Abstract
Scope:
The molecular mechanisms whereby gallates in green tea exert metabolic effects are poorly understood.
Methods and results:
We found that GPRC6A, a multi-ligand-sensing G-protein-coupled receptor that regulates energy metabolism, sex hormone production, and prostate cancer progression, is a target for gallates. Sodium gallate (SG), gallic acid (GA) > ethyl gallate (EG) > octyl gallate (OG) dose dependently activated ERK in HEK-293 cells transfected with GPRC6A but not in non-transfected controls. SG also stimulated insulin secretion in β-cells isolated from wild-type mice similar to the endogenous GPRC6A ligands, osteocalcin (Ocn) and testosterone (T). Side-chain additions to create OG resulted in loss of GPRC6A agonist activity. Another component of green tea, epigallocatechin 3-gallate (EGCG), dose-dependently inhibited Ocn activation of GPRC6A in HEK-293 cells transfected with GPRC6A and blocked the effect of Ocn in stimulating glucose production in CH10T1/2 cells. Using structural models of the venus fly trap (VFT) and 7-transmembrane (7-TM) domains of GPRC6A, calculations suggest that l-amino acids and GA bind to the VFT, whereas EGCG is calculated to bind to sites in both the VFT and 7-TM.
Conclusion:
GA and EGCG have offsetting agonist and antagonist effects on GPRC6A that may account for the variable metabolic effect of green tea consumption.
Keywords: computational modeling, EGCG, gallic acid, GPCR, GPRC6A
1. Introduction
Green tea (Camellia sinensis L.) consumption is reported to have multiple health benefits, including reducing the risks for developing metabolic syndrome (MetS), type 2 diabetes (T2D), and prostate cancer (PCa), as well as many others.[2–9] Gallates represent 50% of the naturally occurring polyphenolic compounds in green tea and are also present in gallnuts, wine, chocolate, fruits, and vegetables.[1,10]
Two important bioactive components of green tea, gallic acid (GA; 3,4,5-trihydroxybenzoic acid) and epigallocatechin 3-gallate [EGCG; (2R,3R)-2-(3,4,5-tryhydroxyphenyl)-3,4-dihydro-1 (2H)-benzopyran-3,5,7-triol 3-(3,4,5-trihydroxybenzoate)], are best studied. GA and EGCG are both reported to have beneficial effects on MetS.[11] GA administration induces weight loss, improves glucose tolerance, lowers triglyceride concentrations, and increases insulin sensitivity after GA in preclinical models of T2D.[12–16] GA administration to humans also has anti-obesity effects.[17] The oral administration of EGCG also decreases blood glucose, improves lipid parameters, and reduces inflammatory cytokines in pre-clinical models of MetS.[18,19] EGCG also regulates mitochondrial biogenesis, ATP production, and apoptosis.[20] GA and EGCG also have chemopreventive effects on multiple cancer cell lines[19,21] and antioxidant properties.[22] EGCG inhibits PCa cell growth in xenograft models and is associated with a reduced risk of PCa in men.[23–25]
Overall, human studies using these green tea–derived compounds have not been as successful as in vitro and animal studies.[4,26] There are issues of stability and bioavailability of EGCG, which undergoes metabolism to GA in the gut, so it is unclear if the beneficial metabolic effects of EGCG are due to EGCG or GA.[27,28] The effects of GA and EGCG on metabolism and PCa are controversial, with some studies showing a benefit and others reporting no effects.[29,30] In order to understand the potential health benefits of green tea, and the variability in the therapeutic effects of green tea, the molecular mechanisms mediating these metabolic and anti-cancer effects of EGCG and GA need to be defined.
GPRC6A, a family C, nutrient G-protein-coupled receptor, is a potential molecular target for components of green tea. GPRC6A is derived from distinct domains created by fusion of a periplasmic nutrient venus fly trap (VFT) motif with a classical 7-transmembrane (7-TM) domain. Computational structural models of the 7-TM domain have, in combination with experiment, defined the structural basis for direct binding of T and Ocn to GPRC6A.[31] In contrast to T and Ocn, l-amino acids and cations are sensed by the VFT.[31,32] The possibility that GPRC6A may be a target for the polyphenolic gallates is suggested by our recent findings that di-phenolic selective androgen modulators (SARMS) activate GPRC6A.[32] Moreover, the biological functions of GPRC6A are remarkably similar to those purported for GA and EGCG. Indeed, GPRC6A has been implicated in a variety of health conditions, ranging from being a potential target for treating T2D and MetS, to playing a role in PCa progression.[33] GPRC6A is expressed in key metabolic tissues, where it regulates glucose and fat metabolism.[34–40] Ablation of Gprc6a in mice results in obesity, glucose intolerance, hepatic steatosis, and insulin resistance,[41] and deletion of Ocn, a natural ligand for GPRC6A, leads to a metabolic phenotype identical to Gprc6a−/− mice.[42] Administration of Ocn improves glucose tolerance, increases insulin sensitivity, β-cell mass, and insulin secretion, reduces fat, increases muscle mass, and reverses hepatosteatosis in mice fed high fat diets.[36,43] Ablation of GPRC6A attenuates, whereas Ocn administration exacerbates, PCa progression in xenograft models.[44]
In this study, we used a combination of functional studies and computational molecular dynamics–based docking using structural models of both VFT and 7-TM domains to investigate the effects of GA and EGCG on GPRC6A activation.
