Abstract
STUDY QUESTION
What are the causal relationships between polycystic ovary syndrome (PCOS) and body mass index (BMI)?
SUMMARY ANSWER
Bidirectional Mendelian randomization analyses suggest that increased BMI is causal for PCOS while the reverse is not the case.
WHAT IS KNOWN ALREADY
The contribution of obesity to the pathogenesis of PCOS is controversial. To date, published genetic studies addressing this question have generated conflicting results and have not utilized the full extent of known single nucleotide polymorphisms associated with body mass index (BMI).
STUDY DESIGN, SIZE, DURATION
This cross-sectional Mendelian randomization (MR) and genetic association study was conducted in 750 individuals of European origin and with PCOS and 1567 BMI-matched controls.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Cases and controls were matched for BMI as well as for distribution of weight categories (normal weight, overweight, obese). Two-sample MR using inverse variance weighting (IVW) was conducted using a 92-SNP instrument variable for BMI with PCOS as the outcome, followed by two-sample MR with a 16-SNP instrument variable for PCOS with BMI as the outcome. Sensitivity analyses included MR-Egger and maximum likelihood methods. Secondary analyses assessed associations of genetic risk scores and individual SNPs with PCOS, BMI and quantitative androgen-related and glucose homeostasis-related traits.
MAIN RESULTS AND THE ROLE OF CHANCE
Each standard deviation genetically higher BMI was associated with a 4.89 (95% CI 1.46–16.32) higher odds of PCOS. Conversely, genetic risk of PCOS did not influence BMI. Sensitivity analyses yielded directionally consistent results. The genetic risk score of 92 BMI SNPs was associated with the diagnosis of PCOS (OR 1.043, 95% CI 1.009–1.078, P = 0.012). Of the 92 BMI risk variants evaluated, none were associated individually with PCOS after considering multiple testing. The association of FTO SNP rs1421085 with BMI was stronger in women with PCOS (β = 0.071, P = 0.0006) than in controls (β = 0.046, P = 0.065).
LIMITATIONS, REASONS FOR CAUTION
The current sample size, while providing good power for MR and genetic risk score analyses, had limited power to demonstrate association of individual SNPs with PCOS. Cases and controls were not matched for age; however, this was mitigated by adjusting analyses for age. Dietary and lifestyle data, which could have been used to explore the greater association of the FTO SNP with BMI in women with PCOS, was not available.
WIDER IMPLICATIONS OF THE FINDINGS
Increasing BMI appears to be causal for PCOS but having PCOS does not appear to affect BMI. This study used the most comprehensive set of SNPs for BMI currently available. Prior studies using fewer SNPs had yielded conflicting results and may have been confounded because cases and controls were not matched for weight categories. The current results highlight the potential utility of weight management in the prevention and treatment of PCOS.
STUDY FUNDING/COMPETING INTEREST(S)
National Institutes of Health Grants R01-HD29364 and K24-HD01346 (to R.A.), Grant R01-DK79888 (to M.O.G.), Grant U54-HD034449 (to R.S.L.), Grant U19-HL069757 (to R.M.K.). The funders had no influence on the data collection, analyses or conclusions of the study. No conflict of interests to declare.
TRIAL REGISTRATION NUMBER
N/A
Keywords: polycystic ovary syndrome, Mendelian randomization, genome wide association, genetic variants, single nucleotide polymorphism
Introduction
Polycystic ovary syndrome (PCOS) affects 6–10% of reproductive aged women, making it the most common endocrinopathy in this age group (Goodarzi et al., 2011). Although PCOS is a heterogeneous disorder with multiple phenotypes, it is most classically characterized by hyperandrogenism and oligoovulation. Women with PCOS are at significantly increased risk for insulin resistance, dyslipidemia, impaired glucose tolerance and type 2 diabetes mellitus (Salley et al., 2007). Women with PCOS have a high prevalence (50–70%) of overweight and obesity, with obesity exacerbating the PCOS phenotype (Lim et al., 2013). Conversely, lifestyle modification and weight loss can improve features of PCOS (Nybacka et al., 2011).
Both PCOS and obesity are highly heritable traits (Stunkard et al., 1990; Vink et al., 2006). Studies evaluating the impact of obesity genes on the susceptibility for PCOS have yielded conflicting results. The fat mass and obesity associated gene (FTO), an important candidate gene for obesity identified in genome wide association studies (GWAS) (Loos, 2012), was associated with PCOS in some studies, whereas other studies did not find an association after controlling for BMI (Barber et al., 2008; Ewens et al., 2011; Li et al., 2013). A recent meta-analysis examining the relationship between FTO variants and PCOS found that the effect of the FTO variant on BMI is larger in women with PCOS compared to normal controls (Wojciechowski et al., 2012), suggesting that PCOS itself may modify the impact of FTO on BMI in women with PCOS.
While there appears to be a relationship between PCOS and obesity, the direction and mechanism underlying this association are unknown. One approach to investigate parameters that are correlated, but not necessarily causally related, has been to determine whether genetic variations in a potential underlying trait are associated with the outcome of interest. In this approach, which ranges from simple association to formal instrumental variable (Mendelian randomization (MR)) analysis, an association between the genetic variants associated with the risk factor and the outcome of interest supports a causal relationship. Existing studies of genetic variants for obesity in PCOS have yielded conflicting results, with one study finding no association of a 12-single nucleotide polymorphism (SNP) BMI genetic risk score (GRS) with PCOS (Louwers et al., 2014) and another reporting positive results using 32 BMI SNPs in a Mendelian randomization analysis (Day et al., 2015). The aim of our study was to conduct the first bidirectional state-of-the-art two-sample MR analyses interrogating causal relationships between BMI and PCOS. We also sought to determine whether the overall genetic burden for obesity, as well as individual BMI susceptibility alleles, predispose to PCOS. We pursued these aims using a comprehensive set of 92 SNPs for BMI, more than that used in prior studies (12 or 32 SNPs) (Louwers et al., 2014; Day et al., 2015). Another unique feature of this study, in contrast to prior studies, is that cases and controls were well-matched for BMI, avoiding any possible confounding effect of higher BMI in the women with PCOS.
