Abstract
Few studies have examined the interaction of human geography, microbial community structure and obesity. We tested obese adult volunteers from France, Saudi Arabia, French Polynesia and from a traditional population in the village of Trois-Sauts in French Guiana by sequencing the V3-V4 region. We also sequenced homemade fermented cachiri beers that were obtained from the traditional Amazonian population and are highly consumed by this population. We found that French and Saudis had significantly less richness and biodiversity in their gut microbiota than Amazonians and Polynesians (p <0.05). Principle coordinate analysis of the overall composition of the genera communities revealed that the microbiomes of Amazonians clustered independently from the other obese individuals. Moreover, we found that Amazonians presented significantly stricter anaerobic genera than the Saudis, French and Polynesians (p < 0.001). Polynesians presented significantly lower relative abundance of Lactobacillus sp. than French (p 0.01) and Saudis (p 0.05). Treponema berlinense and Treponema succinifaciens were only present in the gut microbiome of Amazonians. The cachiri beers presented significantly more bacterial species in common with the gut microbiome of Amazonians (p < 0.005). Obese individuals with different origins present modifications in their gut microbiota, and we provide evidence that the cachiri beers influenced the gut microbiome of Amazonians.
Keywords: Cachiri, gut microbiota, metagenomics, obesity, traditional living
Background
Obesity is a major public health challenge of the twenty-first century and has traditionally been thought of as a state of caloric imbalance, where the intake of calories exceeds the expenditure of calories [1]. The gut microbiota plays an important role in the harvesting, storage and expenditure of energy obtained from the diet, and recently the duodenal microbiota of obese individuals was found to exhibit alterations in fatty acid and sucrose breakdown pathways, probably induced by dietary imbalance [2]. Moreover, dietary habits alter the gut microbiota in a way that causes its metabolic activity to favour energy acquisition from ingested food [3]. The colonization of the gut microbiota begins at birth, and its composition is influenced by changes in diet and by life events [4]. Sequencing surveys of the human intestinal microbiome have revealed differences in the microbiome among people with different origins, and indicate that geography is an important factor affecting the gut microbiome [5], [6], [7], [8].
Several studies have revealed the positive correlation of a western-style dietary pattern with a higher incidence of obesity [9]. However, obesity is not always associated with a western-style diet; as an example, local Hawaiians eating a native diet have been reported to have an increased prevalence of obesity and type 2 diabetes compared with other ethnic groups living in Hawaii [10]. In French Polynesia, 38% of men and 40% of women were reported to be obese in 2002, and 34% and 26%, respectively, were reported to be overweight [11]. Similarly, a high prevalence of obesity has also been reported in French Guiana [11]. There is a gap in our knowledge of how local food influences the gut microbiome of obese individuals. The transition from a hunter–gatherer type of lifestyle to modern western society, with its tremendous technological advances in food processing, led to significant changes in food intake and composition. Comparative studies between non-industrialized rural communities and industrialized western communities have revealed an association of the gut microbiota with their respective lifestyles [8]. However, most studies have tested normal-weight urban and westernized individuals, and only a few have focused on the gut microbiomes of traditional populations [5], [6], [12], [13].
Our aim was to compare for the first time urban and traditionally living obese individuals to determine their gut microbiota modifications. To test this, we used high-throughput 16S ribosomal RNA (rRNA) gene amplicon sequencing to characterize the gut microbiota of obese individuals from France, Saudi Arabia, French Polynesia, and from a traditional population in the village of Trois-Sauts in French Guiana. This local obese population is particular because villagers consume high quantities daily of non-filtered fermented manioc beers called cachiri. We hypothesized that this constant high consumption of cachiri might have an impact on their gut microbiome. To confirm this, we also sequenced these homemade, non-filtered fermented manioc beers and compared them with the gut microbiome of this traditional community living on the Oyapock River in French Guiana.
Methods
Subject selection criteria
We tested obese adult volunteers from France, Saudi Arabia, French Polynesia and from a traditional population from Amazonian French Guiana. The Amerindian population was from the village of Trois-Sauts. This village is located in the southernmost part of the Guianese territory with no modern farming or agricultural plants [14]. The inclusion criteria were obese healthy adults with a body mass index value ≥ 30 kg/m2. The exclusion criteria were individuals under 18 years of age, inflammatory bowel disease, diarrhoea or treatment with an antibiotic in the previous 2 months before sampling. Stool samples were collected under aseptic conditions with clean, dry, screw-top containers and immediately stored at –20°C. We also tested two different homemade fermented manioc beers (cachiri), named umani and hakula, obtained from the traditional Amazonian population. Both beers were collected under aseptic conditions and immediately stored at –20°C.
