Abstract
Background
The occurrence of Salmonella enterica subspecies diarizonae serovar 61: k: 1, 5, (7) (SASd) and other Salmonella organisms in sheep in the German federal state of Thuringia was examined for the first time. Pooled faecal samples from 90 flocks located in this state were monitored.
Results
Only SASd was detected in 74 (82.2%) out of the 90 sheep herds, other Salmonella serovars were not identified. A positive correlation was found between the flock size and the detection probability of SASd. Despite the agent’s high prevalence, clinical symptoms of a disease exclusively due to SASd have not been observed. The SASd strains were characterised by macrorestriction analysis, antimicrobial testing and the biochemical profile. All strains were sensitive to 13 out of 14 antimicrobial substances and resistant to only sulfamethoxazole. The high number of macrorestriction groups of SASd strains indicated a low clonality of the serovar.
Conclusions
Data from sheep derived foods and public health data in Germany strongly suggest that the significance of SASd for public health is considerably lower than that of serovars belonging to Salmonella enterica subspecies enterica. For this reason and because of the low disease-causing potential of SASd in sheep, it is worthwile to consider a reduction in ongoing activities from combating to monitoring serovar 61: k: 1, 5, (7) in the sheep population.
Keywords: Salmonella diarizonae, Sheep, Germany, Epidemiology, Zoonotic potential
Background
Sheep can be infected with a wide range of Salmonella enterica serovars that are not restricted to ovine and do not establish an endemic pattern [1]. The host-specific serovar Salmonella enterica subspecies enterica serovar Abortus-ovis represents a common cause of abortion and mortality in young lambs in endemic areas of Europe and Western Asia [1, 2]. While the sheep associated serovar Salmonella enterica subspecies diarizonae serovar 61: k: 1, 5, (7) (SASd) is also considered as host-adapted [3–6], it displays a very different epidemiological pattern. SASd is able to produce both intestinal and extra-intestinal infections with faecal, vaginal and nasal colonisation as well as shedding, but mostly without clinical disease [7–9]. However, it has also been reported that SASd may occasionally cause rhinitis or chronic nasal inflammation [10, 11], orchitis [12], or aborted foetuses [4]. As these properties deviate from the classical characteristics of host-restricted, host-adapted or ubiquitous serovars [13], the term sheep associated serovar [6] appears most appropriate to characterise serovar 61: k: 1, 5, (7).
A few studies have been undertaken to investigate the prevalence of SASd and other Salmonella serovars in sheep, particularly in the United Kingdom [5, 14], Sweden [6], Norway [9], USA [15], Iceland [16] and Switzerland [17]. Among these countries, SASd prevalence rates in sheep flocks varied from 11% [17] to more than 70% [15] which is very likely due to different study designs. Nevertheless, SASd was the predominant serovar in all these countries [6, 9, 14, 15, 17] except for Iceland [16]. Data specifying the occurrence of Salmonella spp. or SASd in the German sheep population are not available.
Therefore, the aim of this study was to obtain information on the occurrence of Salmonella spp. in sheep throughout the federal state of Thuringia. Furthermore, the isolated Salmonella organisms were characterised phenotypically and genotypically to explore a possible epidemiological connection.
Methods
Sampling of sheep flocks and bacteriology
A total of 90 sheep flocks of different size (9 categories of herd size) located throughout the German federal state of Thuringia (Table 1, Fig. 1) were monitored over a period of 1 year. Herds from each Thuringian region were selected, which are representative in terms of sheep density, size and management system. In each herd, 3 pooled faecal samples (ca. 25–30 g) consisting each of fresh sheep faeces from at least 10 different places, in order to represent at least 10 different animals and several parts of the herd were collected (in total 270 samples from 90 herds). Individual samples were either taken rectally or collected from fresh droppings without environmental contamination. The pooled samples were kept in plastic containers until bacteriological examination started on the day of sampling. Analysis of faecal samples was carried out according to ISO 6579-1 [18]. Buffered peptone water, modified semi-solid Rappaport-Vassiliadis, xylose lysine deoxycholate agar and Rambach agar were applied for the analysis. Furthermore, deoxycholate-citrate agar was used to possibly increase the isolation rate of SASd (all from SIFIN, Berlin Germany). All Salmonella isolates were serotyped using poly- and monovalent anti-O as well as anti-H sera (SIFIN) according to the Kauffmann-White scheme [19]. The biochemical profile of Salmonella strains originating from different regions was determined using the identification system API 20E (bioMerieux, Nürtingen, Germany). Furthermore, sheep flocks were also observed for signs of morbidity and the history of clinical symptoms in the flock was assessed.