Materials and Methods
Subjects
The cohort consisted of 750 subjects with PCOS and 1567 BMI-matched controls, all of European origin. All subjects with PCOS met the 1990 NIH criteria (Azziz et al., 2009) and thus had clinical hyperandrogenism (i.e. hirsutism) and/or hyperandrogenemia and chronic oligoovulation. Parameters for defining hirsutism, hyperandrogenemia, oligoovulation and exclusion of related disorders were previously reported (Jones et al., 2012). The majority (1469) of the control subjects were general community controls from either the Cholesterol and Atherosclerosis Pharmacogenetics (CAP) study (Simon et al., 2006) or the Multi-Ethnic Study of Atherosclerosis (MESA) (Bild et al., 2002), from which only European-origin individuals were selected. These included 287 and 1182 subjects, from 603 CAP and 2685 MESA subjects respectively, selected to match the BMI of the PCOS cases. Clinical characteristics of PCOS and control subjects are displayed in Table I. The cohort was matched not only for mean BMI, but also for distribution between BMI categories: normal weight (BMI < 25 kg/m2), overweight (BMI ≥ 25 kg/m2 and < 30 kg/m2) and obese (BMI ≥ 30 kg/m2) (Table II).
Table I.
Baseline characteristics of PCOS and control subjects.
| PCOS (n = 750) | Controls (n = 1567) | P-value | |
|---|---|---|---|
| Age (yr) | 27 (23–31.9) | 59.6 (51–68) | <0.0001 |
| BMI (kg/m2) | 31.7 (25.3–37.6) | 30.6 (28.6–33.8) | 0.59 |
| Total Testosterone (nmol/L) | 2.39 (1.77–3.08) | 1.14 (0.88–1.72)a | <0.0001 |
| DHEAS (μmol/L) | 6.05 (4.17–8.53) | 2.72 (1.84–4.48)a | <0.0001 |
| Fasting insulin (pmol/L) | 96.0 (54.6–156.0) | 45.0 (27.0–73.2) | <0.0001 |
| Fasting glucose (mmol/L) | 4.77 (4.50–5.15) | 5.00 (4.66–5.50) | <0.0001 |
| HOMA2-IR | 2.1 (1.4–3.3) | 1.1 (0.77–1.79) | <0.0001 |
| HOMA2-%B | 176.2 (128.9–226.1) | 97.2 (74.9–130.2) | <0.0001 |
Values presented as median (interquartile range). Transformed (log or square root) means were compared using unpaired t-tests. aAndrogen levels were available from 98 deeply phenotyped controls.
Table II.
BMI distribution of PCOS and control subjects.
| PCOS (n = 750) | Controls (n = 1567) | Chi square P-value | |
|---|---|---|---|
| Normal weight (BMI < 25 kg/m2) | 23.1% (173) | 23% (361) | 0.9998 |
| Overweight (BMI ≥25 and < 30 kg/m2) | 20.4% (153) | 20.4% (320) | |
| Obese (BMI ≥30 kg/m2) | 56.5% (424) | 56.5% (886) |
The BMI-matched cohort described above was used for all analyses in this report, except the MR analysis in which PCOS was the exposure and BMI was the outcome, as described below.
Ethical approval
The study was approved by the Institutional Review Boards of Cedars-Sinai Medical Center (CSMC) and all other recruiting centers. All participants gave written informed consent.
Genotyping
Genotyping was previously performed at CSMC using Infinium II technology on the Metabochip, following the manufacturer’s protocol (Illumina, San Diego, CA). The Metabochip is a high throughput genotyping platform that was designed to provide a method by which loci associated with a number of traits related to cardiac and metabolic diseases could be rapidly genotyped (Voight et al., 2012). The genotyping and quality control details were previously described, which included a principal component analysis to correct for potential population stratification, allowing identification and removal of subjects with substantial non-Caucasian admixture (Jones et al., 2012).
For this project, we initially focused on 97 SNPs associated with BMI in a large GWAS and Metabochip analysis (Locke et al., 2015). The genotypes for 93 of those 97 SNPs (or SNPs in linkage disequilibrium, r2 > 0.8) were extracted from our Metabochip data. Four of the BMI-associated SNPs, or SNPs in linkage disequilibrium with these four, were not available on the Metabochip and were not included in the analysis. One more SNP did not meet MR quality control, as described below, thus, 92 BMI SNPs were ultimately examined.
Statistical analysis
All continuous parameters with a non-normal distribution were logarithmically or square root transformed. Clinical characteristics between cases and controls were compared using unpaired t-tests. The MR and association analyses described below were adjusted for age.
Mendelian randomization
In the first set of two-sample MR analyses, we considered BMI as the exposure and PCOS as the outcome. Published summary results from the GWAS meta-analysis for BMI by Locke et al. were used to generate a genetic instrument for BMI, focusing on the 93 SNPs that were robustly associated with BMI. One SNP (rs2033529) was removed from the MR analysis due to allele harmonization issues between exposure and outcome data (Hartwig et al., 2016). The remaining 92 SNPs were used to construct the genetic instrument for BMI. SNP to outcome estimates were obtained from our BMI-matched dataset. The primary MR analysis was conducted using the inverse variance weighted (IVW) method, wherein the SNP to outcome estimate is regressed on the SNP to exposure estimate. We used fixed effects IVW given that we did not detect instrument variable assumption violations (neither heterogeneity nor pleiotropy were observed). The causal effect estimates, equivalent to beta coefficients, were calculated and then transformed to odds ratios.
Given that MR using conventional IVW may be affected by directional pleiotropy (in which exposure SNPs may act through alternative traits), we carried out sensitivity analyses wherein we conducted MR using additional techniques that are affected differently by genetic confounding. These included the maximum likelihood and MR-Egger methods (Pierce and Burgess, 2013; Bowden et al., 2015). Since MR-Egger is robust against directional pleiotropy under the InSIDE (Instrument Strength Independent of Direct Effect) assumption, the intercept serves as an indicator of directional pleiotropy (Bowden et al., 2015). All analyses were conducted in R 3.4.2 with the package ‘TwoSampleMR’ (Hemani et al., 2018).