2. Experimental Section
2.1. Reagents
Curcumin, GA, sodium gallate, EG, octyl gallate (OG), catechin, catechin gallate, epigallocatechin, epigallocatechin gallate, testosterone (T), l-arginine, and insulin were purchased from Sigma.
Osteocalcin (Ocn) was purified from bovine tibial bone extracts.[45] Decarboxylated Ocn was produced by treating Ocn in vacuo at 110°C.[46] The purity and decarboxylation state were confirmed by native gel electrophoresis, or by blotting followed by reaction with DBS staining for γ -carboxyglutamic acid.[47]
2.2. Cell Culture
All culture reagents were from Invitrogen. Human embryonic kidney HEK-293 cells were obtained from American Type Culture Collection. HEK-293 cells stably transfected with pcDNA3.mGPRC6A (with Myc-tagged) were created as previously described.[48,49] HEK-293 and HEK-293 transfected with a mouse GPRC6A cDNA cells[49,50] were cultured in DMEM medium supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 1% penicillin/streptomycin (P/S) (Invitrogen).
2.3. Measurement of Total and Phospho-ERK by ERK Elisa Analysis
Briefly, HEK-293 cells transfected with/without mouse GPRC6A cDNA plasmid were starved by overnight incubation in serum-free DMEM/F12 containing 0.1% bovine serum albumin (BSA) and stimulated with various ligands at different doses. ERK activation were assessed 20 min after treatment by using ERK1/2 (phospho-T203/Y204) ELISA Kit (Invitrogen) corrected for the amount of total ERK using ERK1/2 (Total) ELISA Kit (Invitrogen) to measure ERK levels.
2.4. siRNA Suppression of GPRC6A Gene Expression
For GPRC6A knockdown experiments, two short interfering RNAs (siRNAs) (19 nucleotides each) were designed from the human GPRC6A sequence (NM 148963) and mouse GPRC6A sequence (NM 153071).[51] These are human GPRC6A siRNA-514: GCCACAGGTGGGTTATGAA and mouse GPRC6A siRNA-492: GCCACAGGTGAGTTATGAA. Two siRNA hairpins were synthesized and cloned into a pSilencerTM 4.1-CMV neo vector (Ambion). A circular pSilencer 4.1-CMV neo vector that expresses a hairpin siRNA with limited homology to any known sequence was used as a negative control. The constructs of siRNA duplexes were stably transfected into human PCa 22Rv1 and mouse fibroblast C3H10T1/2 cells using Lipofectamine (Invitrogen) and were selected by G418 (Invitrogen). Successful knockdown of GPRC6A were confirmed by assessing RT-PCR analysis of GPRC6A expression.[51]
2.5. Glucose Production Assay
Cells were washed three times with PBS to remove glucose and then incubated for 4 h in 250 μL glucose production medium [glucose- and phenol red-free DMEM containing the gluconeogenic substrates, sodium lactate (20 mm) and sodium pyruvate (2 mm)] in the presence or absence of 1 μm SG. One hundred microliters of medium was sampled for measurement of glucose concentration using a Glucose Assay Kit (Cayman Chemical). Glucose concentration was normalized with cellular protein concentration measured by DC Protein Assay Kit (Bio Rad).
2.6. GPRC6A Structural Modeling
Details of the computational modeling are given in the Supporting Information section. GPRC6A is an evolutionary fusion of a VFT motif with a classical 7-TM GPCR domain that is closely related to mGlurRs and CaSR, the calcium-sensing receptor, for which there is extensive structural information on the binding domains.[52] By combining experiment and computational modeling, we have defined the molecular and structural basis for the multi-ligand sensing of GPRC6A and established the structural basis for the action of orthosteric ligands, allosteric modulators, and antagonists of the receptor.[53,54]
We have developed GPRC6A homology models based on the templates mGluR-1/mGluR-5 for the 7-TM domain.[31,32] We used these 7-TM models to define the structural basis for direct binding of T and Ocn to GPRC6A by combining docking studies and experimental site-directed mutagenesis to identify the residues that contribute to T[31] and Ocn[32] binding to the allosteric binding site in the TM domain.
We also constructed here a model of the VFT domain, to which basic amino acids, such as l-Arg and l-Orn, and various diva-lent (Ca2+ and Zn2+) and trivalent cations (Gd3+ and Au3+) bind. Earlier mutagenesis studies and redocking of these amino acids in the VFT domain allowed the orthosteric binding pocket to be identified.[1]
Molecular dynamics (MD) simulations of both the constructed models of the 7-TM and VFT domains allowed the identification of a conformational ensemble of these constituent domains of GPRC6A. The conformational ensembles were used for further docking studies with the compounds from the GA platform in order to probe the structural basis for the multi-ligand sensing of GPRC6A. All docking studies and the methods used for running the MD simulations are provided in the Supporting Information data.
Based on our computational modeling and empirical observations of the functional effects of modifications of a GA chemical platform, we established an effective computational protocol for generating GPRC6A antagonists. Indeed, we can now transform an agonist GA to an antagonist EGCG in the GA platform by making judicious side chain modifications (Figure 4B), setting the stage for a detailed understanding of the structural basis of agonist and antagonist effects on GPRC6A.