Table 1.
Occurrence of Salmonella serovar 61: k: 1, 5, (7) in sheep flocks of different size in the federal state Thuringia in Germany
| Herd size (number of animals) | Number of herds sampled | Number of positive herds | % of positive herds |
|---|---|---|---|
| 1–30 | 15 | 3 | 20.0 |
| 31–50 | 5 | 4 | 80.0 |
| 51–100 | 7 | 6 | 85.7 |
| 101–300 | 4 | 4 | 100 |
| 301–500 | 23 | 21 | 91.3 |
| 501–750 | 18 | 18 | 100 |
| 751–1.000 | 9 | 9 | 100 |
| 1.001–1.500 | 5 | 5 | 100 |
| 1.501–3.000 | 4 | 4 | 100 |
| Total | 90 | 74 | 82.2 |
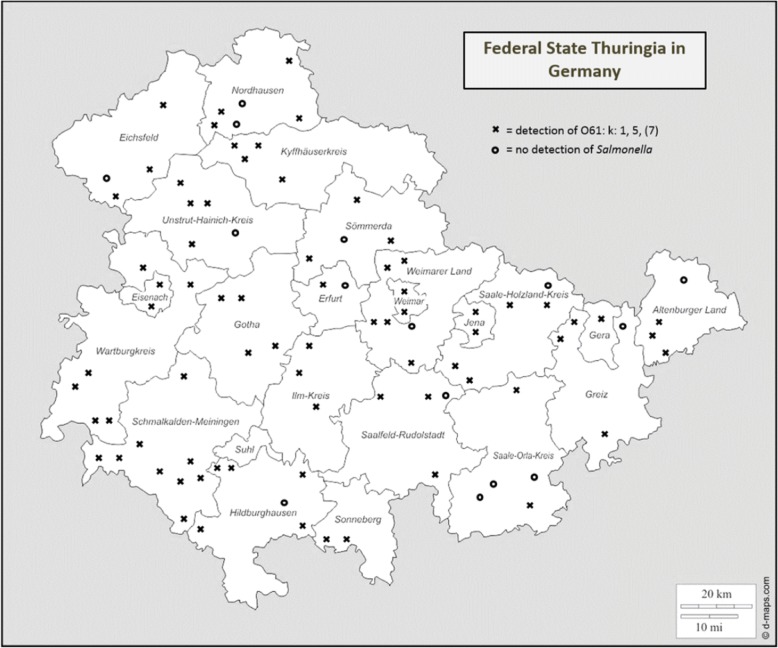
Fig. 1.
Location of sheep flocks sampled for Salmonella in the federal state Thuringia in Germany. (source: https://d-maps.com/carte.php?num_car=24110&lang=de)
Antimicrobial susceptibility testing
Antimicrobial susceptibilities of the Salmonella strains detected in sheep flocks were assessed by determining the minimum inhibitory concentration (MIC) using the broth microdilution method with Sensititre™ EUVSEC plates (Trek Diagnostic Systems Ltd., East Grinstead, United Kingdom). Epidemiological cut-off values according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) were used [20]. Antimicrobial susceptibilities to sulfamethoxazole, trimethoprim, ciprofloxacin, tetracycline, meropenem, azithromycin, nalidixic acid, cefotaxime, chloramphenicol, tigecycline, ceftazidime, colistin, ampicillin and gentamicin were examined.
Genotyping using pulsed-field gel electrophoresis (PFGE)
Macrorestriction analysis was carried out as described [21]. Isolates were considered to be different when a one-band difference between fragments over 70 kb was observed [21, 22].
Statistical analysis
A logistic regression was performed between the two variables infection status as the dichotom dependent variable and herd size (category as nominal variable 1 to 9, Table 1) as independent variable. Statistical evaluation was conducted using MedCalc Software.
Results
Bacteriological analysis
Pooled faecal samples (n = 270) from 90 farms were examined for the occurrence of Salmonella organisms (Fig. 1). Only SASd was detected in 74 (82.2%) out of the 90 sheep flocks, other Salmonella serovars were not found (Table 1). In total, 206 (76.3%) pooled samples were positive for SASd and 64 (23.7%) were negative. The odds ratio for herd size was 2.285 with a 95% confidence interval from 1.878 to 2.779 (p < 0.0001). Therefore, the parameter herd size is a highly significant variable for the prediction of the infection status of the flock.