The second set of MR analyses handled PCOS as the exposure and BMI as the outcome. Summary results from a recent GWAS meta-analysis for PCOS in Europeans were used as the source of data to construct the instrument variable for PCOS (Day et al., 2018). Of the 14 SNPs associated with PCOS at genome-wide significance therein, we included all except rs853854, which was excluded because it is an A/T SNP with an allele frequency close to 50%, thus avoiding ambiguity in analyzing such palindromic SNPs (Hartwig et al., 2016). We also included three SNPs (rs13405728, rs2349415, rs2272046) originally identified in Chinese GWAS that were associated with PCOS in the European meta-analysis and were not in linkage disequilibrium with the 14 SNPs described above. Therefore, the instrument variable for PCOS consisted of 16 SNPs. SNP to outcome estimates were generated in the entire cohort of 2685 European subjects from MESA. Fixed effects IVW, MR-Egger and maximum likelihood MR analyses were conducted as described above.
Genetic risk score and SNP analyses
To test for the combined effect of all BMI risk alleles, we generated an unweighted genetic risk score (GRS) for each subject. We used an unweighted score because effect sizes for BMI have been determined in individuals without PCOS and effect sizes for all SNPs for PCOS are unknown. Also, unweighted scores perform similarly to weighted scores when constituent SNP effect sizes are similar (Burgess and Thompson, 2013), as is the case for BMI SNPs (Loos, 2012). The GRS was the sum of alleles possessed by each individual that were previously associated with increased BMI. The 92 SNPs used in the MR analyses were included in the risk score (possible range 0–184). The association between the GRS and PCOS was evaluated using logistic regression. Given that the cases and controls were matched for BMI, we did not adjust the regression for BMI.
We used linear regression to assess association of the GRS with BMI in the entire cohort as well as within cases and controls separately. Within the subjects with PCOS, we assessed the impact of the GRS on additional continuous quantitative traits (total testosterone, dehydroepiandrosterone sulfate (DHEAS), fasting glucose, fasting insulin and homeostasis model assessments of insulin resistance (HOMA2-IR) and beta-cell function (HOMA2-%B) (Levy et al., 1998)).
The individual association of the 92 SNPs (additive model) with PCOS was tested using logistic regression and with BMI using linear regression.
Significance was taken at a P-value of < 0.05 for the GRS association analysis with PCOS. Analyses of the GRS against quantitative traits used a Bonferroni multiple testing corrected P-value of 0.007 (0.05/7) because seven quantitative traits were examined. For analyses of individual SNPs, we used a multiple testing corrected P-value of 0.00054 (0.05/92) based on 92 independent SNPs genotyped.
Generalized linear models were used to statistically compare the regression coefficients from the regression of BMI on the GRS between the PCOS cases and controls. A similar comparison was carried out comparing the association of the FTO SNP with BMI in cases and controls.
Power analysis
For Mendelian randomization, the F statistic is an indicator of the strength of the instrument variable, with values over 10 reflecting strong instruments (Pierce et al., 2011). The 92-SNP instrument variable for BMI had an F statistic of 63.4, and the 16-SNP instrument variable for PCOS had an F statistic of 241.4. Thus, both instruments are well powered to estimate the causal effect of the exposure on the outcome.
We used the PROC Power Procedure with LOGISTIC statement in SAS (SAS Institute, Cary, NC) to determine the power to detect association between the GRS and PCOS in our cohort. With a mean of 88.9 and standard deviation of 6.1 for the GRS in our cohort, the sample size of 750 cases and 1567 controls has excellent power (≥97%) to detect association with PCOS at odds ratio ≥1.03 and moderate power (72%) to detect association at odds ratio as low as 1.02.
Results
The mean BMI was not significantly different between PCOS and control subjects in our BMI-matched cohort (Table I). In addition, controls and PCOS subjects were evenly distributed between three BMI categories – normal weight, overweight and obese (Table II).
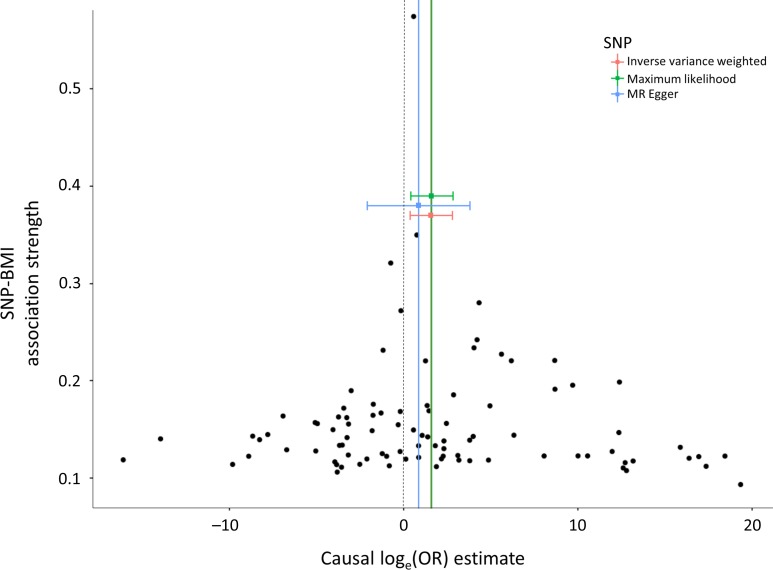
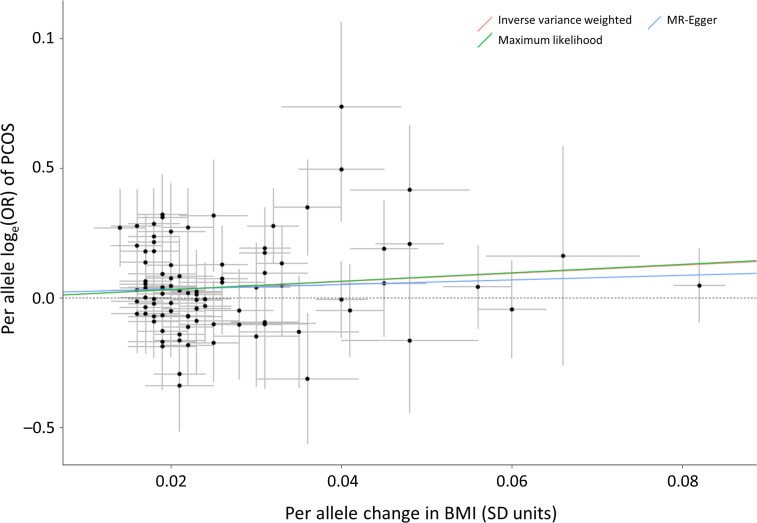
Causal effect estimates of BMI, as represented by the 92-SNP instrument, on PCOS are displayed in Table III. The primary MR analysis by IVW showed a significant causal effect, wherein each standard deviation genetically higher BMI was associated with a 4.89 (95% CI 1.46–16.32) higher odds of PCOS. The association of the 92-SNP instrument with PCOS was also evaluated using the maximum likelihood and MR-Egger techniques. As seen in Table III and Fig. 1, directionally consistent results were observed, suggesting that the findings are relatively unaffected by violation of MR assumptions. Scatterplots (Fig. 2) demonstrate that SNPs with greater effect on BMI have a greater effect on the risk of PCOS. The intercept of the MR-Egger regression was not significantly different from zero (P = 0.61), suggesting that genetic pleiotropy is not driving the MR results.