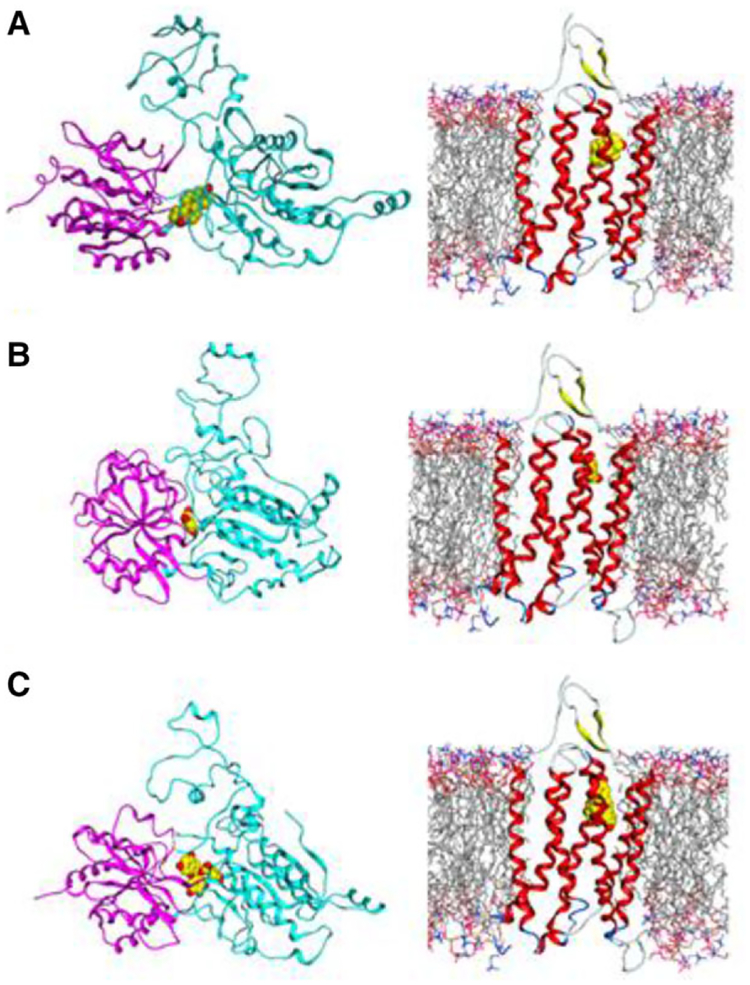
Figure 4.
Schematic of GPRC6A domains and structural models of the VFT and TM showing: A) Testosterone (T) agonist docked in the VFT and TM domains; B) Gallate (GA) agonist docked in the closed conformation of VFT and TM; C) EGCG antagonist docked in open VFT conformation and in TM domain.
3. Results
3.1. Sodium Gallate and Gallic Acid Are GPRC6A Agonists
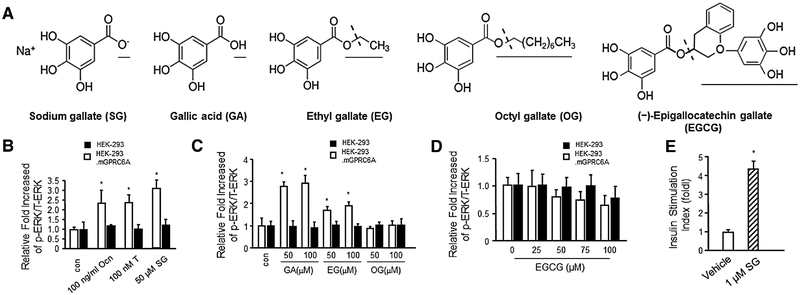
We tested the effects of a series of gallate compounds, including SG,[1] GA, EG, OG, and EGCG, for their ability to activate GPRC6A in HEK-293 cells transfected with either GPRC6A or a control vector. SG is a sodium salt of GA (3,4,5-trihydroxybenzoic acid),[1] EG is the ethyl ester of GA (ethyl 2,4,5-trihydroxybenzoate), OG is a 1-octanol ester of GA (octyl 3,4,5-trihydroxybezoate), and EGCG is ester of epigalocatechin and GA (2R,3R)-2-(3,4,5-tryhydroxyphenyl)-3,4-dihydro-1 (2H)-benzopyran-3,5,7-triol 3-(3,4,5-trihydroxybenzoate) (Figure 1A).
Figure 1.
Natural compounds gallic acid, sodium gallate, and EGCG are agonists or antagonists of GPRC6A. A) Chemical structures of gallates and EGCG. B) Gallic acid and C) ethyl gallate stimulated GPRC6A mediated ERK phosphorylation, but not C) octyl gallate and D) EGCG in HEK-293 stably transfected with GPRC6A cDNA, but the stimulation of SG, GA, and EG were not appealed in control HEK-293 without GPRC6A cDNA. The bar graph in the bottom panels depicts the fold-increase in ERK activation in response to Ocn, T, GA, EG, OG, and EGCG in control HEK-293 and HEK-293 transfected with GPRC6A cDNA. E) SG (1 μm) stimulated insulin secretion in mouse isolated islets. Values represent the mean ± SEM. * indicates significant differences from control and stimulated groups at p < 0.05 (n ≥ 4).
First, we compared SG with the known GPRC6A agonists, T and Ocn, for their ability to stimulate GPRC6A using ERK activity as a readout (Figure 1B). We observed ERK activation by 50 μm SG, 100 nm T, and 100 ng mL−1 Ocn in HEK-293 cells over-expressing GPRC6A (Figure 1B), but not in untransfected HEK-293 control (Figure 1B). There was no significant difference in the magnitude of ERK activation by SG, T, and Ocn (Figure 1B). We have also shown the dose dependent stimulation of GA and SG in Supporting Information Figure 1A.