Extraction of DNA from stool samples and 16S rRNA sequencing using MiSeq technology
Faecal DNA was extracted from samples using the NucleoSpin® Tissue Mini Kit (Macherey Nagel, Hoerdt, France) as previously described [15]. The 16s amplicon sequencing targeting the surrounding conserved regions V3-V4 primers with overhang adapters was performed using Illumina MiSeq 250bp pair end reads as previously described [2].
Bioinformatic analysis
A total of 14 428 091 sequences were found after Illumina paired end reads were assembled using FLASH [16]. QIIME version 1.8.0 software pipeline was used for the rest of the analysis by choosing ChimeraSlayer [17] for removing chimeric sequences, UCLUST [18] for operational taxonomic unit (OTU) extraction as described in our previous work [13], [15]. These OTUs represent a total of 9 676 207 sequences. All the raw sequences of fastq files have been submitted to EMBL-EBI [19] with the accession number PRJEB17702. The Silva SSU and LSU database [20] for release 119 was used for taxonomic assignment of the OTUs as previously described [13], [21] using blast [22].
Database of obligate anaerobes
Each phylotype was assigned as ‘aerotolerant’, ‘obligate anaerobe’ or ‘unknown’ according to its oxygen tolerance. These data are available online at https://www.mediterranee-infection.com/.
Statistical analysis
Rarefaction and Principal coordinate analysis with unifrac distance were performed in QIIME [17] by considering all the OTUs. Normalization and Adonis [23] tests were performed as described previously [13]. The OTUs, Chao1 and Shannon index from sequence reads were analysed by one-way analysis of variance and Tukey's honestly significant difference tests. The identification of significantly different bacterial taxa was performed by the non-parametric Kruskal–Wallis test along with Mann–Whitney analyses. We used Dunn's multiple comparisons test when comparing three or more groups.
Results
Participants
Overall, we tested 40 healthy obese volunteers, including 12 individuals from France, 9 from Saudi Arabia, 7 from Amazonia and 12 Polynesians (Table 1). The volunteers from France and Saudi Arabia had been previously reported [24]. For ethical reasons, all individuals from Saudi Arabia were male.
Table 1.
Characteristics of participants
| Age, years (median ±SD) | Sex (% male) | |
|---|---|---|
| French | 39 ± 13 | 58% |
| Saudis | 26 ± 3 | 100% |
| Polynesians | 45 ± 12 | 92% |
| Amazonians | 33 ± 17 | 57% |
Composition and common core of gut microbiota
The analysis of the high-quality trimmed reads revealed that the gut microbiota of French, Polynesians and Saudis contained sequences mostly belonging to Firmicutes, followed by Actinobacteria (see Supplementary material, Fig. S1), whereas for the gut microbiota of Amazonians, most of the sequences belonged to Firmicutes, followed by Bacteroidetes. Spirochaetae were only present in the stools of Amazonians, Armatimonadetes only in Polynesians and Gemmatimonadetes only in French. The relative abundance of Fusobacteria was significantly higher in Amazonians than Saudis and Polynesians (p 0.02 and p 0.005, respectively). Moreover, Amazonians presented significantly more relative abundance of Bacteroidetes in their stools than French, Saudis and Polynesians (p 0.0005, p 0.0002 and p 0.0005, respectively), significantly less relative abundance of Actinobacteria than French, Saudis and Polynesians (p 0.0006, p 0.0002 and p 0.002, respectively) and less relative abundance of Firmicutes than Saudis and French (p 0.0007 and p 0.05, respectively). Finally, Saudis presented significantly less relative abundance of Proteobacteria than Amazonians (p 0.02).
Obese individuals presented 240 different genera in their microbiome. Indeed, 197 different genera were present in the microbiome of French, 138 different genera in Amazonians, 154 in Saudis and 109 in Polynesians (see Supplementary material, Fig S2). We then examined the existence of a common bacterial core among the different populations tested and found that 40 different bacteria genera were present in at least 50% of the entire obese population tested.
