’s L4 strains. This is the case for L3, which is hypothesized to have been dispersed via the overland Silk Road16, and that is concentrated in Northwest China (Fig 1a). In contrast, the three indigenous L4 sublineages (L4.2, L4.4 and L4.5) were rarely detected in a large sample from countries including India, Afghanistan, and Uzbekistan (Supplementary Table 6). Importantly, isolates from these countries were nested among samples from China, suggesting L4.2, L4.4 and L4.5 diversified in China before spreading to other countries in Southern and Central Asia (Supplementary Fig. 5). The emergence of the three L4 sublineages (1160–1268 AD) is coincident with the period of peak activity for the Maritime Silk Road in China, during the Song dynasty (960–1279 AD). Trading in this period was intense and extensive between multiple ports in Europe and ports in South China such as Guangzhou and Quanzhou46,47 (Fig. 6f). The southern Chinese origin and timing for the origin of the indigenous L4 lineages are consistent with an emergence in the context of the Silk Road era maritime trade between Europe and China (Fig. 6f).
DISCUSSION
Through reconstruction of the evolutionary history of MTBC strains circulating in China, we demonstrate that the current TB epidemic stems almost entirely from historical human migration events that established L2 and three sublineages of L4 starting around 1000 years before present. Our data show that these strains underwent massive expansion over the past 1,000 years. This is remarkable in that there is no historical record of a severe TB outbreak in China11,12, unlike the situation in Europe where the epidemic of 18th to 19th centuries constitutes a clearly defined and documented epidemiological phenomenon with marked cultural and demographic impacts10. A potential explanation for this disparity is differences in the timing and pace of industrialization in China and Europe. The devastating TB epidemic in Europe is believed to have been triggered by transitions in social conditions such as overcrowding and malnutrition linked to the industrial revolution13. The environment in China differed markedly in this period, as China did not participate in the industrial revolution48 and the historical urbanization rate rarely exceeded ten percent of the total population49. Historical TB expansions in China could reflect cryptic but frequent transmission, as reflected by the numerous descriptions of “Lao-bing” (TB) in historical medical texts12. Our results indicate there was a period of four centuries during which the four indigenous Chinese sublineages expanded simultaneously, suggesting that growth of the pathogen population was driven by common ecological transitions such as growth of the host population and an increase of urbanization (urbanization rate increased from 5% in Tang dynasty (618–907 AD) to 10~13% in Song dynasty (960–1279 AD))50.
Why so little genetic diversity?
A simple explanation for the limited number of introductions of MTBC to China is its distance from the African continent, where MTBC appears to have first emerged16,31. However, there is evidence to suggest that MTBC dispersed readily between Southeast Asia and the African continent16, which argues against a simple model in which migration events scale with distance. Historically isolationist policies may be of greater relevance: for example, during the Ming and Qing Dynasties (1368–1912 AD), feudal rulers adopted a policy of seclusion that hampered exchanges with the outside world51,52. Interestingly, we did not identify any indigenous sublineages that were introduced during that time interval. Alternatively, past genetic diversity may have been higher than it is at present, if, for example, multiple lineages were displaced by a sweep of L2.3 or if they failed to survive the bacterial population reduction that followed the widespread implementation of anti-TB therapy. The newly discovered sublineage L4.11 could support this notion, as its wide geographical distribution at low frequency suggests that the population has been through expansion and contraction.
Propagation of the TB epidemic in China
TB in China continues to be characterized by substantial ongoing transmission, with a reported recent transmission rate of ~30%53. Our analyses suggest that the current TB situation in China represents the waning era of an epidemic that expanded over the past millennium. As discussed above, historically isolationist policies may have limited the incursion of foreign MTBC in China, thereby contributing to the minimal genetic diversity observed here. Our results also suggest that historical patterns of mixing were uneven, and enabled a “winner take all” dynamic in which a small subset of the MTBC strains introduced to China came to dominate the population. The contrast between the indigenous genotypes and L3 or L1 is illustrative: L3 and L1 remain at low frequencies and are geographically restricted to the Northwest (Xinjiang province) and Southeast (Taiwan), respectively. We hypothesize that the disparate fates of these lineages and sublineages reflect historical variation in mobility and/or growth among sub-populations of hosts.