Table III.
Mendelian randomization results for causality of BMI on PCOS
| IVW | MR-Egger | Maximum likelihood | |||||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | Intercept | P value for intercept | OR | 95% CI |
| 4.89 | 1.46–16.32 | 2.41 | 0.13–46.0 | 0.02 | 0.61 | 5.07 | 1.50–17.10 |
Figure 1.
Funnel plots showing the causal effect estimates of BMI on PCOS. Black dots represent the causal effect estimates for each of the 92 SNPs. The three colored squares represent the causal effect estimates from the three MR methods, with corresponding horizontal lines displaying the 95% confidence intervals.
Figure 2.
Individual effects of 92 genetic variants on BMI and the odds of PCOS. The effect size in the entire cohort of each of the 92 SNPs on BMI (in standard deviation (SD) units) is on the X-axis. The loge odds ratio of each SNP for PCOS is on the Y-axis. The trend lines represent the results of the three MR analyses based on the 92 SNPs.
MR analyses assessing causality in the opposite direction were also performed. None of the three MR methods found evidence of a causal effect of having PCOS on BMI (Table IV).
Table IV.
Mendelian randomization results for causality of PCOS on BMI.
| IVW | MR-Egger | Maximum likelihood | |||||
|---|---|---|---|---|---|---|---|
| Beta | 95% CI | Beta | 95% CI | Intercept | P value for intercept | Beta | 95% CI |
| 0.003 | −0.008–0.013 | −0.13 | −0.06–0.03 | 0.003 | 0.46 | 0.003 | −0.008–0.013 |
Consistent with the MR results, the GRS composed of BMI SNPs was associated with the diagnosis of PCOS (OR 1.043, 95% CI 1.009–1.078, P = 0.012) (Table V), indicating a 4.3% increase in odds of PCOS per increment in GRS. The mean GRS was 88.7 for controls and 89.4 for PCOS.
Table V.
Association analysis for BMI-increasing GWAS risk alleles with the diagnosis of PCOS.
| Chr | Position | SNP | Nearest gene(s) | Risk increasing allele | Risk allele frequency | OR | 95%_CI | P value |
|---|---|---|---|---|---|---|---|---|
| - | - | Genetic risk score | - | - | - | 1.043 | 1.01–1.08 | 0.012 |
| 1 | 47457264 | rs977747 | TAL1 | T | 0.392 | 0.96 | 0.73–1.28 | 0.80 |
| 1 | 49210592 | rs3127553a | AGBL4 | G | 0.372 | 0.96 | 0.73–1.26 | 0.77 |
| 1 | 50332407 | rs11583200 | ELAVL4 | C | 0.386 | 1.00 | 0.76–1.32 | 0.98 |
| 1 | 72523773 | rs3101336 | NEGR1 | C | 0.642 | 1.14 | 0.87–1.51 | 0.34 |
| 1 | 74764232 | rs1514175a | FPGT, TNNI3K | T | 0.435 | 1.00 | 0.75–1.32 | 0.97 |
| 1 | 78219349 | rs12401738 | FUBP1 | A | 0.327 | 0.85 | 0.64–1.13 | 0.26 |
| 1 | 96696685 | rs11165643 | PTBP2 | T | 0.573 | 0.93 | 0.71–1.23 | 0.62 |
| 1 | 109956211 | rs17024393 | GNAT2 | C | 0.028 | 1.18 | 0.53–2.76 | 0.70 |
| 1 | 176156103 | rs543874 | SEC16B | G | 0.192 | 1.23 | 0.88–1.73 | 0.22 |
| 1 | 200050910 | rs2820292 | NAV1 | C | 0.539 | 1.14 | 0.86–1.49 | 0.36 |
| 2 | 622348 | rs13021737 | TMEM18 | G | 0.804 | 0.96 | 0.66–1.38 | 0.81 |
| 2 | 25003800 | rs10182181 | ADCY3 | G | 0.478 | 1.19 | 0.91–1.56 | 0.20 |
| 2 | 26782315 | rs11126666 | KCNK3 | A | 0.261 | 0.87 | 0.63–1.20 | 0.39 |
| 2 | 59159129 | rs1016287 | LINC01122 | T | 0.289 | 1.02 | 0.75–1.40 | 0.88 |
| 2 | 62906552 | rs11688816 | EHBP1 | G | 0.511 | 1.03 | 0.79–1.37 | 0.78 |
| 2 | 142759755 | rs2121279 | LRP1B | T | 0.121 | 1.37 | 0.90–2.11 | 0.14 |
| 2 | 164275935 | rs1460676 | FIGN | C | 0.167 | 1.29 | 0.90–1.87 | 0.17 |
| 2 | 181259207 | rs1528435 | UBE2E3 | T | 0.623 | 1.20 | 0.90–1.60 | 0.22 |
| 2 | 207963763 | rs17203016 | CREB1 | G | 0.178 | 0.71 | 0.50–1.01 | 0.057 |
| 2 | 213121476 | rs7599312 | ERBB4 | G | 0.736 | 0.83 | 0.61–1.13 | 0.26 |
| 2 | 219057996 | rs492400 | USP37 | C | 0.427 | 1.03 | 0.78–1.37 | 0.83 |
| 2 | 226824609 | rs2972143a | LOC646736 | A | 0.35 | 1.31 | 0.98–1.76 | 0.069 |
| 3 | 25081441 | rs6804842 | RARB | G | 0.583 | 0.94 | 0.71–1.24 | 0.64 |
| 3 | 61211502 | rs2365389 | FHIT | C | 0.598 | 1.05 | 0.79–1.39 | 0.75 |
| 3 | 85912107 | rs7622475a | CADM2 | C | 0.224 | 1.04 | 0.74–1.46 | 0.81 |
| 3 | 142788703 | rs2035935a | RASA2 | C | 0.066 | 0.84 | 0.49–1.48 | 0.56 |