In nearly all flocks with more than 100 animals the sheep associated SASd was detected. Indeed, even in flocks with only 30–100 animals, 80 -86% of the flocks were positive for SASd. All herds with more than 500 animals showed a prevalence of 100%. The prevalence value of 100% for the herd size 101–300 is probably an outlier because only 4 herds were sampled. In all flocks with proven SASd in faecal samples no signs of morbidity exclusively due to this serovar were observed and the history of the flocks did not reveal any evidence for clinical symptoms caused by SASd.
Antimicrobial susceptibilities
All 74 SASd strains tested were sensitive to 13 out of 14 antimicrobials. The organisms were all resistant to sulfamethoxazole (MIC > 512 μg/ml). Level of susceptibility against the other antimicrobial substances did not reveal differences between the strains, the resistance patterns of the SASd were nearly identical. MIC values (μg/ml) of the strains were as follows: trimethoprim (1–2), ciprofloxacin (0.06), tetracycline (4–8), meropenem (< 0.03), azithromycin (8–16), nalidixic acid (8), cefotaxime (< 0.25), chloramphenicol (< 8), tigecycline (1–2), ceftazidime (< 0.05), colistin (< 1), ampicillin (2) and gentamicin (1). Further differentiation among the SASd strains based on the resistance pattern was not possible.
Biochemical characterisation
SASd strains were tested using API 20E and identified as Salmonella. They all showed a positive ONPG reaction due to their β-galactosidase production which is characteristic for Salmonella enterica subspecies arizonae and diarizonae [19] and might, however, complicate the isolation of SASd organisms [22]. Also, tests for lysine decarboxylase, production of hydrogen sulphide and production of acid from sorbitol varied between the SASd strains examined. Therefore, “Analytical Profile Indices” of the SASd strains were I for 7,704,552 (lysine decarboxylase +), II for 3,704,552 (lysine decarboxylase -) and III for 3,704,152 (lysine decarboxylase -, sorbitol -) (Table 2). The different API 20E indices I, II, or III were not used to generate a further distinction in addition to the macrorestriction groups, since a correlation between both features was missing. Because of the biochemical characteristics of SASd, the use of more selective media than required by ISO 6579-1 [19] (e.g. deoxycholate-citrate agar) very likely raised the number of SASd strains found in sheep flocks.