Our analyses here suggest that L2.3 spread out of Beijing, and we posit that the dominance of this sublineage reflects the centralization of the city within China over the past 700 years. Specific historical phenomena that are likely to have contributed to L2.3’s dominance include growth and urbanization of the Beijing population and the city’s role as a hub for migration within China36. Phenomena that are extrinsic to the pathogen – such as crowding and host malnutrition – can be powerful drivers of TB transmission and can lead to the dominance of specific MTBC strains that land under favorable conditions17,54. Our results also suggest that L2.3 strains spread more rapidly than do strains of the other extant sublineages. Hence, the rapid expansion of the L2.3 population may also have been facilitated by relatively high rates of disease progression following infection, and/or larger numbers of secondary cases per source case55,56. Recent studies have shown increased virulence of “modern” Beijing (L2.3) strains in mouse infection models and induction of lower levels of proinflammatory cytokines than “ancient” Beijing (L2.2) strains57–59; more recently, a study shows that mutations of ppe38 in L2.3 strains could completely block the secretion of two large subsets of ESX-5 substrates and lead to hypervirulent phenotype60; enhanced virulence could plausibly lead to more rapid disease progression following infection and consequently high rates of spread. We posit that the success of the L2.3 sublineage results both from factors that are intrinsic to the bacteria (e.g., increased virulence leading to rapid onset of transmissible disease) as well as extrinsic conditions, such as an expanding and mobile host population. It is possible that these phenomena interacted to make L2.3 a dominant sublineage: e.g., the large size and high density of the human population may have better sustained ongoing transmission of rapidly progressive forms of TB than low-density communities of hosts.
Genetic continuity of MTBC population
A recent study of MTBC genomes from eighteenth-century Hungary found them to be nested within a phylogeny of contemporary strains, which points to continuity of MTBC lineages over the past two centuries in Europe61. We inferred that MTBC strains currently circulating in China trace to introductions that occurred around 1,000 years before present. These findings demonstrate that the MTBC lineages that become established in favorable environments can persist for centuries. We hypothesize that the capacity of MTBC to establish long-term latent infections contributes to this continuity, which contrasts with bacterial pathogens such as Salmonella typhi, Vibrio cholerae and Yersinia pestis, for which massive lineage replacements have been observed62–65. (See additional discussion in Supplementary Appendix).
In conclusion, we demonstrate that China’s TB epidemic is a historical heritage that stems from just a handful of introductions of its causative agent, M. tuberculosis. These introduced strains expanded in parallel, presumably in response to common ecological drivers of epidemic TB. The long-term genetic continuity and distinctive structure of local MTBC populations in this high incidence region highlight the potential for MTBC population dynamics to guide TB epidemiologic surveillance.
ONLINE METHODS
Countrywide sampling of MTBC isolates
The MTBC isolates analyzed in this study consist of two sample sets. 1) Whole country data set (70 random sites). In 2007, a total of 3,929 culture positive MTBC isolates were collected from 70 counties for the purpose of surveillance for drug-resistance. These counties covered 31 out of the 34 provincial regions of China (Supplementary Table 2)66. The number of isolates collected from each province was proportional to the number of smear-positive cases reported in that province relative to the total number of cases nationwide. Those isolates were recovered on Lowenstein-Jensen medium from stored MTBC samples that were previously kept in −80°C freezer. However, 702 isolates failed in recovery, possibly due to multiple freeze and thaw cycles. 2) Whole population data set (six population sites). From 1 June 2009 to 31 December 2010, a total of 1,375 MTBC isolates were collected at six county sites from six different provinces in China in a population-based molecular epidemiologic study. 1,351 of them were available for genotyping in this study (Supplementary Table 2)55. These six county sites represented different geographic settings spreading south to north and west to east, and covered a total population of about 5.8 million inhabitants. All the county sites’ locations were marked on the map using Tableau (v10.4) (https://www.tableau.com).