Gut microbiota differences among the different groups
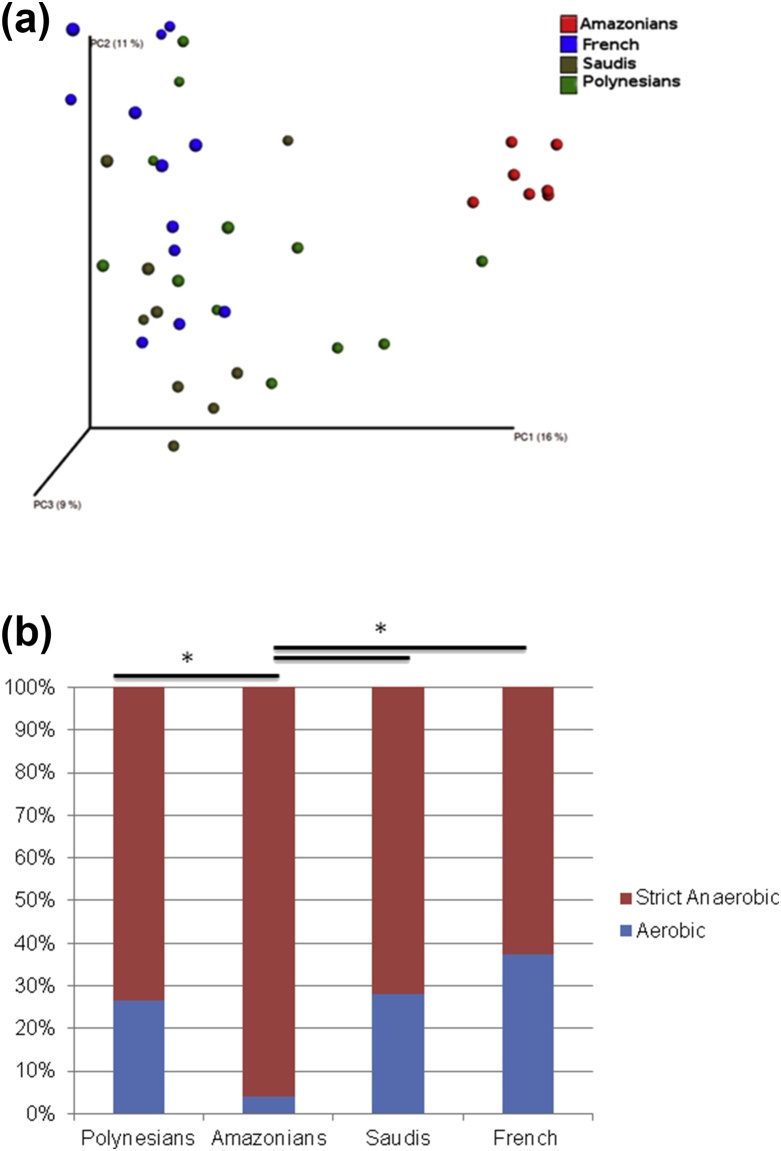
Principal coordinate analysis of the overall composition revealed that the microbiomes of Amazonians clustered independently from the other obese individuals (Fig. 1a). The adonis test using weighted unifrac distance gave a p value of 0.001 and R2 value of 0.26, which indicates that at least 26% of the variation in distances is explained by this grouping. The microbial richness, revealed that the gut microbiomes of French and Saudis had significantly less richness and biodiversity (p 0.05) than those of Amazonians and Polynesians (see Supplementary material, Fig. S3).
Fig. 1.
(a) Principal coordinate analysis comparison of microbial community composition among the obese individuals; (b) relative abundance of anaerobic and aerobic genera in the microbiome of obese French, Saudis, Polynesians and Amazonians.
We then investigated the distribution of aerobic and anaerobic strict genera and detected 108 aerobic and 130 strict anaerobic genera in the gut microbiome of obese individuals. Specifically, Saudis presented 96 different strict anaerobic and 57 aerobic genera, French 109 anaerobic and 87 aerobic genera, Amazonians 72 anaerobic and 36 aerobic genera, and Polynesians 97 anaerobic and 40 aerobic genera. Chi-square testing revealed no significant difference between the existence of different strict anaerobic and aerobic genera in the gut microbiome among the different groups. Moreover, no difference was found in the relative abundance of strict anaerobic genera between Saudis, French and Polynesians. In contrast, Amazonians presented significantly stricter anaerobic genera than Saudis, French and Polynesians (p < 0.001) (Fig. 1b).