Table 2.
Characteristics of strains of Salmonella serovar 61: k: 1, 5, (7)
| Strain | Analytical profile index | Macrorestriction pattern | Macro-restriction groupc | |
|---|---|---|---|---|
|
Xba I Xa |
Spe I Sb |
|||
| 1478 | II | 5 | 2 | E |
| 1513 | II | 1 | 4 | A |
| 1515 | I | 2 | 1 | B |
| 1520 | I | 2 | 1 | B |
| 1521 | III | 1 | 4 | A |
| 1529 | II | 3 | 2 | C |
| 1530 | II | 3 | 2 | C |
| 1544 | II | 5 | 2 | E |
| 1546 | III | 3 | 2 | C |
| 1547 | III | 3 | 2 | C |
| 1552 | III | 5 | 6 | K |
| 1561 | II | 6 | 2 | L |
| 1563 | I | 2 | 1 | B |
| 1569 | II | 4 | 5 | D |
| 1570 | III | 5 | 2 | E |
| 1575 | II | 9 | 2 | M |
| 1588 | III | 3 | 2 | C |
| 1641 | III | 5 | 2 | E |
| 1645 | I | 8 | 3 | H |
| 1646 | III | 3 | 2 | C |
| 1649 | II | 6 | 6 | F |
| 1650 | III | 7 | 2 | G |
| 1651 | III | 7 | 2 | G |
| 1674 | II | 8 | 3 | H |
| 1675 | III | 5 | 2 | E |
| 1676 | II | 9 | 3 | I |
| 1677 | I | 2 | 1 | B |
| 1679 | I | 2 | 4 | J |
| 1680 | III | 1 | 4 | A |
Biochemical index: I: 7704552 (lysine decarboxylase +), II: 3704552 (lysine decarboxylase -), III: 3704152 (lysine decarboxylase -, sorbitol -)
a,Pattern numbers correspond to lane numbers in Fig. 2
bPattern numbers correspond to lane numbers in Fig. 3
cMacrorestriction group as combination of macrorestricion patterns
Macrorestriction analysis
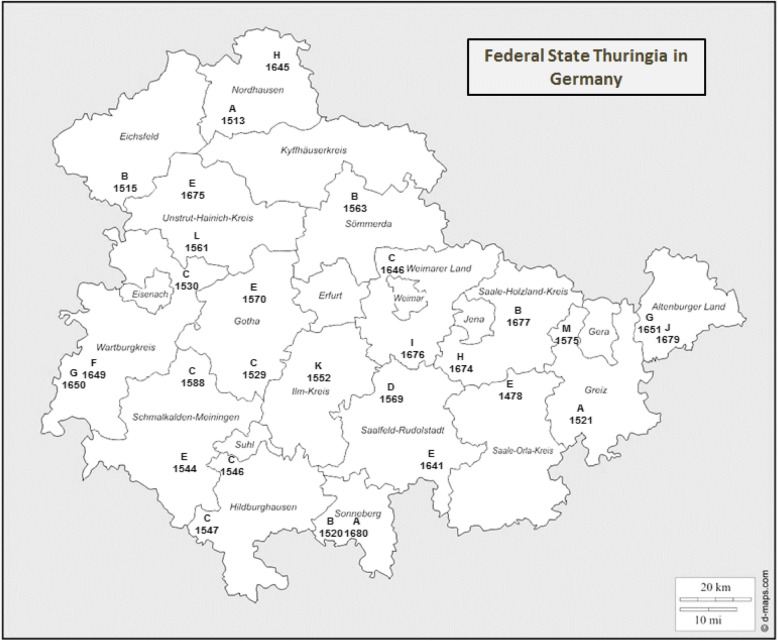
Two different restriction endonucleases, XbaI and SpeI (Figs. 2 and 3) were used to cleave whole-cell DNA of 29 SASd isolates originating from different regions and flocks in Thuringia (Table 2, Fig. 4). XbaI digestion yielded nine (X1 to X9) different patterns, SpeI digestion resulted in six (S1 to S6) patterns. Therefore, the SASd strains tested revealed a high genetic diversity. Results of macrorestriction analysis allowed the assignment of the 29 strains examined in this study to 13 macrorestriction groups A to M (Table 2). In most cases, strains belonging to the same macrorestriction group (e.g. A for strains 1513, 1680 or B for strains 1515, 1520, 1563, 1677) were located apart from each other in very different regions of Thuringia. Only group C occurred more frequently in different related counties of the federal state (Fig. 4).
Fig. 2.
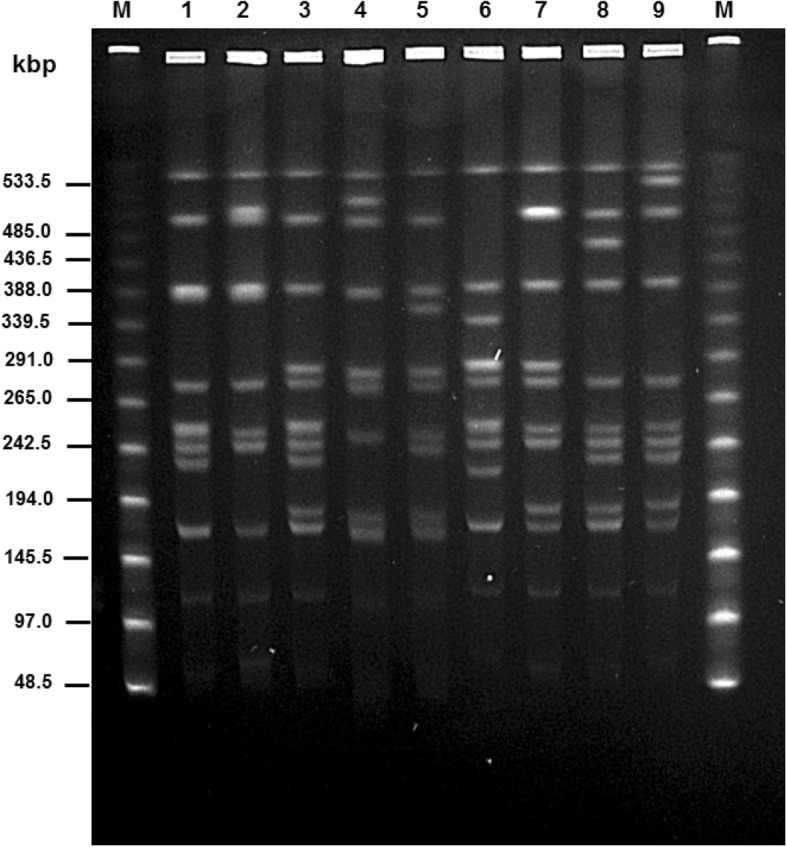
XbaI (X1-X9) macrorestriction patterns of Salmonella strains 61: k: 1, 5, (7)
Fig. 3.