DNA extraction and SNP typing
Genomic DNA of MTBC isolates was extracted through the boiling method for lineage and sublineage genotyping67. As MTBC does not appear to engage in lateral gene transfer and homoplastic single nucleotide polymorphisms (SNPs) are rare23,68, SNPs are ideal markers for typing MTBC sublineages69. L2 strains were classified into L2.1 (proto-Beijing), L2.2 (“ancient” Beijing or atypical Beijing) and L2.3 (“modern” Beijing or typical Beijing) sublineages30,70,71, while L4 strains were classified according to the previously defined 10 sublineages21,30. We developed six real-time PCR melting curve assays for SNP typing using the well-characterized sublineage-specific SNP markers (Supplementary Table 7). The principle of this SNP typing assay is similar to the drug-resistant mutation detection assay we developed previously67. Briefly, one dually labeled probe was designed for each sublineage-specific phylogenetic SNP. For each single strand nucleotide probe, one end was labeled with a fluorophore (FAM or ROX) and the other end was labeled with a quencher. As the targeted sublineage strains differed by one nucleotide in the detecting region compared with other strains, the melting curve analysis would show different Tm values due to the altered annealing ability67. The targeted sublineage is thereby differentiated from the remaining sublineages. One probe was designed to differentiate L2 strains from the remaining isolates. For L2 sublineage typing, two probes were designed to detect L2.2 and L2.3, while the remaining isolates were whole-genome sequenced to detect L2.1. For L4 sublineage typing, three probes were designed to detect the common L4 sublineages (L4.5, L4.4, and L4.2) based on our pilot typing results, while the remaining L4 isolates and those showing ambiguous typing results were further whole-genome sequenced. A total of thirty-two isolates that could not be assigned to any known sublineage or that showed ambiguous typing results underwent whole-genome sequencing. All the real-time PCR melting curve analysis assays were performed on a Bio-Rad CFX96 platform.
Public data collection
Spoligotyping data collection.
As spoligotyping results are generally concordant with lineage classifications based on whole-genome sequence data72, we first collected spoligotyping data from MTBC isolates sampled throughout China to obtain an initial picture of the MTBC population structure in the country. We searched for research articles that published spoligotyping data of MTBC isolates collected from China on PubMed. We identified a total of 96 articles in a preliminary search, a large proportion of which did not provide detailed spoligotyping results or reported previously published data. A total of 26 articles provided valid spoligotyping results for 16,621 MTBC isolates with either original typing records or summarized typing results (Supplementary Table 1). Each isolate was assigned to the relevant MTBC lineage according to previously identified links between spoligotypes and phylogenetic lineages72. A total of 15,217 isolates can be successfully assigned to known lineages, among the remaining isolates in the sample, 1,030 (6.2%) were classified as “Orphan”, 371 (2.2%) as “Manu2” and 3 (<0.01%) as “M. bovis”. Orphan strains could be variant types from any lineage that have not been recorded in the SpolDB4.0 database73, whereas Manu2 types could be DNA samples that are a mixture of L2 and L4 strains74. These two types were excluded from our subsequent analyses. The number of MTBC isolates from each province was normalized to the relative human demographic data when calculating the total prevalence of each MTBC lineage (Supplementary Table 1).
WGS data collection.
To identify whole genome sequencing data from global MTBC isolates, we searched for articles with WGS data published in PubMed and downloaded the original sequencing reads from European Nucleotide Archive (EMBL-EBI). The geographic origin and year of collection for each isolate was extracted from the relevant article. We sent an inquiry to study authors for papers that did not include this information. A total of 15,047 MTBC isolates’ sequencing data were downloaded, and we obtained geographic information for 12,596 of them (Supplementary Table 1).