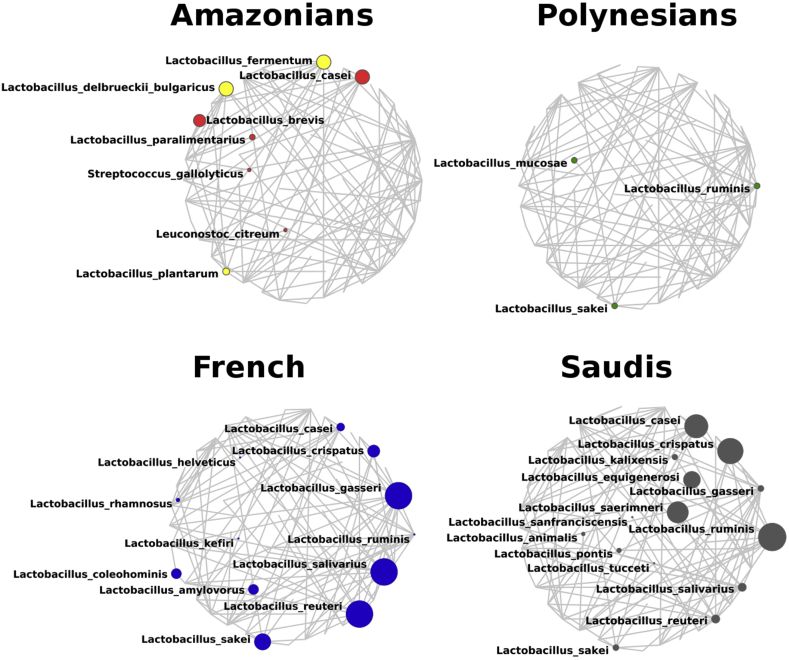
Obese individuals presented differences in the presence of Lactobacillus sp. in their gut microbiota. Polynesians presented significantly lower relative abundance of Lactobacillus sp. than French (p 0.01) and Saudis (p 0.05) (Fig. 2). Moreover, extensive differences were found at the Lactobacillus species level. As a result, the relative abundances of Lactobacillus fermentum and Lactobacillus plantarum were significantly greater in the gut microbiome of Amazonians compared with French (p 0.03 and p < 0.001, respectively) and Saudis (p 0.01 and p < 0.001, respectively), whereas Polynesians presented none of these Lactobacillus species. In contrast, their relative abundance showed that Lactobacillus reuteri, Lactobacillus salivarius, Lactobacillus gasseri and Lactobacillus sakei were significantly more common in French compared with the other populations (p < 0.01). Polynesians presented only Lactobacillus ruminis in their gut microbiota; L. ruminis was not detected in the gut microbiomes of French and Amazonians. Moreover, sequences of Treponema berlinense and Treponema succinifaciens were only present in the gut microbiome of Amazonians (see Supplementary material, Fig. S1).
Fig. 2.
Network of Lactobacillus spp. present in the gut microbiome of obese individuals. Larger circle size indicates the higher abundance and smaller circle indicates the lower abundance; yellow colour indicates Lactobacillus spp. present in homemade beers.
Impact of human microbiome on cachiri beer
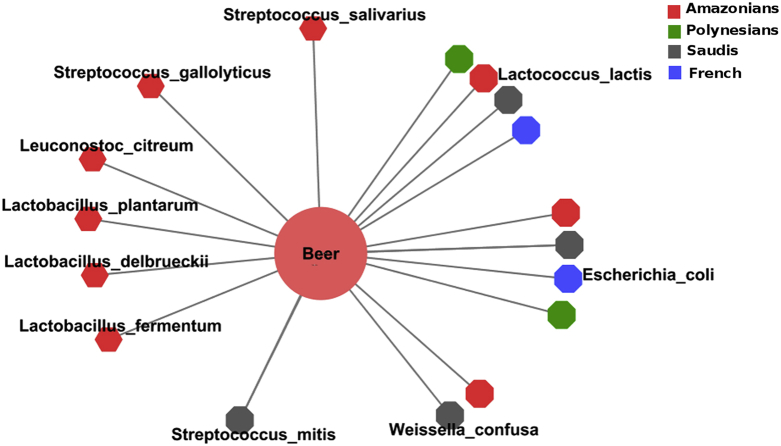
We tested two samples of umani and two of hakula manioc beers obtained from the traditional Amazonian population and their analysis revealed that sequences mostly belonged to Firmicutes, followed by Proteobacteria. We found 51 different genera, and most of the sequences belonged to Lactococcus, Streptococcus, Weissella and Lactobacillus. In addition, we found 71 different species, and most of the sequences belonged to Lactococcus lactis, Weissella confusa and Streptococcus mitis. We then examined the existence of a common bacterial core among the different obese populations and the ten most common bacterial species present in the homemade fermented beers (Fig. 3). We found that the homemade beers presented significantly more bacteria species in common with the gut microbiome of Amazonians than the other populations (p 0.005 compared with Polynesians and French, p 0.056 compared with Saudis). Moreover, L. fermentum, L. plantarum, Lactobacillus bulgaricus, Leuconostoc citreum and Streptococcus macedonicus were only present in the homemade beers and in the gut microbiome of the Amazonian population, but were absent from the other obese populations.