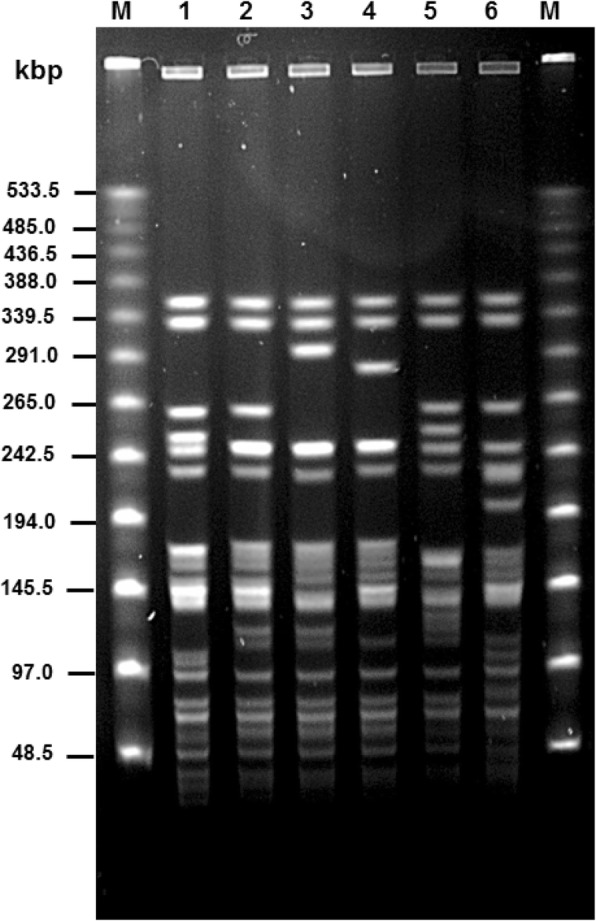
Spe I (S1-S6) macrorestriction patterns of Salmonella strains 61: k: 1, 5, (7)
Fig. 4.
Distribution of macrorestriction groups (A-M) of Salmonella serovar 61: k: 1, 5, (7) strains in sheep flocks in the federal state Thuringia in Germany. (source: https://d-maps.com/carte.php?num_car=24110&lang=de)
Discussion
In the sheep population of the federal state of Thuringia, only the serovar 61: k: 1, 5, (7) was detected in more than 80% of the examined flocks. The dominant appearance of SASd in sheep confirms study results from other countries. However, the detected prevalence of 82% in this study is high compared to data from other countries, such as Norway (20%) [9], Switzerland (11%) [17], USA (72%) [15] or Sweden (12–40%) [6]. Real differences probably exist between countries, but the variation will also be heavily influenced by different study designs, sampling methods or sampling material. In view of the moderate to high prevalence of SASd in different countries, the occurrence of this serovar seems to be due to a long lasting persistence in sheep flocks because of its host associated characteristics. Other Salmonella serovars that are not confined to sheep are obviously rare and do not establish an epidemic pattern, and their occurrence appears to be related to pastures contaminated with wild bird faeces [2].
Despite the considerable detection rate of SASd in sheep flocks, it might be possible that the real incidence in sheep is even higher because of the biochemical features of this serovar [14]. The lactose positive SASd organisms might constitute a special diagnostic problem, therefore, serovars of Salmonella subspecies diarizonae may slip through undetected [19]. Hence, in addition to method ISO 6579-1 [18], it is recommended to use further indicator media when SASd strains are suspected [23]. This is supported by this study as the variation also in the production of hydrogen sulphide between SASd strains observed hampers the detection when xylose lysine deoxycholate agar is used.
Antimicrobial testing revealed that SASd organisms from 74 sheep flocks in Thuringia were sensitive to 13 of 14 antimicrobials. All strains were resistant to only sulfamethoxazole, confirming results from earlier studies [6, 22] that did not find resistant SASd strains. Therefore, the risk of transferring antimicrobial resistances via SASd strains from sheep to other animal hosts or humans might be considered as negligible.
Although a wide distribution of SASd was found in this study, clinical symptoms of a disease exclusively due to SASd have not been detected in that period. Despite signs of rhinitis, nasal inflammation or aborted foetuses which were noted occasionally in different studies [4, 10, 11], it cannot be ruled out that factors other than SASd were involved in producing these clinical signs. This occasional occurrence of clinical symptoms is also supported by the observation that experimental infections of sheep with SASd indeed resulted in intestinal or nasal colonisation but not in the induction of clinical signs of a disease [24, 25]. The lack of clinical symptoms in sheep flocks harbouring SASd [6, 9] has been regularly observed.