Next, we tested the effects of other members of this chemical series in activating GPRC6A in vitro. We found that GA and EG, similar to SG, were able to activate ERK phosphorylation through GPRC6A-mediated mechanism, but OG, with the ester formed from 1-octanol did not activate GPRC6A at concentrations of up to 100 μm (Figure 1C). GA was more potent than EG, as evidenced by a nearly threefold greater increase in ERK activation at 50 μm concentrations (Figure 1C). None of the gallates stimulated ERK activity in HEK-293 cells not transfected with mouse GPRC6A cDNA (Figure 1C).
Finally, we examined the effects of the green tea phenol EGCG in activating GPRC6A-mediated ERK phosphorylation in HEK-293 cells transfected with GPRC6A cDNA. EGCG at concentrations ranging from 25 to 100 μm did not stimulate ERK activity in GPRC6A expressing HEK-293 cells (Figure 1D). In fact, high concentrations of EGCG inhibited constitutive stimulation of ERK activity by GPRC6A in the absence of an agonist (Figure 1D).
To assess the biological effects of gallate activation of GPRC6A, we examined insulin secretion in pancreatic islets that express endogenous GPRC6A and increase insulin secretion in response to Ocn. We found that 1 μm SG significantly stimulated insulin secretion compared to the vehicle in cultured pancreatic islets (Figure 1E), a value similar to that reported for Ocn.[32]
3.2. EGCG Is a GPRC6A Antagonist
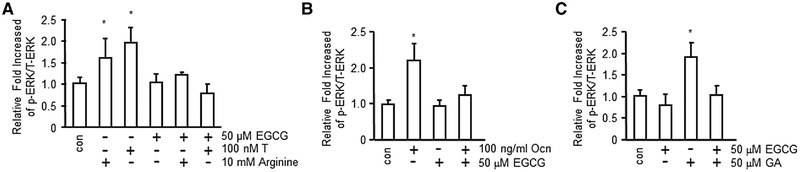
Since EGCG is an ester of epigallocatechin and GA without agonist activity, we explored the alternative possibility that EGCG might block the effects of GPRC6A agonists. We found that EGCG at 50 μm concentration inhibited the effects of known GPRC6A ligands, T (100 nm), l-arginine (Arg; 10 mm), and Ocn (100 ng mL−1) in stimulating ERK activity in GPRC6A expressing HEK-293 cells (Figure 2A, B). 50 μm EGCG also inhibited GA-stimulated GPRC6A-mediated ERK phosphorylation in HEK-293 cells transfected GPRC6A (Figure 2C).
Figure 2.
Green tea polyphenyl EGCG is antagonist of GPRC6A. EGCG inhibited GPRC6A ligands, A) T and Arg, B) Ocn-stimulated ERK phosphorylation in HEK-293 cells transfected GPRC6A cDNA. C) GA-stimulated ERK phosphorylation was also blocked by EGCG in HEK-293 cells transfected GPRC6A cDNA. The bar graph in bottom panels depicts fold-increase in ERK activation in response to GA, GE, and OG in HEK-293 transfected with GPRC6A cDNA. Values represent the mean ± SEM. * indicates significant differences from control and stimulated groups at p < 0.05 (n ≥ 4).
We also examined activities of other polyphenols from green tea (Supporting Information Figure B). Similar to EGCG, we found that catechin gallate inhibited, but catechin (Cat) and epigallocatechin (EGC) did not block, agonist stimulation of GPRC6A (Supporting Information Figure 1C, D).
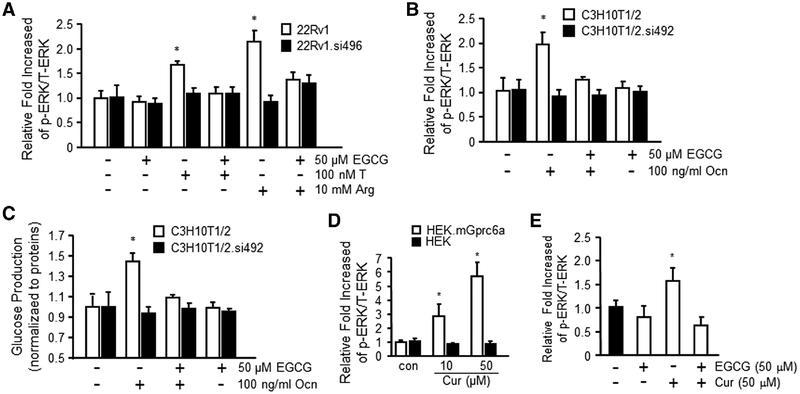
To further study the effects of EGCG on GPRC6A-mediated ERK activation in endogenous expressed GPRC6A cell lines, we accessed the human PCa cell, 22Rv1, and mouse fibroblast cell line, C3H10T1/2 (Figure 3A, B). We found that 50 μm EGCG significantly blocked the effects of GPRC6A agonists 10 mm Arg, 100 nm T, and 100 ng mL−1 Ocn to stimulate ERK phosphorylation in both 22Rv1 (Figure 3A) and C3H10T1/2 cells (Figure 3B), consistent with antagonist effects. To confirm the importance of GPRC6A in mediating EGCG antagonist effects, we used small interfering RNAs to knock down GPRC6A expression in 22Rv1 or C3H10T1/2 cells, respectively.[51] Knockdown of GPRC6A was associated with loss of both agonist response to Arg, T, and Ocn, and antagonist effects of EGCG (Figure 3A, B).
Figure 3.