Fig. 3.
Network of the most common bacterial species present in homemade beers and in the gut microbiome of each obese population tested.
Discussion
In this study, we found that obese individuals with different origins show differences in their gut microbiota. The quality of DNA extraction was verified for all samples before proceeding to sequencing, while the V3-V4 region has been commonly used for the characterization of the gut microbiome. Moreover, differences in sample preservation and DNA isolation protocols as well as sequencing of different 16S rRNA gene regions impact the representation of the microbial community [25], [26], [27], [28], [29], [30]. Here, we used the same sample preservation and DNA extraction protocol, as well as sequencing of the V3-V4 region, for all the analysed samples.
We found that the obese individuals with a westernized diet presented similarities in their gut microbiota independently of their origin. It was previously reported that non-western populations have more microbial richness than western populations [7], and our analyses of microbial richness yielded similar results. Polynesians and Amazonians presented higher microbial diversity than French and Saudis. Their diet is mostly composed of many complex polysaccharides, such as starch, cellulose, xylan, xyloglucan and pectin. Indeed, the diet of Amazonians is predominantly vegetarian, low in fat and animal protein, and rich in starch, fibre and plant polysaccharides, whereas Polynesians although commonly consuming imported westernized foods, also have an important level of intake from food plants, hunting and fishing. As a result, our findings confirm that the human gut microbiota is influenced by the different dietary regimens of humans and by meal-to-meal variations [31], [32]. The exposure to the large variety of environmental microbes associated with a high-fibre diet was previously associated with an increase of the potentially beneficial bacterial genomes, enriching the microbiome [31], [32]. The diet can influence the bacterial diversity from carnivore to omnivore to herbivore [33] and a high-fibre diet has been associated with an enrichment of the microbiome [6]. The microbial diversity of the gut microbiota was possibly decreased during human civilization [34], and recent lifestyle changes in humans have depleted the human microbiome of microbial diversity that was present in wild-living ancestors [35] such as the Amazonian population analysed here. In addition, for the digestion of plant material through fermentation an anaerobic environment in the gut is critical [32]. As a result it is possible that the gut microbiota of Amazonians is adapted and enriched with more anaerobic bacteria to depredate the increased uptake of starch, fibre and plant polysaccharides.
Similar to previous studies on traditional populations [12], the traditional Amazonian population had spirochaetes in their gut microbiome, particularly T. berlinense and T. succinifaciens. Spirochaetes have been commonly reported in the gut microbiome of primates such as wild apes [36], macaques [37] and wild hominids [38]. Spirochaetes were also detected in ancient human populations [39] and in traditional populations [5], [12]. It was found that Treponema strains identified in the gut microbiota of traditional populations clustered with other treponemes reported from termites [12]. In our study, T. berlinense and T. succinifaciens were previously isolated from the gastrointestinal tract of swine [40], [41]. Recently, T. succinifaciens was also detected in traditional populations from Peru [12]. We believe that spirochaetes are not associated with obesity but in this Amerindian community, where animals and humans live in very close proximity, this finding suggests possible cross-transmission of spirochaetes between humans and animals.