| 3 | 187301576 | rs4234589a | ETV5 | A | 0.863 | 1.05 | 0.71–1.58 | 0.78 |
| 4 | 44877284 | rs10938397 | GNPDA2 | G | 0.424 | 0.99 | 0.75–1.33 | 0.97 |
| 4 | 77315142 | rs17001561a | SCARB2 | A | 0.166 | 0.90 | 0.62–1.32 | 0.60 |
| 4 | 103407732 | rs13107325 | SLC39A8 | T | 0.083 | 1.52 | 0.93–2.50 | 0.093 |
| 4 | 145878514 | rs11727676 | HHIP | T | 0.902 | 0.73 | 0.45–1.19 | 0.21 |
| 5 | 75050998 | rs2112347 | POC5 | T | 0.632 | 1.14 | 0.85–1.53 | 0.39 |
| 5 | 153518086 | rs7715256 | GALNT10 | G | 0.431 | 0.94 | 0.70–1.27 | 0.69 |
| 6 | 34671142 | rs205262 | C6orf106 | G | 0.291 | 0.93 | 0.69–1.26 | 0.65 |
| 6 | 50944238 | rs734597a | TFAP2B | A | 0.168 | 1.21 | 0.84–1.75 | 0.30 |
| 6 | 109084356 | rs9400239 | FOXO3 | C | 0.713 | 1.37 | 1.00–1.86 | 0.048 |
| 6 | 120227364 | rs9374842 | LOC285762 | T | 0.758 | 0.83 | 0.60–1.15 | 0.26 |
| 6 | 137717234 | rs13201877 | IFNGR1 | G | 0.128 | 0.92 | 0.62–1.36 | 0.67 |
| 6 | 162953340 | rs13191362 | PARK2 | A | 0.889 | 0.90 | 0.60–1.36 | 0.62 |
| 7 | 75001105 | rs1167827 | HIP1 | G | 0.572 | 1.07 | 0.81–1.43 | 0.60 |
| 7 | 95007450 | rs6465468 | ASB4 | T | 0.296 | 0.94 | 0.70–1.27 | 0.69 |
| 8 | 76969139 | rs17405819 | HNF4G | T | 0.7 | 1.31 | 0.98–1.76 | 0.070 |
| 8 | 81538012 | rs16907751 | ZBTB10 | C | 0.888 | 0.88 | 0.57–1.33 | 0.54 |
| 8 | 85242264 | rs2033732 | RALYL | C | 0.749 | 1.10 | 0.80–1.50 | 0.56 |
| 9 | 15624326 | rs4740619 | C9orf93 | T | 0.544 | 1.77 | 1.03–3.04 | 0.036 |
| 9 | 28404339 | rs10968576 | LINGO2 | G | 0.31 | 0.84 | 0.62–1.13 | 0.26 |
| 9 | 110972163 | rs6477694 | EPB41L4B | C | 0.341 | 1.06 | 0.80–1.40 | 0.71 |
| 9 | 119418304 | rs1928295 | TLR4 | T | 0.548 | 1.02 | 0.77–1.35 | 0.91 |
| 9 | 128500735 | rs10733682 | LMX1B | A | 0.49 | 1.00 | 0.76–1.33 | 0.99 |
| 10 | 87400884 | rs7899106 | GRID1 | G | 0.052 | 2.09 | 1.11–3.99 | 0.022 |
| 10 | 102385430 | rs17094222 | HIF1AN | C | 0.216 | 0.90 | 0.65–1.26 | 0.55 |
| 10 | 104859028 | rs11191560 | NT5C2 | C | 0.091 | 1.10 | 0.68–1.81 | 0.70 |
| 10 | 114748339 | rs7903146 | TCF7L2 | C | 0.704 | 1.01 | 0.75–1.37 | 0.93 |
| 11 | 8644592 | rs7113874a | TRIM66 | C | 0.638 | 1.03 | 0.77–1.37 | 0.84 |
| 11 | 27656701 | rs7103411a | BDNF | T | 0.782 | 0.95 | 0.67–1.35 | 0.79 |
| 11 | 43820854 | rs2176598 | HSD17B12 | T | 0.264 | 0.98 | 0.71–1.35 | 0.91 |
| 11 | 47607569 | rs3817334 | MTCH2 | T | 0.409 | 1.08 | 0.82–1.42 | 0.59 |
| 11 | 114527614 | rs12286929 | CADM1 | G | 0.54 | 0.89 | 0.68–1.18 | 0.43 |
| 12 | 48533735 | rs7138803 | BCDIN3D | A | 0.388 | 1.32 | 1.00–1.76 | 0.054 |
| 12 | 121347850 | rs11057405 | CLIP1 | G | 0.921 | 0.91 | 0.55–1.47 | 0.69 |
| 13 | 26908262 | rs1885988a | MTIF3 | G | 0.17 | 0.86 | 0.59–1.26 | 0.44 |
| 13 | 53000207 | rs12429545 | OLFM4 | A | 0.132 | 1.05 | 0.72–1.54 | 0.80 |
| 13 | 65103705 | rs9540493 | MIR548X2 | A | 0.424 | 1.15 | 0.87–1.51 | 0.32 |
| 13 | 78478920 | rs1441264 | MIR548A2 | A | 0.625 | 0.91 | 0.69–1.20 | 0.52 |
| 14 | 24998019 | rs10132280 | STXBP6 | C | 0.687 | 0.99 | 0.74–1.33 | 0.96 |
| 14 | 28806589 | rs12885454 | PRKD1 | C | 0.666 | 0.75 | 0.56–1.00 | 0.049 |
| 14 | 78969207 | rs7141420 | NRXN3 | T | 0.519 | 0.97 | 0.73–1.29 | 0.83 |
| 15 | 49535902 | rs3736485 | DMXL2 | A | 0.467 | 1.24 | 0.94–1.64 | 0.13 |
| 15 | 65869870 | rs2241420a | MAP2K5 | G | 0.754 | 0.91 | 0.66–1.25 | 0.57 |
| 15 | 70881044 | rs7164727 | LOC100287559 | T | 0.684 | 1.27 | 0.94–1.71 | 0.12 |
| 16 | 3567359 | rs758747 | NLRC3 | T | 0.288 | 0.96 | 0.71–1.30 | 0.79 |
| 16 | 19842890 | rs12446632 | GPRC5B | G | 0.855 | 1.64 | 1.10–2.43 | 0.014 |
| 16 | 28240912 | rs2650492 | SBK1 | A | 0.293 | 1.09 | 0.82–1.45 | 0.57 |
| 16 | 28796987 | rs3888190 | ATP2A1 | A | 0.377 | 1.21 | 0.92–1.60 | 0.17 |
| 16 | 29922838 | rs4787491 | INO80E | G | 0.541 | 0.99 | 0.75–1.30 | 0.93 |
| 16 | 31037396 | rs9925964 | KAT8 | A | 0.619 | 1.03 | 0.78–1.37 | 0.81 |
| 16 | 47620091 | rs2080454 | CBLN1 | C | 0.356 | 1.07 | 0.80–1.42 | 0.65 |
| 16 | 52358455 | rs1421085a | FTO | C | 0.43 | 1.05 | 0.79–1.39 | 0.74 |
| 17 | 1951886 | rs9914578 | SMG6 | G | 0.196 | 0.95 | 0.68–1.34 | 0.78 |