EGCG inhibited GPRC6A-mediated biological function. A) EGCG (50 μm) inhibited 10 mm Arg and 100 nm T-stimulated ERK phosphorylation in human prostate cancer cell line 22Rv1 that endogenous expressed GPRC6A. Result shows GPRC6A.siRNA492 (si496, human GPRC6A) knockdown 22Rv1 cells did not respond to Arg, T, and EGCG. B) EGCG (50 μm) inhibited GPRC6A ligand, Ocn (100 ng mL−1) stimulated ERK phosphorylation in mouse fibroblast, C3H10T1/2 cells. C) EGCG (50 μm) blocked 100 ng mL−1 Ocn-stimulated glucose production in C3H10T1/2 cells, but not in GPRC6A.siRNA496 (si492, mouse GPRC6A) knockdown C3H10T1/2 cells. Curcumin (Cur) stimulates D) GPRC6A-mediated ERK phosphorylation and blocked by E) EGCG in HEK-293 cells transfected with GPRC6A cDNA or control HEK-293 cells. Values represent the mean ± SEM. * indicates significant differences from control and stimulated groups at p < 0.05 (n ≥ 4).
To investigate the effects of EGCG on GPRC6A-mediated regulation of glucose production, we performed glucose production studies in mouse embryo fibroblast C3H10T1/2 and C3H10T1/2 GPRC6A knock downed by siRNA cells (C3H10T1/2.si492) (Figure 3C). We found that EGCG attenuated Ocn, a known ligand of GPRC6A, and stimulated glucose production in C3H10T1/2 cells (Figure 3C). We previously showed that the di-phenyl SARM compounds activate GPRC6A.[31] These compounds resemble the structure of curcumin, an active ingredient of turmeric, which has been shown to elevate serum insulin level and improve insulin resistance and glucose homeostasis in db/db mice.[55] We demonstrated that curcumin activates GPRC6A in a heterologous cell expression system, and that EGCG, a GPRC6A antagonist, blocked curcumin stimulation of GPRC6A activity (Figure 3D, E).
3.3. Structural Basis of Agonists and Antagonistic Effects of GA and EGCG
To examine the structural basis for the observed functional effects of GA and EGCG on GPRC6A, we performed computational MD and docking to structural models of GPRC6A.[31,32] For these studies, we developed a new model of l-amino acid binding to the VFT motif (Supporting Information Figures 2 and 3). Residues Ser-149, Ser-171, Thr-172, Tyr-220, and Asp-303 present in the binding pocket (Supporting Information Table 1) are found to be highly conserved in all family C GPCRs. Out of these, Ser-149 and Thr-172 have already been shown using mutagenesis to be important for binding these amino-acids in GPRC6A.[1] Non-conserved Gly-147, Tyr-148, Glu-170, Ala-173, Thr-216, Asp-218, Leu-278, Arg-279, Gln-280, Asn-304, and Leu-411 are other residues predicted to be important from the present docking studies, interacting with all three amino acids known to activate GPRC6A (Supporting Information Table 1 and Supporting Information Figure 4). Out of these binding pocket residues, charged Glu-170 is specific to GPRC6A, whereas in other receptors it is present as a hydrophobic residue (alanine in CaSR) or a polar residue (serine in mGluR1). Similarly, Thr-216 is conserved in GPRC6A and glutamate receptors but not in CaSR where an alanine residue is present instead.[56]
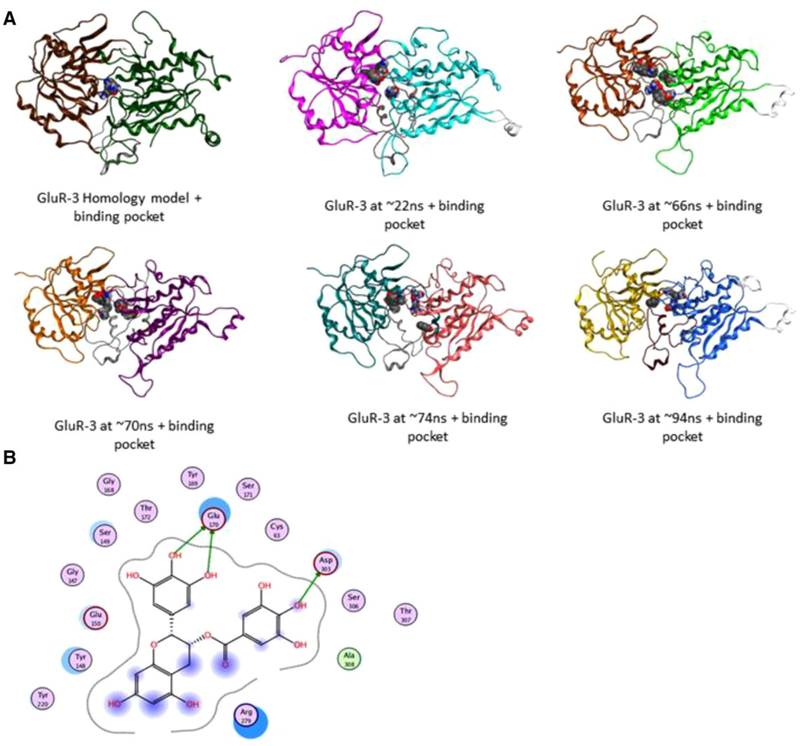
Using an MD-derived conformational ensemble of the GPRC6A VFT together with the previously constructed models of the 7-TM of GPRC6A, we computationally docked GA and the antagonist EGCG. For this, GA and EGCG were docked to each of the 18 selected structures of the VFT and 7-TM domains of GPRC6A (Figure 4 and Supporting Information Figure 4). The MD simulations showed that the VFT domain undergoes closed-to-open conformational changes, and that the open conformation (indicated in cyan and purple on Figure 4A) increases the size of the binding pocket.