In this study, SASd was detected in nearly all flocks with more than 100 sheep and even in 80 -86% of sheep flocks with only 30–100 animals. A positive correlation was found between the increasing flock size and the increasing probability of detecting SASd. A higher SASd prevalence in larger sheep flocks was also found in earlier studies [26] which might be due to the more successful and long-lasting persistence of SASd in such flocks. Others [9] found a low within-flock prevalence regardless of the flock size, indicating that the transmission rate of the organism is limited. However, comprehensive information on the infection routes for both within the flock and from herd to herd are still missing. The role of the “ram circle”, the exchange of rams between different farms, in the transmission of SASd between flocks has been discussed [9, 26], though, clear evidence is not available. Even the mode of spreading of the organism between single animals is not completely known, and detailed studies on infection routes are still needed.
To obtain information on a possible epidemiological connection and on the distribution of SASd in the Thuringian sheep population macrorestriction analysis of a representative number of isolates originating from different regions and flocks was carried out. Because of the high degree of similarity in antimicrobial resistance pattern of SASd and the missing correlation between biochemical index and macrorestriction pattern, only results of the macrorestriction analysis were used to generate macrorestriction groups [21] for discrimination among SASd strains. The high number of genotypes revealed after digestion with both XbaI and SpeI resulted in a high number of macrorestriction groups, indicating low clonality of SASd. As most macrorestriction groups were dispersed throughout the federal state and because of the lack of epidemiological data on risk factors for transmission routes it was not possible to make conclusions on an epidemiological context of SASd organisms in Thuringia. Despite the more frequent occurrence of macrorestriction group C in related counties, reasons for an exchange of this SASd group between the sheep herds could not be identified. Comparable data from macrorestriction studies of SASd are rare [17, 22, 27] and cannot be compared directly with results of this study.
The wide distribution of SASd in sheep flocks in Thuringia also raises the question on its zoonotic potential. How to deal with findings of SASd in sheep flocks? In contrast to poultry, cattle and pigs, there is no regulation in place on the control of Salmonella infections in sheep in Germany. Despite the likely moderate to high prevalence of SASd in the German sheep population, meat, meat products and cheese from sheep are very rarely contaminated with Salmonella organisms, SASd has not been isolated from these foods [28]. Human infections caused by serovar 61: k: 1, 5, (7) have not been notified in Germany since 2000, so that the significance of SASd for public health is negligible compared to that of Salmonella enterica subspecies enterica serovars [29].
Therefore, SASd seemed merely to be a commensal and colonising inhabitant of the intestine and the upper respiratory tract in healthy sheep, which may occasionally become invasive only in debilitated animals. The pathogenic significance of SASd as monocausal agent is considered as low since clinical signs in sheep flocks harbouring SASd are, consistent to results of this study, not regularly observed [6, 9]. It is also concluded that control measures applied upon findings of SASd in sheep have very little impact on reducing risks to human health and that measures to eradicate this serovar from sheep herds will probably not reduce its prevalence in the sheep population [6]. For these reasons, Sweden was the first country to make an exception for serovar SASd in Salmonella control [6], thus reducing activities from combating to monitoring serovar SASd in sheep. Despite the limited but confirming data on the disease-causing and zoonotic potential of SASd, the Swedish strategy could also be a guideline for Germany, which is supported also by the results of this study. Nevertheless, given the special and interesting characteristics of SASd and the persisting lack of knowledge on the infection, further studies on the pathogenicity and transmission routes of this organism in sheep will be most valuable.
Conclusions
In the sheep population of the German federal state of Thuringia, only Salmonella enterica subspecies diarizonae serovar 61: k: 1, 5, (7) (SASd) was detected in more than 80% of the examined flocks. Antimicrobial testing revealed that all SASd organisms from sheep flocks in Thuringia were sensitive to 13 of 14 antimicrobials, therefore, the risk of transferring antimicrobial resistances via SASd to humans can be considered as low. Despite the high prevalence of this agent, clinical symptoms of a disease exclusively due to SASd have not been observed. A high number of macrorestriction groups of SASd strains were found indicating a low clonality of the serovar. Data from sheep derived foods and public health data in Germany strongly suggest that the significance of SASd for public health is considerably lower than that of Salmonella enterica subspecies enterica serovars. For this reason and because of the low disease-causing potential of SASd in sheep, in accordance with strategies in other countries, it is worthwile to consider a reduction in the ongoing activities from combating to monitoring SASd in the sheep population also in Germany.