Whole-genome sequencing and SNP calling
A minimum spanning tree was constructed based on 15-locus MIRU-VNTR data of all the MTBC isolates genotyped here. We selected MTBC isolates from each of those clades to represent countrywide genetic diversity (a perl script was written to sample isolates from each clade randomly). We purposefully did not select L2 isolates because previous studies have generated an abundance of whole genome sequences of this lineage from China31,70,75. Genomic DNA was extracted from the 306 MTBC isolates (three of L1, 23 of L3 and 280 of L4) following theCetyltrimethylammonium bromide-lysozyme (CTAB) method as described before70. A 300bp fragment length library was constructed for each DNA sample, and whole genome sequencing was performed on an Illumina Hiseq 2500 system with either single-end or paired-end strategy. We used a previously validated pipeline for the mapping of short sequencing reads to the reference genome57. In brief, the Sickle76 tool was used for trimming whole-genome sequencing data. Sequencing reads with Phred base quality above 20 and read length longer than 30 were kept for analysis. The whole genome sequence of M. tuberculosis H37Rv strain (NC_000962.2) was used as the reference template for read mapping. Sequencing reads were mapped to the reference genome using Bowtie2 (v2.2.9)77. SAMtools (v1.3.1)78 was used for SNP calling with mapping quality greater than 30. Fixed mutations (frequency ≥ 75%) were identified using VarScan (v2.3.9)79 with at least 10 reads supporting and the strand bias filter option on. We excluded all SNPs that were located in repetitive regions of the genome (e.g., PPE/PE-PGRS family genes, phage sequence, insertion or mobile genetic elements) that are difficult to characterize with short read sequencing technologies. Small insertions or deletions (INDELs) identified by VarScan (v2.3.9) were also excluded.
Phylogenetic reconstruction
Mixed infection isolates were excluded for phylogenetic reconstruction by investigating the genotype heterozygosity of SNPs as described previously80. For all phylogenetic reconstruction, SNPs of MTBC isolates were combined into a single consensus and non-redundant list while those nucleotide positions with gaps in more than 5% of the taxa were excluded (possibly due to INDELs, low coverage or mapping quality at those sites). The alignments of polymorphic positions from all strains were used for phylogeny reconstruction using MEGA 6.081. The neighbor-joining method was used for initial inference of phylogeny structure when the taxa numbers were large. But for the final estimation of phylogenies, Maximum Likelihood (ML) method was applied under the general time reverse (GTR) model with at least 100 replicates for bootstrapping confidence levels. Phylogeny trees were visualized in FigTree (v1.4.3) (http://tree.bio.ed.ac.uk/software/figtree/) or ITOL (v3)82. We adapted a recently described hierarchical nomenclature to define nodes and sub-clades within the tree in the definition of sublineages30.
Population genetic analyses
Pairwise SNP distance.
We wrote a Perl script to calculate the number of pairwise SNP distances. For those countries with a large number of MTBC isolates, we randomly subsampled to 200 isolates in the calculation of pairwise SNP distances so as to be able to compare them with other countries with samples closer to 100 isolates. The distribution and mean pairwise SNP distance for each country was plotted with ggplot2 in RStudio (v3.4.0)83. As the pairwise SNP distances from the country samples were not normally distributed, Wilcoxon rank-sum test was used to test the differences between countries.
Nucleotide diversity (π).
Average pairwise nucleotide diversities per site (π) were calculated using the nuc.div function in “pegas” library, which uses the equation by Nei, M et al84. As sample sizes varied among countries, we generated 100 subsamples, each equal to 200 isolates, and thereby calculated the confidence interval for those countries with samples greater than 200 MTBC isolates. The random subsampling process was completed through rand command in Perl. We used all isolates for analysis of data from countries with 50–200 isolates. We did not include data from countries with fewer than 50 isolates available.
Rarefaction analysis.
Rarefaction is an ecological technique designed to estimate the species richness expected for a given number of individual samples, which is based on the construction of rarefaction curves. We used this method to evaluate sublineage diversity of MTBC isolates from different countries. Rarefaction analysis was performed in RStudio (v3.4.0) using the library “iNEXT”85.
Phylogeographic analysis
We used RASP86, which implements both Bayesian and parsimony (S-DIVA) approaches, to estimate the ancestral geographic ranges of L2 and the three major L4 sublineages in China. In order to estimate the geographic origins of those MTBC sublineages, we divided the world map into five broad geographic areas and used them as a proxy for the most likely origin of each strain (Supplementary Fig. 1). The reason we treated China independently from Asia was because we wanted to test whether the sublineages originated and diversified locally as opposed to being imported from other countries in Asia. The Maximum Likelihood phylogenies of both lineages and the corresponding geographic regions of origin for those isolates were loaded as a distribution. RASP reconstruction was performed without the outgroup. For the parsimony-based analyses, a maximum of two ancestral areas per node were allowed for range reconstruction. For the Bayesian-based analyses, five different chains during 500 thousand generations were run. For contour plots in Fig. 6, the prevalence of each sublineage was determined by the MTBC isolates we genotyped in each province. Sufer (v12) software (http://www.goldensoftware.com/) was used for contour plotting of each sublineage’s frequency throughout the county.