| 17 | 5223976 | rs1000940 | RABEP1 | G | 0.305 | 1.38 | 1.02–1.88 | 0.037 |
| 17 | 76230166 | rs12940622 | RPTOR | G | 0.582 | 0.98 | 0.74–1.30 | 0.88 |
| 18 | 19358886 | rs1808579 | C18orf8 | C | 0.545 | 1.20 | 0.92–1.57 | 0.19 |
| 18 | 38401669 | rs7239883 | LOC284260 | G | 0.385 | 1.22 | 0.92–1.63 | 0.16 |
| 18 | 55034299 | rs7243357 | GRP | T | 0.818 | 1.02 | 0.71–1.45 | 0.92 |
| 18 | 55980115 | rs6567160 | MC4R | C | 0.235 | 1.04 | 0.76–1.43 | 0.79 |
| 19 | 18315825 | rs17724992 | PGPEP1 | A | 0.736 | 1.04 | 0.76–1.42 | 0.79 |
| 19 | 39001372 | rs29941 | KCTD15 | G | 0.701 | 0.93 | 0.69–1.26 | 0.65 |
| 19 | 50087459 | rs2075650 | TOMM40 | A | 0.861 | 1.06 | 0.72–1.57 | 0.76 |
| 19 | 50894012 | rs2287019 | QPCTL | C | 0.805 | 1.42 | 0.99–2.03 | 0.059 |
| 19 | 52281735 | rs2303108a | ZC3H4 | G | 0.695 | 0.95 | 0.70–1.30 | 0.77 |
| 20 | 50521269 | rs6091540 | ZFP64 | C | 0.723 | 0.84 | 0.62–1.14 | 0.28 |
| 21 | 39213610 | rs2836754 | ETS2 | C | 0.621 | 1.32 | 1.00–1.75 | 0.051 |
aSNPs rs3127553, rs1514175, rs2972143, rs7622475, rs2035935, rs4234589, rs17001561, rs734597, rs7113874, rs7103411, rs1885988, rs2241420, rs1421085, rs2303108 are in linkage disequilibrium (r2 > 0.8) with published (Locke et al., 2015) SNPs, rs657452, rs12566985, rs2176040, rs13078960, rs16851483, rs1516725, rs17001654, rs2207139, rs4256980, rs11030104, rs12016871, rs16951275, rs1558902, rs3810291, respectively. Gene names are those given in the publication that identified these SNPs (Locke et al., 2015). Association analyses are adjusted for age.
In the entire cohort, the GRS for the BMI risk alleles was positively associated with BMI (β = 0.080, P = 0.0001) with every unit increase in GRS resulting in a 0.080 kg/m2 increase in BMI. In women with PCOS and controls examined separately, the GRS association with BMI was similar, with no difference between the regression coefficients (P = 0.99).
In the women with PCOS, no significant associations were observed between the GRS and any of the other quantitative traits assessed, including total testosterone (P = 0.51), DHEAS (P = 0.09), fasting glucose (P = 0.76), fasting insulin (P = 0.20), HOMA2-IR (P = 0.52) and HOMA2-%B (P = 0.32).
Of the 92 BMI risk variants evaluated, none were associated individually with PCOS after considering multiple testing (Table V).
FTO SNP rs1421085 (r2 = 1 with rs1558902 associated with BMI (Locke et al., 2015)) was not associated with the diagnosis of PCOS (OR 1.05, 95% CI 0.79–1.39, P = 0.74); however, it was associated with BMI in the entire cohort (β = 0.071, P = 0.0006) and within the women with PCOS (β = 0.11, P = 0.0019). In the controls, rs1421085 did not quite attain a significant association with BMI (β = 0.046, P = 0.065). The association between FTO SNP rs1421085 and BMI was significantly different between PCOS subjects and controls (P = 0.033).
Discussion
This is the first study to conduct bidirectional MR between BMI and PCOS. Our MR results indicate that increased BMI is causal for PCOS, while having PCOS does not have a causal impact on BMI. In line with these results, we found that a genetic risk score based on BMI-increasing alleles was associated with PCOS. We also report detailed results centered on the FTO SNP, which is of considerable interest to PCOS researchers.
A previous study by Louwers et al., which did not include formal MR analysis, did not find an association between BMI risk alleles and PCOS independent of BMI (Louwers et al., 2014). This study also used a genetic risk score, but the score was constructed from only 12 SNPs. We similarly did not find an association between the genetic risk score and the diagnosis of PCOS when we used only the same 12 SNPs for the risk score (data not shown). In addition, compared to the NIH criteria used to diagnose PCOS in our cohort, the Rotterdam criteria used in the previous study may result in reduced power due to greater heterogeneity of cases and a higher proportion of undiagnosed cases within population based controls (up to 20%) (Yildiz et al., 2012).