Acknowledgments
The skilled technical assistance of Silke Keiling and Lisa Rüter is gratefully acknowledged. We particularly thank Dr. Roland Diller, Dr. Timo Homeier-Bachmann and Susanne Amler for their advice and evaluation of the statistical analysis. Special thanks are due to Prof. Dr. Konrad Sachse for critical reading of the manuscript and language editing.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Abbreviations
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- MIC
Minimum inhibitory concentration
- S.
Salmonella
- SASd
Salmonella enterica subspecies diarizonae serovar 61: k: 1, 5, (7)
Authors’ contributions
UM1 coordinated the study, analysed the sample material, performed data evaluation and wrote the manuscript. UM2 coordinated and carried out sampling and health evaluation of the flocks. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was performed in accordance with the German Animal Protection Act and approved by an ethical committee (registration number: 2684-04-15-BFI-18-101). The collection of specimens was in accordance with local legislation. Consent to collect the samples was obtained from the animal owners verbal. No methods of euthanasia/ anesthesia were employed.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ulrich Methner, Phone: + 493641-804-2267, Email: ulrich.methner@fli.de.
Udo Moog, Email: umoog@thueringertierseuchenkasse.de.
References
- 1.Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernhard S, Casadesus J, Platt DJ, Olson JE. Host adapted serotypes of Salmonella enterica. Epidemiol Infect. 2000;125:229–255. doi: 10.1017/S0950268899004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uzzau S. Salmonella infections in sheep. In: Barrow PA, Methner U, editors. Salmonella in domestic animals. Second Ed. CABI international. 2013. pp. 295–304. [Google Scholar]
- 3.Greenfield J, Greenway JA, Bigland CH. Arizona infections in sheep associated with gastroenteritis and abortion. Vet Rec. 1973;92:400–401. doi: 10.1136/vr.92.15.400. [DOI] [PubMed] [Google Scholar]
- 4.Long JR, Finley GG, Clark MH, Rehmtulla AJ. Ovine fetal infection due to Salmonella arizonae. Can Vet J. 1978;19:260–263. [PMC free article] [PubMed] [Google Scholar]
- 5.Hall MLM, Rowe B. Salmonella arizonae in the United Kingdom from 1966 to 1990. Epidemiol Infect. 1992;108:59–65. doi: 10.1017/S0950268800049505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sören K, Lindblad M, Jernberg C, Eriksson E, Melin L, Wahlström H, Lundh M. Changes in the risk management of Salmonella enterica subspecies diarizonae serovar 61: k: 1, 5, (7) in Swedish sheep herds and sheep meat due to the results of a prevalence study 2012. Acta Vet Scand. 2015;57:1–7. doi: 10.1186/s13028-015-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sojka WJ, Wray C, Shreeve JE, Bell JC. The incidence of Salmonella infection in sheep in England and Wales, 1975 to 1981. Br Vet J. 1983;139:386–392. doi: 10.1016/S0007-1935(17)30383-4. [DOI] [PubMed] [Google Scholar]
- 8.Pritchard J. Salmonella arizonae in sheep. Can Vet J. 1990;31:42. [PMC free article] [PubMed] [Google Scholar]
- 9.Sandberg M, Alvseike O, Skjerve E. The prevalence and dynamics of Salmonella enterica IIIb 61: k: 1, 5, (7) in sheep flocks in Norway. Prev Vet Med. 2002;52:267–275. doi: 10.1016/S0167-5877(01)00250-1. [DOI] [PubMed] [Google Scholar]
- 10.Lacastra D, Ferrer LM, Ramos JJ, Buesco JP, Borobia M, Ruiz de Arcaute M, Figueras L, Gonzalez-Sainz JM, De las Heras M. Chronic proliferative rhinitis associated with Salmonella enterica subspecies diarizonae serovar 61:k:1, 5, (7) in sheep in Spain. J Comp Pathol. 2012;147:406–409. doi: 10.1016/j.jcpa.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Stokar-Regenscheit N, Overesch G, Giezendanner R, Roos S, Gurtner C. Salmonella enterica subspecies diarizonae serotype 61:k:1, 5, (7) associated with chronic proliferative rhinitis and high nasal colonization rates in a flock of Texel sheep in Switzerland. Prev Vet Med. 2017;145:78–82. doi: 10.1016/j.prevetmed.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Ferreras MC, Munoz M, Perez V, Benavides J, Garcia-Pariente C, Fuertes M, Aduriz G, Garcia-Marin JF. Unilateral orchitis and epididymitis caused by Salmonella enterica subspecies diarizonae infection in a ram. J Vet Diagn Investig. 2007;19:194–197. doi: 10.1177/104063870701900211. [DOI] [PubMed] [Google Scholar]