Bayesian based coalescent analysis
Dating analysis.
We selected 96 MTBC strains from a published study to represent the global diversity of MTBC lineages31. These 96 strains together with the 23 L2 strains and 41 L4 strains (10 L4.2 strains, 12 L4.4 strains, 11 L4.5 strains and 8 L4.11 strains) sampled from China were used for phylogenetic reconstruction. A total of 24,792 concatenated genome-wide variable positions were used for phylogenetic analyses. We estimated the dates of the most common recent ancestors of L2 and L4 and their sublineages using BEAST (v1.8.0)87. The XML input file was modified to specify the number of invariant sites in the MTBC genomes. For the MTBC genome substitution rate, we imposed a normal distribution for the substitution rate of MTBC with a mean of 4.6×10−8 substitutions per genome per site per year (3.0×10−8 to 6.2×10−8, 95% HPD interval) as in a previous study1. We used an uncorrelated lognormal distribution for the substitution rate, and we used a constant population size for the tree priors. We ran three chains of 5×107 generations sampled every 10,000 to assure independent convergence of the chains, the first 10% of which were discarded as a burn-in. Convergence was assessed using Tracer (v1.6.0), ensuring all relevant parameters reached an effective sample size >100. Phylogenetic trees were visualized using Figtree (v1.4.3).
Bayesian Skyline Plot (BSP) analysis.
BSP analysis was applied to estimate the past effective population size dynamics of L2 and L4 sublineages based on the substitution rate model indicated above. Ideally, one should use all the samples for BSP analysis, however, this was not computationally feasible. In addition, the number of isolates that were whole-genome sequenced for each sublineage was not proportional to their epidemiological prevalence, as some sublineages (L4.2, L4.4 and L4.5) were oversampled while others (L2.2 and L2.3) were undersampled. To reduce this bias and improve tractability of our analyses, we subsampled isolates from the original collection and constructed a whole genome sequence dataset containing 500 MTBC isolates. The isolates from different sublineages in this dataset were randomly sampled from the original sequenced genomes and the number was proportional to their prevalence in China. Sublineage-based skyline analysis was performed, and the ages of the most recent common ancestors from dating analysis were used as the tree heights. For the MTBC effective population size curve in Fig. 4b, we integrated all isolates above for the BSP analysis. In each case, three chains of 5×107 generations sampled every 10,000 to assure independent convergence of the chains. For the past human population size changes, the demographic data were obtained from historical investigations and records32. For the population dynamics plot in Fig. 4c, the relative prevalence of each sublineage in the past was estimated from the effective population growth curves generated by BSP analysis (each sublineage was estimated separately) and was plotted using the “streamgraph” package in RStudio (v3.4.0).
Population growth rate estimation
The population growth rate per year was calculated using the effective population growth curves generated from BSP analysis88. Each skyline plot consisted of 100 smoothed data points, at ≈5~11 y intervals. For L2.2 and L4.5, the effective population size increase was preceded by a brief period of stationary size. The initial population size N0 was set as the minimum population size during the period immediately preceding population growth. The population size became stationary or even decreased at later stages. Thus, we estimated the effective population growth rate for the increasing interval in our data88. The exponential growth equation was chosen for this analysis:
In this equation, r represents the population growth rate per year, N0 is the initial population size and t is the duration of time since growth began.