We found important changes in the presence of Lactobacillus sp. among the obese individuals. Lactobacillus sp. are associated with weight modifications [42], [43], as they possess an ability to breakdown fructose or glucose and to reduce ileal brake effects [44]. Lactobacillus reuteri, L. gasseri, L. fermentum, L. plantarum and L. sakei have been previously associated with obesity [45], [46] and were frequently detected in the gut microbiomes of our individuals. Differences in Lactobacillus species in the gut microbiome of obese individuals with different origins can be explained by the consumption of different fermented dairy products and probiotics [47], [48]. Interestingly, the traditional Amazonian population consumes high quantities of cachiri. Its preparation is usually done by women, who chew the manioc and then spit it in a jar and leave it for fermentation. Making these fermented beers is based upon old empirical knowledge, which is transferred from generation to generation. Previous studies on similar fermented drinks revealed that they are dominated by lactic acid bacteria including L. plantarum and L. fermentum [49]. We found that these beers were highly contaminated by several bacteria, mostly by L. fermentum, L. plantarum and L. bulgaricus. Indeed, Lactobacillus spp. are the most prevalent beer-fermenting bacteria, but sterile filtration is used to physically remove microorganisms to ensure microbiological stability [50]. These lactobacilli were absent from the other obese populations and only existed in the gut microbiome of Amazonians, indicating that the increased beer consumption could explain the presence of these Lactobacillus spp. uniquely in the gut microbiome of Amazonians. Differences in cultural traditions and food influence the composition of the gut microbiome [7], and our results reinforced the fact that the very high consumption of cachiri has an impact on the gut microbiome of Amazonians. However, a limitation of our study was that we did not examine other aspects of diet aside from beer that can influence the gut microbiome.
Conclusions
We found that the gut microbiome of the obese Amerindian population, living isolated in a very specific environment, is different to that of obese populations living in industrialized areas probably because of the increased complex polysaccharides and cachiri consumption. To understand how the cultural and dietary habits influence the gut microbiota, it is important to continue sampling populations from different geographical regions with different lifestyles. The gut microbiome responds to what people eat and we reinforce the fact that it is very difficult to draw conclusions about the association of the gut microbiota and obesity because of the discrepancy among different populations, as individuals with the same obese phenotype present modifications in their gut microbiota.
Transparency declaration
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This study protocol was approved by the Ethics Committee of the King AbdulAziz University under agreement number 014-CEGMR-2-ETH-P and by the Ethics Committee of the Institut Fédératif de Recherche IFR48, Faculty of Medicine, Marseille, France. The agreement of the Ethics Committee of the IFR48 (Marseille, France) was obtained under reference 09-022 and the agreement of the Ethics Committee of the Institute Louis Malarde was obtained under reference 67-CEPF. All methods in this study were carried out in accordance with the approved guidelines. Informed consent forms were provided to all participants and obtained at the time of sample collection.
Availability of data and material
All the raw sequences of fastq files have been submitted to EMBL-EBI [19] with the accession number PRJEB17702. The data sets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Funding
No funding was obtained for this study.
Author contributions
EA analysed the data, organized the study and wrote the manuscript, DB analysed the sequencing data and wrote the manuscript, MY found the participants and collected samples in Saudi Arabia, DM found the participants, collected clinical information and samples and organized the study in French Polynesia, FD found the participants and organized the study in French Guiana, CM collected clinical information and samples in French Guiana and wrote the manuscript, CR realized the Illumina MiSeq deep sequencing, BD organized the study in French Guiana, took the cachiri samples and wrote the manuscript, BG found the obese participants in France, collected their stool samples and wrote the manuscript, EIA organized the study in Saudi Arabia, FB collected samples in Saudi Arabia, AD collected stool samples in France and wrote the manuscript, DR organized the study and wrote the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2018.11.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Fig. S1: The relative abundance of the gut microbiota phyla among the obese groups tested.
Fig. S2: Bacteria genera presented in the gut microbiota of (a) at least one obese individual; (b) at least 50% of the tested population.
Fig. S3: Boxplots of the observed operational taxonomic units, the Chao1 indexes and the non-parametric Shannon indices; *p < 0.05.