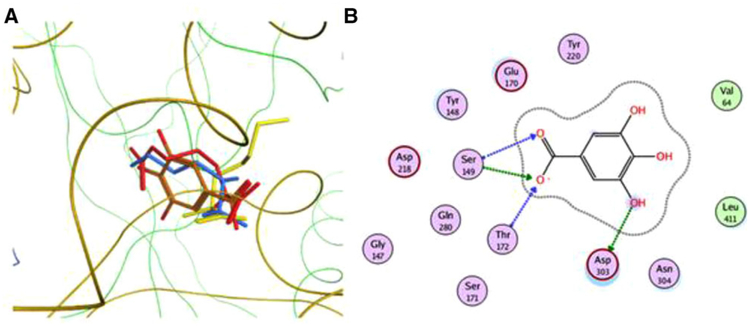
The agonist GA is predicted to bind to the VFT in the same site where the l-amino acids and gallate have also been predicted to bind (Figure 5A). In the homology model of the VFT, Ser-149, Thr-172, and Asp-303 are important residues binding to GA in the pocket (Figure 5B). Glu-170 and Asp-218 are also present in this GA binding pocket. Computational modeling of GPRC6A also permits the in silico binding of compounds that may inhibit GPRC6A, so we examined whether EGCG would be predicted to bind to GPRC6A in our models (Figure 6). Hence, EGCG was also docked to the conformational ensemble of the VFT and 7-TM domains. The binding sites found in VFT conformational ensemble are shown in Figure 6A, and the binding pocket residues of the conformation showing the best docking score are shown in Figure 6B. The docking scores and the binding pocket residues in these sites are given in Tables 1 and 2.
Figure 5.
A) Docking gallate and amino acids in the homology model of the VFT domain of GPRC6A. Red: Arg, yellow: Lys, blue: ornithine, and orange: gallate. These ligands share close binding modes in the small binding pocket. B) Orthosteric binding pocket residues.
Figure 6.
A) Docking of EGCG in starting VFT homology model and different snapshots obtained from MD. The two subdomains of the VFT transition from “closed” to “open” conformation in these snapshots. B) Orthosteric binding pocket residues in 2nd MD snapshot at ≈ 66 ns present in the open conformation.
Table 1.
Autodock Vina binding free energies of T, GA, and EGCG in different models.
| Ligand | Binding free energy score [kcal mol−1] |
|---|---|
| Testosterone | TM homology model: −11.2 |
| TM MD snapshot: −9.6 | |
| VF (closed conformation): +3.2 | |
| VF MD snapshot (open conformation): −6.6 | |
| EGCG | TM homology model: −5.9 |
| TM MD snapshot: −10.8 | |
| VF (closed conformation): −1.8 | |
| VF (open conformation): −7.6 | |
| Gallate | TM homology model: −6.0 |
| TM MD snapshot: −5.8 | |
| VF (closed conformation): −6.9 | |
| VF MD snapshot (open conformation): −5.5 |
Table 2.
Calculated binding site for T, GA, and EGCG based on best Vina docking scores.
| Ligand | Vina docking score (preferred binding sites) | Binding site residues |
|---|---|---|
| Testosterone | TM: −11.2 | F-666, G-667, F-670, A-751, F-752, M-755, L-756, I-759, A-794, W-795, F-798, E-814, V-817, I-818 |
| Gallate | VF (closed conformation): −6.9 | V-64, G-147, Y-148, S-149, E-170, S-171, T-172, D-218, Y-220, L-278, R-279, Q-280, D-303, N-304, L-411 |
| EGCG | TM MD snapshot: −10.8 | R-662, Q-663, T-664, Q-715, I-718, C-719, W-722, L-723, F-725, A-726, P-728, E-746, G-747, S-748, A-751, F-752, M-755, E-814 |
| EGCG | VF (open conformation): −7.6 | C-61, G-147, T-148, S-149, E-150, G-168, Y-169, E-170, S-171, T-172, Y-220, R-279, T-302, D-303, S-306, A-308 |
We found that EGCG binds well in those MD snapshots in which the VFT domain is in the open conformation. Residues Glu-170 and Asp-303 are important for binding/recognizing EGCG (Figure 6B). Ser-149 and Thr-172 are also present in this binding pocket (Figure 6B). These four residues along with Asp-218 form a common group of residues present in the orthosteric binding pocket shown to be interacting with all ligands—amino acids, gallates, and EGCG.
Docking of the antagonist EGCG is particularly interesting in that binding to the closed conformation of the VFT is calculated to be very poor (score of −1.8 kcal mol−1), but binding to the open conformation of the VFT model (score −7.6 kcal mol−1) and in a MD snapshot of the TM domain (score of −10.8 kcal mol−1) are significantly better (Table 1). Table 2 shows binding sites identified to potentially bind GA and EGCG compared to T. The binding site in the VFT domain is present at the interface of the two constituent subdomains that becomes open during the MD simulation, and the other binding site is located in the allosteric pocket of the 7-TM region. Different molecules are calculated to bind more strongly to one or the other site (Table 1).