Data availability
Sequencing reads have been submitted to the European Nucleotide Archive (EMBL-EBI) under study accession PRJEB23157. The geographic Information for individual isolates are listed in Supplementary Table 3. The analysis scripts used in this study are available online at GitHub (https://github.com/StopTB/China_TB_Evolutionary_History).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Timothy M. Walker for sharing the geographic information of 3,651 MTBC isolates of multiple continents’ origin. We also thank Yun-Xin Fu and Xiaoming Liu for advice on effective population size calculation and fruitful discussions. We also thank Daniela Brites and Chuanchao Wang for their help in clarifying technical details of data analysis during this work. This work was supported by Natural Science Foundation of China (91631301 and 81661128043 to Q.G., 81701975 to Q.L.); C.S.P was supported by the National Institutes of Health (grant 1R01AI113287–01A1); S.G. was supported by the Swiss National Science Foundation (grants IZRJZ3_164171, 310030_166687, IZLSZ3_170834 and CRSII5_177163); this work was also supported by MINECO research grant SAF2016–77346-R European Research Council (ERC) (638553-TB-ACCELERATE to) and National Science and Technology Major Project of China (2017ZX10201302 to Q.G) and Sanming project of Medicine in Shenzhen (SZSM201611030 to Q.G.) and JSGG20170413142559220.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
ETHICS STATEMENT
The analysis of anonymous MTBC isolates collected in China in this study was approved by the Tuberculosis Research Ethics Review Committee of the China CDC and the Institutional Review Board of the Institutes of Biomedical Sciences, Fudan University.
Reference
- 1.Bos KI et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 514, 494–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Global Tuberculosis Report 2017. (Geneva: World Health Organization, 2017). [Google Scholar]
- 3.Narain JP, Raviglione MC & Kochi A HIV-associated tuberculosis in developing countries: epidemiology and strategies for prevention. Tuber Lung Dis 73, 311–21 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Steffen R, Rickenbach M, Wilhelm U, Helminger A & Schar M Health problems after travel to developing countries. J Infect Dis 156, 84–91 (1987). [DOI] [PubMed] [Google Scholar]
- 5.Fusegawa H et al. Outbreak of tuberculosis in a 2000-year-old Chinese population. Kansenshogaku Zasshi 77, 146–9 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Prasad PV General medicine in Atharvaveda with special reference to Yaksma (consumption/tuberculosis). Bull Indian Inst Hist Med Hyderabad 32, 1–14 (2002). [PubMed] [Google Scholar]
- 7.Suzuki T & Inoue T Earliest evidence of spinal tuberculosis from the Aneolithic Yayoi period in Japan. International Journal of Osteoarchaeology 17, 392–402 (2007). [Google Scholar]
- 8.Li X et al. Archaeological and palaeopathological study on the third/second century BC grave from Turfan, China: Individual health history and regional implications. Quaternary international 290, 335–343 (2013). [Google Scholar]
- 9.Packard RM White plague, black labor: Tuberculosis and the political economy of health and disease in South Africa, (Univ of California Press, 1989). [Google Scholar]
- 10.Dubos RJ & Dubos J The white plague: tuberculosis, man, and society, (Rutgers University Press, 1952). [Google Scholar]
- 11.Stead WW The origin and erratic global spread of tuberculosis. How the past explains the present and is the key to the future. Clin Chest Med 18, 65–77 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z Epidemic chronology of ancient China (in Chinese), (Fujian science and Technology Press, 2007). [Google Scholar]
- 13.Bates JH & Stead WW The history of tuberculosis as a global epidemic. Medical Clinics of North America 77, 1205–1217 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Perry EJ & Selden M Chinese society: Change, conflict and resistance, (Routledge, 2003). [Google Scholar]
- 15.Wang F & Zuo X Inside China’s cities: Institutional barriers and opportunities for urban migrants. The American Economic Review 89, 276–280 (1999). [Google Scholar]
- 16.O’Neill MB et al. Lineage specific histories of Mycobacterium tuberculosis dispersal in Africa and Eurasia. bioRxiv (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pepperell CS et al. Dispersal of Mycobacterium tuberculosis via the Canadian fur trade. Proc Natl Acad Sci U S A 108, 6526–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershberg R et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol 6, e311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirth T et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog 4, e1000160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagneux S & Small PM Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis 7, 328–37 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Stucki D et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet 48, 1535–1543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linz B et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pepperell CS et al. The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog 9, e1003543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjorn-Mortensen K et al. Tracing Mycobacterium tuberculosis transmission by whole genome sequencing in a high incidence setting: a retrospective population-based study in East Greenland. Sci Rep 6, 33180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee RS et al. Population genomics of Mycobacterium tuberculosis in the Inuit. Proc Natl Acad Sci U S A 112, 13609–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comas I et al. Population Genomics of Mycobacterium tuberculosis in Ethiopia Contradicts the Virgin Soil Hypothesis for Human Tuberculosis in Sub-Saharan Africa. Curr Biol 25, 3260–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holt KE et al. Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat Genet 50, 849–856 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Soolingen D et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol 33, 3234–8 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang Y et al. Spoligotyping and drug resistance analysis of Mycobacterium tuberculosis strains from national survey in China. PLoS One 7, e32976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coll F et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun 5, 4812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comas I et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet 45, 1176–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge J China Population History (Zhongguo Renkou Shi). (Shanghai: Fudan University Press, 2000). [Google Scholar]
- 33.Wang L et al. Tuberculosis prevalence in China, 1990–2010; a longitudinal analysis of national survey data. Lancet 383, 2057–2064 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Neher RA & Hallatschek O Genealogies of rapidly adapting populations. Proc Natl Acad Sci U S A 110, 437–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magiorkinis G et al. Integrating phylodynamics and epidemiology to estimate transmission diversity in viral epidemics. PLoS Comput Biol 9, e1002876 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guang-Hui H Historical population geography of Beijing (in Chinese), (Peking University Press, Beijing, 1996). [Google Scholar]
- 37.Hou Ren-Zhi TX-F Historical geography of Beijing city (in Chinese), (Beijing Yanshan Press, Beijing, 2000). [Google Scholar]
- 38.HUANG Q. s. & YANG G.-h. The Placename of Immigration in Sichuan and Huguang People Migrate into Sichuan [J]. Journal of Southwest China Normal University (Philosophy & Social Sciences Edition) 3, 023 (2005). [Google Scholar]
- 39.Millward JA Eurasian crossroads: a history of Xinjiang, (Columbia University Press, 2007). [Google Scholar]
- 40.Poston DL Jr, Mao MX & Yu M-Y The global distribution of the overseas Chinese around 1990. Population and Development Review, 631–645 (1994). [Google Scholar]
- 41.Li PS The Rise and Fall of Chinese Immigration to Canada: Newcomers from Hong Kong Special Administrative Region of China1 and Mainland China, 1980–20002. International Migration 43, 9–34 (2005). [Google Scholar]
- 42.King H & Locke FB Chinese in the United States: A century of occupational transition. International Migration Review, 15–42 (1980). [PubMed] [Google Scholar]
- 43.Gagneux S et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103, 2869–73 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNeill WH Human migration in historical perspective. Population and Development Review, 1–18 (1984). [Google Scholar]
- 45.Gan F Ancient glass research along the Silk Road, (World Scientific, 2009). [Google Scholar]
- 46.Kauz R Aspects of the Maritime Silk Road: From the Persian Gulf to the East China Sea, (Otto Harrassowitz Verlag, 2010). [Google Scholar]
- 47.McPherson K China and the Maritime Silk Route in Proceedings of the UNESCO Quanzhou International Seminar on China and the Maritime Routes of the Silk Roads. Quanzhou: Fujian People’s Publishing House; 55–60 (1991). [Google Scholar]
- 48.Lin JY The Needham puzzle: Why the industrial revolution did not originate in China. Economic development and cultural change 43, 269–292 (1995). [Google Scholar]
- 49.Jones EL, Frost L & White C Coming Full Circle: An Economic History of the Pacific Rim, (Westview Pr, 1993). [Google Scholar]
- 50.Yusuf S & Saich A China urbanizes: consequences, strategies, and policies, (World Bank Publications, 2008). [Google Scholar]
- 51.Millward J, Dunnell RW, Elliott MC & Forêt P New Qing Imperial History Making of Inner Asian Empire at Qing Chengde. New York: RoutledgeCurzon; (2004). [Google Scholar]
- 52.Mote FW, Twitchett D & Fairbank JK The Cambridge History of China: Volume 7, The Ming Dynasty, 1368–1644, (Cambridge University Press, 1988). [Google Scholar]