References
- 1.Angelakis E., Merhej V., Raoult D. Related actions of probiotics and antibiotics on gut microbiota and weight modification. Lancet Infect Dis. 2013;13:889–899. doi: 10.1016/S1473-3099(13)70179-8. [DOI] [PubMed] [Google Scholar]
- 2.Angelakis E., Armougom F., Carriere F., Bachar D., Laugier R., Lagier J.C. A metagenomic investigation of the duodenal microbiota reveals links with obesity. PLoS One. 2015;10(9):e0137784. doi: 10.1371/journal.pone.0137784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spor A., Koren O., Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 4.Angelakis E., Armougom F., Million M., Raoult D. The relationship between gut microbiota and weight gain in humans. Future Microbiol. 2012;7:91–109. doi: 10.2217/fmb.11.142. [DOI] [PubMed] [Google Scholar]
- 5.Schnorr S.L., Candela M., Rampelli S., Centanni M., Consolandi C., Basaglia G. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Filippo C., Cavalieri D., Di P.M., Ramazzotti M., Poullet J.B., Massart S. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escobar J.S., Klotz B., Valdes B.E., Agudelo G.M. The gut microbiota of colombians differs from that of Americans, Europeans and Asians. BMC Microbiol. 2014;14:311. doi: 10.1186/s12866-014-0311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidemann C., Schulze M.B., Franco O.H., van Dam R.M., Mantzoros C.S., Hu F.B. Dietary patterns and risk of mortality from cardiovascular disease, cancer, and all causes in a prospective cohort of women. Circulation. 2008;118:230–237. doi: 10.1161/CIRCULATIONAHA.108.771881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandinetti A., Chang H.K., Mau M.K., Curb J.D., Kinney E.K., Sagum R. Prevalence of glucose intolerance among Native Hawaiians in two rural communities. Native Hawaiian Health Research (NHHR) Project. Diabetes Care. 1998;21:549–554. doi: 10.2337/diacare.21.4.549. [DOI] [PubMed] [Google Scholar]
- 11.Daigre J.L., Atallah A., Boissin J.L., Jean-Baptiste G., Kangambega P., Chevalier H. The prevalence of overweight and obesity, and distribution of waist circumference, in adults and children in the French Overseas Territories: the PODIUM survey. Diabetes Metab. 2012;38:404–411. doi: 10.1016/j.diabet.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Obregon-Tito A.J., Tito R.Y., Metcalf J., Sankaranarayanan K., Clemente J.C., Ursell L.K. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun. 2015;6:6505. doi: 10.1038/ncomms7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angelakis E., Yasir M., Bachar D., Azhar E.I., Lagier J.C., Bibi F. Gut microbiome and dietary patterns in different Saudi populations and monkeys. Sci Rep. 2016;6:32191. doi: 10.1038/srep32191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angebault C., Djossou F., Abelanet S., Permal E., Ben S.M., Diancourt L. Candida albicans is not always the preferential yeast colonizing humans: a study in Wayampi Amerindians. J Infect Dis. 2013;208:1705–1716. doi: 10.1093/infdis/jit389. [DOI] [PubMed] [Google Scholar]
- 15.Zoetendal E.G., Booijink C.C., Klaassens E.S., Heilig H.G., Kleerebezem M., Smidt H. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006;1:954–959. doi: 10.1038/nprot.2006.143. [DOI] [PubMed] [Google Scholar]
- 16.Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 19.Stoesser G., Sterk P., Tuli M.A., Stoehr P.J., Cameron G.N. The EMBL nucleotide sequence database. Nucleic Acids Res. 1997;25:7–14. doi: 10.1093/nar/25.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillou L., Bachar D., Audic S., Bass D., Berney C., Bittner L. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41(Database issue):D597–D604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 24.Yasir M., Angelakis E., Bibi F., Azhar E.I., Bachar D., Lagier J.C. Comparison of the gut microbiota of people in France and Saudi Arabia. Nutr Diabetes. 2015;5:e153. doi: 10.1038/nutd.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirsepasi H., Persson S., Struve C., Andersen L.O., Petersen A.M., Krogfelt K.A. Microbial diversity in fecal samples depends on DNA extraction method: easyMag DNA extraction compared to QIAamp DNA stool mini kit extraction. BMC Res Notes. 2014;7:50. doi: 10.1186/1756-0500-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson G., Cox F., Kittelmann S., Miri V.H., Zethof M., Noel S.J. Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS One. 2013;8(9):e74787. doi: 10.1371/journal.pone.0074787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McOrist A.L., Jackson M., Bird A.R. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J Microbiol Methods. 2002;50:131–139. doi: 10.1016/s0167-7012(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 28.Smith B., Li N., Andersen A.S., Slotved H.C., Krogfelt K.A. Optimising bacterial DNA extraction from faecal samples: comparison of three methods. Open Microbiol J. 2011;5:14–17. doi: 10.2174/1874285801105010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vo A.T., Jedlicka J.A. Protocols for metagenomic DNA extraction and Illumina amplicon library preparation for faecal and swab samples. Mol Ecol Resour. 2014;14:1183–1197. doi: 10.1111/1755-0998.12269. [DOI] [PubMed] [Google Scholar]