In turn, a GWAS for PCOS by Day et al. (2015) in European individuals found in Mendelian randomization analysis that a GRS based on 32 BMI-raising alleles (Speliotes et al., 2010) was strongly associated with PCOS in a large sample size. In our cohort, a sub-analysis using a GRS based on the 24 available SNPs of these 32 was also associated with PCOS (OR 1.054, 95% CI 1.024–1.084, P = 0.0003), providing confirmation of the results of Day et al. in an independent cohort (no overlap in subjects).
Our study, conducting MR and GRS analysis using 92 SNPs, is the most comprehensive to date. This work is the first to conduct two-sample MR in the PCOS genetics field. Two-sample MR, in which the instrument-exposure and instrument-outcome associations are calculated in non-overlapping samples, has emerged as the preferred method for conducting MR, avoiding biases that may arise when the associations are assessed in a single sample (Hartwig et al., 2016). The large number of SNPs increased the power of the GRS, allowing us to detect association with BMI despite having a modest sample size by GWAS standards.
Another feature distinguishing our study is that we conducted sensitivity analyses with multiple MR methods, an increasingly common practice in high-quality MR studies (Wang et al., 2017; Au Yeung et al., 2018). IVW is usually employed as the primary MR method because it has the greatest power to detect effects. Though less powerful, MR-Egger is included among the sensitivity analyses because it is resistant to pleiotropy and provides a test to detect pleiotropy (intercept versus zero) (Burgess and Thompson, 2017). The finding of directionally consistent results from multiple MR methods increases the robustness of our results.
Another advantage of our study was that our cases and controls were matched for BMI, avoiding the common scenario (as seen in the GWAS (Day et al., 2015)) where cases are more obese than controls, which could confound MR and GRS or SNP association with BMI variants. In the earlier study of 12 SNPs, cases and controls were matched for mean BMI; however, unlike our study, cases and controls differed in distribution across weight groups (normal, overweight, obese) (Louwers et al., 2014).
The FTO SNP rs1421085 did not attain independent significant association with PCOS in our sample, but it was associated with obesity in the entire cohort and within the PCOS subjects. Furthermore, we found the effect size of FTO on BMI was stronger in PCOS subjects than in controls. This is consistent with prior evidence from a meta-analysis demonstrating that FTO plays a greater role in BMI in PCOS than in the general population (Wojciechowski et al., 2012). PCOS may modify the influence of FTO on BMI. This might also reflect lifestyle patterns in PCOS, as adverse diets have been found to strengthen the association of FTO variation with BMI (Qi et al., 2014). Indeed, the balance of evidence suggests greater energy consumption in women with PCOS than in unaffected women (Lin and Lujan, 2014). Whether this explains the differential association of FTO variation with BMI deserves investigation.
Recent studies suggest that the magnitude of association with BMI of genetic variants (GRS and select individual SNPs including FTO) for BMI may be stronger in younger individuals, possibly related to the environment becoming more obesogenic in recent decades (Rosenquist et al., 2015; Winkler et al., 2015; Walter et al., 2016). As the women with PCOS were younger than the controls, this could artifactually increase the magnitude of the association of genetic variants with BMI in the women with PCOS. While we did not observe this with the GRS, it is a concern for the FTO variant. This concern is mitigated by the fact that our results are similar with and without adjustment for age. Similarly, in the meta-analysis cited above, after age adjustment the effect of FTO variation on BMI remained greater in PCOS than in general cohorts (Wojciechowski et al., 2012). Large-scale studies of longitudinal cohorts of women with PCOS would be needed to fully answer whether age drives the stronger association of the FTO SNP with BMI in PCOS.
In summary, we report the first two-sample, bidirectional MR analysis examining the relationship between PCOS and BMI in cases and controls well-matched for BMI to avoid any confounding effects of higher BMI in the women with PCOS. The MR results, backed by the GRS results and the observations of others (Day et al., 2015), suggests that obesity influences the development of PCOS. On the other hand, having PCOS appears not to increase the risk of obesity. These results have significant implications for patient counseling and suggest that anti-obesity measures should be tested for their ability to prevent or treat PCOS.
Authors’ roles
M.A.B. and M.O.G. conceived and designed the study. Y.H., M.A.B., M.R.J. and X.G. contributed to analysis of the data. Y.I.C., J.I.R., R.M.K., R.S.L., and R.A. acquired the data. M.A.B. and M.O.G. drafted the article. Y.H., M.R.J., X.G., Y.I.C., J.I.R., R.M.K., R.S.L., and R.A. critically revised the article. All authors gave final approval of the version to be published.
Funding
This work was supported by National Institutes of Health Grants R01-HD29364 and K24-HD01346 (to R.A.), Grant R01-DK79888 (to M.O.G.), Grant U54-HD034449 (to R.S.L.), Grant U19-HL069757 (to R.M.K.); National Center for Research Resources Grant M01-RR00425 (to the Cedars-Sinai General Clinical Research Center), Grant U54-RR026071 (to Penn State Clinical and Translational Science Institute); Grants to the Reproductive Medicine Network (U10-HD038998, U10-HD055925, U10-HD039005, U10-HD027049, U10-HD055944, U10-HD055942, U10-HD055936, U10-HD038992); the Eris M. Field Chair in Diabetes Research (to M.O.G.); and an endowment from the Helping Hand of Los Angeles, Inc. MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420. MESA Cardiometabochip genotyping data was supported in part by grants and contracts R01HL98077, N02-HL-64278, HL071205, UL1TR001881, P30-DK063491, RD831697, and P50 ES015915. Also supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant P30-DK063491 to the Southern California Diabetes Endocrinology Research Center. Although the research described in this presentation has been funded in part by the United States Environmental Protection Agency through RD831697 to the University of Washington, it has not been subjected to the Agency’s required peer and policy review and therefore does not necessarily reflect the views of the Agency and no official endorsement should be inferred.
Conflict of interest
The funders had no influence on the data collection, analyses or conclusions of the study. There are no conflict of interests to declare.