- 53.Yang C et al. Transmission of Mycobacterium tuberculosis in China: a population-based molecular epidemiologic study. Clin Infect Dis 61, 219–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ackley SF, Liu F, Porco TC & Pepperell CS Modeling historical tuberculosis epidemics among Canadian First Nations: effects of malnutrition and genetic variation. PeerJ 3, e1237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang C et al. Mycobacterium tuberculosis Beijing strains favor transmission but not drug resistance in China. Clin Infect Dis 55, 1179–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Jong BC et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J Infect Dis 198, 1037–43 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Q et al. Genetic features of Mycobacterium tuberculosis modern Beijing sublineage. Emerg Microbes Infect 5, e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Laarhoven A et al. Low induction of proinflammatory cytokines parallels evolutionary success of modern strains within the Mycobacterium tuberculosis Beijing genotype. Infect Immun 81, 3750–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ribeiro SC et al. Mycobacterium tuberculosis strains of the modern sublineage of the Beijing family are more likely to display increased virulence than strains of the ancient sublineage. J Clin Microbiol 52, 2615–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ates LS et al. Mutations in ppe38 block PE_PGRS secretion and increase virulence of Mycobacterium tuberculosis. Nat Microbiol 3, 181–188 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Kay GL et al. Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe. Nat Commun 6, 6717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wirth T Massive lineage replacements and cryptic outbreaks of Salmonella Typhi in eastern and southern Africa. Nat Genet 47, 565–7 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Wagner DM et al. Yersinia pestis and the plague of Justinian 541–543 AD: a genomic analysis. Lancet Infect Dis 14, 319–26 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Mutreja A et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477, 462–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vagene AJ et al. Salmonella enterica genomes from victims of a major sixteenth-century epidemic in Mexico. Nat Ecol Evol 2, 520–528 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Zhao Y et al. National survey of drug-resistant tuberculosis in China. N Engl J Med 366, 2161–70 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Liu Q, Luo T, Li J, Mei J & Gao Q Triplex real-time PCR melting curve analysis for detecting Mycobacterium tuberculosis mutations associated with resistance to second-line drugs in a single reaction. J Antimicrob Chemother 68, 1097–103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farhat MR et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet 45, 1183–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Comas I, Homolka S, Niemann S & Gagneux S Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4, e7815 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo T et al. Southern East Asian origin and coexpansion of Mycobacterium tuberculosis Beijing family with Han Chinese. Proc Natl Acad Sci U S A 112, 8136–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Merker M et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet 47, 242–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barbier M & Wirth T The Evolutionary History, Demography, and Spread of the Mycobacterium tuberculosis Complex. Microbiol Spectr 4(2016). [DOI] [PubMed] [Google Scholar]
- 73.Brudey K et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol 6, 23 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Viegas SO et al. Molecular diversity of Mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in Mozambique. BMC Microbiol 10, 195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet (2013). [DOI] [PubMed] [Google Scholar]
- 76.Joshi NA & Fass JN Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. (2011). [Google Scholar]
- 77.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koboldt DC et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22, 568–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gan M, Liu Q, Yang C, Gao Q & Luo T Deep Whole-Genome Sequencing to Detect Mixed Infection of Mycobacterium tuberculosis. PLoS One 11, e0159029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tamura K, Stecher G, Peterson D, Filipski A & Kumar S MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, 2725–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Letunic I & Bork P Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44, W242–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Team R RStudio: Integrated Development for R. RStudio, Inc., Boston, MA: (2015). [Google Scholar]
- 84.Paradis E pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–20 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Hsieh T, Ma K & Chao A iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods in Ecology and Evolution 7, 1451–1456 (2016). [Google Scholar]
- 86.Yu Y, Harris AJ, Blair C & He X RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Molecular Phylogenetics and Evolution 87, 46–49 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Drummond AJ, Suchard MA, Xie D & Rambaut A Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29, 1969–73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gignoux CR, Henn BM & Mountain JL Rapid, global demographic expansions after the origins of agriculture. Proc Natl Acad Sci U S A 108, 6044–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads have been submitted to the European Nucleotide Archive (EMBL-EBI) under study accession PRJEB23157. The geographic Information for individual isolates are listed in Supplementary Table 3. The analysis scripts used in this study are available online at GitHub (https://github.com/StopTB/China_TB_Evolutionary_History).