4. Discussion
We show for the first time that GPRC6A is a molecular target for chemical components of green tea. GPRC6A is activated by gallates, with the rank-order of potency of SG, GA > EG > OG. The basic gallates SG and GA were the most potent agonists and limited structure-activity studies suggest side chain additions decrease activity. The estimated EC50 for GA activation of GPRC6A is in the μm range, well within the GA blood concentrations attained after oral administration of GA or consumption of green tea.[57] In addition, GA stimulated insulin secretion in pancreatic islets that express GPRC6A, similar to the effects of the known GPRC6A ligands, l-Arg, Ocn, and T.[43,58] The reported metabolic effects of GA are consistent with known effects of GPRC6A activation energy metabolism.[41]
In contrast, we found that EGCG acts as a GPRC6A neutral antagonist. EGCG inhibited both agonist-induced GPRC6A signaling in PCa and C3H10T1/2 cells. The IC50 of EGCG inhibition of GPRC6A is consistent with circulating levels of EGCG measured in pharmacokinetic studies.[59] In addition, since EGCG decreased constitutive activity of GPRC6A, it might also be considered an inverse agonist at high concentrations. Since GPRC6A regulates T production and PCa progression,[44,60] the inhibitory effects on GPRC6A are consistent with known effects of EGCG to suppress T,[60] and would also provide a mechanism to account for EGCG reduction in PCa risks. Future studies in preclinical models will be needed prove that the beneficial effects of green tea on PCa progression is mediated by EGCG inhibition of GPRC6A.
Although EGCG inhibited Ocn activation of GPRC6A, we would expect that EGCG administration would inhibit GPRC6A leading to impaired glucose homeostasis, since loss-of-GPRC6A leads to glucose intolerance and hepatic steatosis.[41] However, oral administration of EGCG to animal models is paradoxically associated with improved glucose tolerance, improve insulin sensitivity and reduced hepatic steatosis,[61,62] effects consistent with GPRC6A activation rather than inhibition. This suggests that conversion of EGCG to the GPRC6A agonist GA by degalloylation may account for the observation that EGCG, like GA and Ocn, can improve glucose metabolism in vivo.[28] Additional in vivo studies are also needed to examine whether GA effects on glucose and fat metabolism and EGCG effects preventing PCa progression are lost in Gprc6a−/− mice.
The constructed computational models show differences in the binding of the GA and EGCG that may account for their respective agonists and antagonist effects. It has been suggested that there is an equilibrium between an inactive open–open (dimeric receptor with open VFT in both monomers), and an active open–closed/closed–closed (dimeric receptor with closed VFT in one or both monomers) conformation of GPRC6A.[56] Our VFT conformational ensemble also provides a tentative relationship between the opening of the VFT hinge and antagonist–agonist behavior.
The agonist GA is found to bind well to a conformation of the VFT in which the two subdomains of the VFT are relatively close to each other. In contrast, if the two subdomains are separated, that is, if the VFT hinge is open, GA is calculated to not bind well. In contrast, EGCG, a larger molecule exhibiting inhibitory properties for GPRC6A, is calculated to bind well to the “open” VFT conformation, but not to the “closed” species (Figure 6A). This illustrates how internal protein dynamics plays an important role in ligand binding strength and specificity. We hypothesize that binding to the closed structure of VFT is associated with agonist properties of ligands, while binding to the open structure is associated with antagonist activity (Figures 5 and 6).
EGCG is predicted to bind to both the gallate binding site and the T binding site. These two binding sites form the conserved orthosteric[56] and allosteric binding sites,[63] respectively, observed in the family C GPCRs. Differences in binding/interactions of this molecule in these two sites compared with the binding of the agonist molecules (Table 2) may allow it to act as an antagonist by stabilizing the inactive forms of the VFT or the 7-TM domains that prevent downstream signaling.
Finally, based on our computational modeling of the GA chemical platform, we have now established insights in to how to transition from agonists to the antagonist EGCG by making judicious side chain modifications (Figure 5). Table 1 shows binding sites identified to potentially bind T, GA, and EGCG.
These observations allow some speculation regarding understanding and optimizing the medicinal effects of green tea. First, it suggests that green tea is a mixture of bioactive compounds with opposite physiological effects on glucose homeostasis. If so, the composition of GA and EGCG may determine the net biological effects of different tea preparations[64] and account for the disappointing clinical effects of green tea consumption.[65] Second, the conversion of EGCG to GA by the gut microbiome may also contribute to the variable metabolic responses to green tea observed in clinical studies, and inhibition of EGCG metabolism may be an alternative way to regulate the ratio of EGCG and GA.[28] Potential interactions with endogenous ligands could also influence the timing of green tea consumption to optimize its biological effects.
Finally, new agonists and antagonists for GPRC6A may be designed based on GA and EGCG pharmacophores with greater efficacy, specificity, and reduced toxicity. Indeed, GA derivatives can have mutagenic and cytotoxic effects that can be mitigated by alkyl gallates with side chains varying from five (pentyl gallate) to eight carbons (OG).[66] We found, however, that OG lost GPRC6A agonist activity, but that a biphenyl series of 3-phenoxy-2-hydroxy-N-[4-nitro or cyano-3-(trifluoromethyl) phenyl] propanamide compounds acts as agonists for GPRC6A.[32] These new compounds may also be more selective by lacking biological effects of green tea that are GPRC6A-independent.[67–70]
In conclusion, we have identified that GPRC6A is a molecular target for GA and EGCG components of green tea. The different agonist and antagonist actions of GA and EGCG on GPRC6A activity may account for the differential biological effects of green tea in improving energy metabolism, regulating sex hormone production, and reducing the risk of PCa. Knowing that green tea contains chemical components with opposite effects on GPRC6A activity, and emerging knowledge of GPRC6A’s role in multiple biological processes may help guide the design of more robust clinical studies to evaluate the beneficial effects of green tea and its components. Knowledge of the structure–activity relationship between green tea components and natural ligands sets the stage for the development of novel, more effective and safe compounds that activate and inhibit GPRC6A.