- 30.Milani C., Hevia A., Foroni E., Duranti S., Turroni F., Lugli G.A. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One. 2013;8(7):e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantarel B.L., Lombard V., Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS One. 2012;7(6):e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El K.A., Armougom F., Gordon J.I., Raoult D., Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 33.Ley R.E., Hamady M., Lozupone C., Turnbaugh P.J., Ramey R.R., Bircher J.S. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moeller A.H., Degnan P.H., Pusey A.E., Wilson M.L., Hahn B.H., Ochman H. Chimpanzees and humans harbour compositionally similar gut enterotypes. Nat Commun. 2012;3:1179. doi: 10.1038/ncomms2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blaser M.J., Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bittar F., Keita M.B., Lagier J.C., Peeters M., Delaporte E., Raoult D. Gorilla gorilla gorilla gut: a potential reservoir of pathogenic bacteria as revealed using culturomics and molecular tools. Sci Rep. 2014;4:7174. doi: 10.1038/srep07174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna P., Hoffmann C., Minkah N., Aye P.P., Lackner A., Liu Z. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008 Feb 8;4(2):e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochman H., Worobey M., Kuo C.H., Ndjango J.B., Peeters M., Hahn B.H. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8(11):e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tito R.Y., Knights D., Metcalf J., Obregon-Tito A.J., Cleeland L., Najar F. Insights from characterizing extinct human gut microbiomes. PLoS One. 2012;7(12):e51146. doi: 10.1371/journal.pone.0051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cwyk W.M., Canale-Parola E. Treponema succinifaciens sp. nov., an anaerobic spirochete from the swine intestine. Arch Microbiol. 1979;122:231–239. doi: 10.1007/BF00411285. [DOI] [PubMed] [Google Scholar]
- 41.Nordhoff M., Taras D., Macha M., Tedin K., Busse H.J., Wieler L.H. Treponema berlinense sp. nov. and Treponema porcinum sp. nov., novel spirochaetes isolated from porcine faeces. Int J Syst Evol Microbiol. 2005;55:1675–1680. doi: 10.1099/ijs.0.63388-0. [DOI] [PubMed] [Google Scholar]
- 42.Angelakis E., Raoult D. The increase of Lactobacillus species in the gut flora of newborn broiler chicks and ducks is associated with weight gain. PLoS One. 2010;5(5):e10463. doi: 10.1371/journal.pone.0010463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angelakis E., Bastelica D., Ben A.A., El F.A., Dutour A., Mege J.L. An evaluation of the effects of Lactobacillus ingluviei on body weight, the intestinal microbiome and metabolism in mice. Microb Pathog. 2012;52:61–68. doi: 10.1016/j.micpath.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Drissi F., Merhej V., Angelakis E., El K.A., Carriere F., Henrissat B. Comparative genomics analysis of Lactobacillus species associated with weight gain or weight protection. Nutr Diabetes. 2014;4:e109. doi: 10.1038/nutd.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Million M., Thuny F., Angelakis E., Casalta J.P., Giorgi R., Habib G. Lactobacillus reuteri and Escherichia coli in the human gut microbiota may predict weight gain associated with vancomycin treatment. Nutr Diabetes. 2013;3:e87. doi: 10.1038/nutd.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlsson F.H., Tremaroli V., Nookaew I., Bergstrom G., Behre C.J., Fagerberg B. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 47.Saulnier D.M., Spinler J.K., Gibson G.R., Versalovic J. Mechanisms of probiosis and prebiosis: considerations for enhanced functional foods. Curr Opin Biotechnol. 2009;20:135–141. doi: 10.1016/j.copbio.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angelakis E., Million M., Henry M., Raoult D. Rapid and accurate bacterial identification in probiotics and yoghurts by MALDI-TOF mass spectrometry. J Food Sci. 2011;76:M568–M572. doi: 10.1111/j.1750-3841.2011.02369.x. [DOI] [PubMed] [Google Scholar]
- 49.Almeida E.G., Rachid C.C., Schwan R.F. Microbial population present in fermented beverage 'cauim' produced by Brazilian Amerindians. Int J Food Microbiol. 2007;120:146–151. doi: 10.1016/j.ijfoodmicro.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 50.Garofalo C., Osimani A., Milanovic V., Taccari M., Aquilanti L., Clementi F. The occurrence of beer spoilage lactic acid bacteria in craft beer production. J Food Sci. 2015;80:M2845–M2852. doi: 10.1111/1750-3841.13112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw sequences of fastq files have been submitted to EMBL-EBI [19] with the accession number PRJEB17702. The data sets used and/or analysed during the current study are available from the corresponding author on reasonable request.