- 13.Sanderson KE, Nair S. Taxonomy and species concepts in the genus Salmonella. In: Barrow PA, Methner U, editors. Salmonella in domestic animals. Second Ed. CABI international. 2013. pp. 1–19. [Google Scholar]
- 14.Davies RH, Evans SJ, Preece BE, Chappell S, Kidd S, Jones YE. Increase in Salmonella enterica subspecies diarizonae serovar 61: k: 1, 5, (7) in sheep. Vet Rec. 2001;149:555–557. doi: 10.1136/vr.149.18.555. [DOI] [PubMed] [Google Scholar]
- 15.Dargatz DA, Marshall KL, Fedorka-Cray P, Erdman MM, Kopral CA. Salmonella prevalence and antimicrobial susceptibility from the national animal health monitoring system sheep 2011 study. Foodborne Pathogens Dis. 2015;12:953–957. doi: 10.1089/fpd.2015.2016. [DOI] [PubMed] [Google Scholar]
- 16.Hjartardottir S, Gunnarson E, Sigvaldadottir J. Salmonella in sheep in Iceland. Acta Vet Scand. 2002;43:43–48. doi: 10.1186/1751-0147-43-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zweifel C, Zychowska MA, Stephan R. Prevalence and characteristics of Shiga toxin-producing Escherichia coli, Salmonella spp. and Campylobacter spp. isolated from slaughtered sheep in Switzerland. Int J Food Microbiol. 2004;92:45–53. doi: 10.1016/j.ijfoodmicro.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 18.ISO 6579-1. Microbiology of the food chain – horizontal method for the detection, Enumeration and serotyping of Salmonella - part 1: detection of Salmonella spp. 2017. [DOI] [PubMed]
- 19.Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella. Paris: Institute Pasteur; 2007. [Google Scholar]
- 20.EUCAST. Breakpoints tables for interpretation of MICs and zone diameters. Version 3.1. 2013. http://www.eucast.org
- 21.Methner U, Heller M, Bocklisch H. Salmonella enterica subspecies enterica serovar Choleraesuis in a wild boar population in Germany. Eur J Wildl Res. 2010;56:493–402. doi: 10.1007/s10344-009-0339-3. [DOI] [Google Scholar]
- 22.Bonke R, Wacheck S, Bumann C, Thum C, Stüber E, König M, Stephan R, Frederiksson-Ahomaa M. High prevalence of Salmonella enterica subsp. diarizonae in tonsils of sheep at slaughter. Food Res Int. 2012;45:880–884. doi: 10.1016/j.foodres.2011.01.050. [DOI] [Google Scholar]
- 23.Alvseike O, Skjerve E. Probability of detection of Salmonella using different analytical procedures, with emphasis on subspecies diarizonae serovar 61: k: 1, 5, (7) [S. IIIb 61: k: 1, 5, (7)] Int J Food Microbiol. 2000;58:49–58. doi: 10.1016/S0168-1605(00)00261-0. [DOI] [PubMed] [Google Scholar]
- 24.Hannam DAR, Wray C, Harbourne JF. Experimental Salmonella arizonae inection of sheep. Br Vet J. 1986;142:458–466. doi: 10.1016/0007-1935(86)90048-5. [DOI] [PubMed] [Google Scholar]
- 25.Brogden KA, Meehan JT, Lehmkuhl HD. Salmonella arizonae infection and colonisation of the upper respiratory tract of sheep. Vet Rec. 1994;135:410–411. doi: 10.1136/vr.135.17.410. [DOI] [PubMed] [Google Scholar]
- 26.Alvseike O, Skjerve E. Prevalence of Salmonella subspecies diarizonae in Norwegian sheep herds. Prev Vet Med. 2002;52:277–285. doi: 10.1016/S0167-5877(01)00252-5. [DOI] [PubMed] [Google Scholar]
- 27.Alvseike O, Vardund T, Lindstedt B, Heir E, Eriksson E, Kapperud G. Molecular epidemiology and population genetics of Salmonella subspecies diarizonae in sheep in Norway and Sweden. Epidemiol Infect. 2004;132:253–261. doi: 10.1017/S0950268803001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.BfR. Agents of zoonoses in Germany in 2014. BfR Wissensch 2016;06/2016:275 pp. www.bfr.bund.de.
- 29.www.rki.de/SurvStat/. Data on notifiable diseaes according the German law on prevention and control of infectious diseases.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.