References
- Au Yeung SL, Luo S, Schooling CM. The impact of glycated hemoglobin (HbA1c) on cardiovascular disease risk: a Mendelian randomization study using UK Biobank. Diabetes Care 2018;41:1991–1997. [DOI] [PubMed] [Google Scholar]
- Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, Janssen OE, Legro RS, Norman RJ, Taylor AE et al. The Androgen Excess and PCOS Society criteria for the polycystic ovary syndrome: the complete task force report. Fertil Steril 2009;91:456–488. [DOI] [PubMed] [Google Scholar]
- Barber TM, Bennett AJ, Groves CJ, Sovio U, Ruokonen A, Martikainen H, Pouta A, Hartikainen AL, Elliott P, Lindgren CM et al. Association of variants in the fat mass and obesity associated (FTO) gene with polycystic ovary syndrome. Diabetologia 2008;51:1153–1158. [DOI] [PubMed] [Google Scholar]
- Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR Jr, Kronmal R, Liu K et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol 2002;156:871–881. [DOI] [PubMed] [Google Scholar]
- Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol 2013;42:1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 2017;32:377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Hinds DA, Tung JY, Stolk L, Styrkarsdottir U, Saxena R, Bjonnes A, Broer L, Dunger DB, Halldorsson BV et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun 2015;6:8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day F, Karaderi T, Jones MR, Meun C, He C, Drong A, Kraft P, Lin N, Huang H, Broer L et al. Large-scale genome-wide meta analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. bioRxiv 2018:290502 doi:https://doi.org/290510.291101/290502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens KG, Jones MR, Ankener W, Stewart DR, Urbanek M, Dunaif A, Legro RS, Chua A, Azziz R, Spielman RS et al. FTO and MC4R gene variants are associated with obesity in polycystic ovary syndrome. PLoS One 2011;6:e16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi MO, Dumesic DA, Chazenbalk G, Azziz R. Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nat Rev Endocrinol 2011;7:219–231. [DOI] [PubMed] [Google Scholar]
- Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol 2016;45:1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Chua AK, Mengesha EA, Taylor KD, Chen YD, Li X, Krauss RM, Rotter JI. Reproductive Medicine Network, Legro RS et al.. Metabolic and cardiovascular genes in polycystic ovary syndrome: a candidate-wide association study (CWAS). Steroids 2012;77:317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JC, Matthews DR, Hermans MP. Correct homeostasis model assessment (HOMA) evaluation uses the computer program. Diabetes Care 1998;21:2191–2192. [DOI] [PubMed] [Google Scholar]
- Li T, Wu K, You L, Xing X, Wang P, Cui L, Liu H, Cui Y, Bian Y, Ning Y et al. Common variant rs9939609 in gene FTO confers risk to polycystic ovary syndrome. PLoS One 2013;8:e66250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SS, Norman RJ, Davies MJ, Moran LJ. The effect of obesity on polycystic ovary syndrome: a systematic review and meta-analysis. Obes Rev 2013;14:95–109. [DOI] [PubMed] [Google Scholar]
- Lin AW, Lujan ME. Comparison of dietary intake and physical activity between women with and without polycystic ovary syndrome: a review. Adv Nutr 2014;5:486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ. Genetic determinants of common obesity and their value in prediction. Best Pract Res Clin Endocrinol Metab 2012;26:211–226. [DOI] [PubMed] [Google Scholar]
- Louwers YV, Rayner NW, Herrera BM, Stolk L, Groves CJ, Barber TM, Uitterlinden AG, Franks S, Laven JS, McCarthy MI. BMI-associated alleles do not constitute risk alleles for polycystic ovary syndrome independently of BMI: a case-control study. PLoS One 2014;9:e87335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybacka A, Carlstrom K, Stahle A, Nyren S, Hellstrom PM, Hirschberg AL. Randomized comparison of the influence of dietary management and/or physical exercise on ovarian function and metabolic parameters in overweight women with polycystic ovary syndrome. Fertil Steril 2011;96:1508–1513. [DOI] [PubMed] [Google Scholar]
- Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 2011;40:740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol 2013;178:1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Chu AY, Kang JH, Huang J, Rose LM, Jensen MK, Liang L, Curhan GC, Pasquale LR, Wiggs JL et al. Fried food consumption, genetic risk, and body mass index: gene-diet interaction analysis in three US cohort studies. BMJ 2014;348:g1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist JN, Lehrer SF, O’Malley AJ, Zaslavsky AM, Smoller JW, Christakis NA. Cohort of birth modifies the association between FTO genotype and BMI. Proc Natl Acad Sci USA 2015;112:354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salley KE, Wickham EP, Cheang KI, Essah PA, Karjane NW, Nestler JE. Glucose intolerance in polycystic ovary syndrome—a position statement of the Androgen Excess Society. J Clin Endocrinol Metab 2007;92:4546–4556. [DOI] [PubMed] [Google Scholar]
- Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, Rotter JI, Nickerson DA, Yang H, Saad M et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol 2006;97:843–850. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010;42:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkard AJ, Harris JR, Pedersen NL, McClearn GE. The body-mass index of twins who have been reared apart. N Engl J Med 1990;322:1483–1487. [DOI] [PubMed] [Google Scholar]
- Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab 2006;91:2100–2104. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, Burtt NP, Fuchsberger C, Li Y, Erdmann J et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet 2012;8:e1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S, Mejia-Guevara I, Estrada K, Liu SY, Glymour MM. Association of a genetic risk score with body mass index across different birth cohorts. JAMA 2016;316:63–69. [DOI] [PubMed] [Google Scholar]
- Wang Q, Holmes MV, Davey Smith G, Ala-Korpela M. Genetic support for a causal role of insulin resistance on circulating branched-chain amino acids and inflammation. Diabetes Care 2017;40:1779–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S, Czajkowski J, Esko T, Fall T, Kilpelainen TO et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet 2015;11:e1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski P, Lipowska A, Rys P, Ewens KG, Franks S, Tan S, Lerchbaum E, Vcelak J, Attaoua R, Straczkowski M et al. Impact of FTO genotypes on BMI and weight in polycystic ovary syndrome: a systematic review and meta-analysis. Diabetologia 2012;55:2636–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz BO, Bozdag G, Yapici Z, Esinler I, Yarali H. Prevalence, phenotype and cardiometabolic risk of polycystic ovary syndrome under different diagnostic criteria. Hum Reprod 2012;27:3067–3073. [DOI] [PubMed] [Google Scholar]