Supplementary Material
Acknowledgments
M.P. and K.K. contributed equally to this work. M.P., K.K., J.B., and L.D.Q. designed the nonclinical studies. M.P., R.Y., and K.K. performed the studies. M.P., K.K., J.S., J.B., and L.D.Q. analyzed the data and wrote the manuscript. The Myc-tagged mouse GPRC6A cDNA was kindly provided by Dr. Yao and Dr. Hampson from Department of Pharmaceutical Sciences, University of Toronto. This work was supported by National Institutes of Health Grant R01-AR37308 and Americans Diabetes Association Grant 1–13-BS-149-BR (to L.D.Q.).
Abbreviations
- EGCG
epigallocatechin gallate
- ERK
extracellular signal-regulated kinase
- GA
gallic acid
- GPCR
G-protein-coupled receptor
- GPRC6A
G-protein-coupled receptor family C member A
- Ocn
osteocalcin
- PCa
prostate cancer
- SG
sodium gallate
- T
testosterone
- TM
transmembrane domain
- VFT
venus fly trap
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Min Pi, Department of Medicine, University of Tennessee Health Science Center, 19 S Manassas St., Memphis, TN 38163, USA.
Karan Kapoor, UT/ORNL Center for Molecular Biophysics, Oak Ridge, TN 37830, USA.
Ruisong Ye, Department of Medicine, University of Tennessee Health Science Center, 19 S Manassas St., Memphis, TN 38163, USA.
Jeremy C. Smith, UT/ORNL Center for Molecular Biophysics, Oak Ridge, TN 37830, USA
Jerome Baudry, UT/ORNL Center for Molecular Biophysics, Oak Ridge, TN 37830, USA; Department of Biochemistry and Cellular and Molecular Biology, University of Tennessee, Knoxville, TN 37996, USA.
Leigh D. Quarles, Department of Biochemistry and Cellular and Molecular Biology, University of Tennessee, Knoxville, TN 37996, USA Department of Medicine, University of Tennessee Health Science Center, 19 S Manassas St., Memphis, TN 38163, USA.
References
- [1].Wellendorph P, Hansen KB, Balsgaard A, Greenwood JR, Egebjerg J, Brauner-Osborne H, Mol. Pharmacol 2005, 67, 589. [DOI] [PubMed] [Google Scholar]
- [2].Afzal M, Safer AM, Al-Bloushi S, BioFactors 2005, 25, 255. [DOI] [PubMed] [Google Scholar]
- [3].Legeay S, Rodier M, Fillon L, Faure S, Clere N, Nutrients 2015, 7, 5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Onakpoya I, Spencer E, Heneghan C, Thompson M, Nutr. Metab. Cardiovasc. Dis 2014, 24, 823. [DOI] [PubMed] [Google Scholar]
- [5].Wang D, Dong H, Zhang H, Zhang Y, Xu Y, Zhao C, Sun Y, Zhou N, ACS. Appl. Mater. Interfaces 2016, 8, 19524. [DOI] [PubMed] [Google Scholar]
- [6].Yang HY, Yang SC, Chao JC, Chen JR, Br. J. Nutr 2012, 107, 749. [DOI] [PubMed] [Google Scholar]
- [7].Li Y, Wang C, Huai Q, Guo F, Liu L, Feng R, Sun C, Diabetes Metab. Res. Rev 2016, 32, 2. [DOI] [PubMed] [Google Scholar]
- [8].Sampath C, Rashid MR, Sang S, Ahmedna M, Biomed. Pharmacother 2017, 87, 73. [DOI] [PubMed] [Google Scholar]
- [9].Yang CS, Wang X, Lu G, Picinich SC, Nat. Rev. Cancer 2009, 9, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dewick PM, Haslam E, Biochem. J 1969, 113, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hsu CL, Yen GC, Br. J. Nutr 2007, 98, 727. [DOI] [PubMed] [Google Scholar]
- [12].Bak EJ, Kim J, Jang S, Woo GH, Yoon HG, Yoo YJ, Cha JH, Scand. J. Clin. Lab. Invest 2013, 73, 607. [DOI] [PubMed] [Google Scholar]
- [13].Sutra T, Oiry C, Azay-Milhau J, Youl E, Magous R, Teissedre PL, Cristol JP, Cros G, J. Agric. Food. Chem 2008, 56, 11683. [DOI] [PubMed] [Google Scholar]
- [14].Totani N, Tateishi S, Takimoto T, Maeda Y, Sasaki H, J. Oleo. Sci 2011, 60, 457. [DOI] [PubMed] [Google Scholar]
- [15].Yang CS, Zhang J, Zhang L, Huang J, Wang Y, Mol. Nutr. Food. Res 2016, 60, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Doan KV, Ko CM, Kinyua AW, Yang DJ, Choi YH, Oh IY, Nguyen NM, Ko A, Choi JW, Jeong Y, Jung MH, Cho WG, Xu S, Park KS, Park WJ, Choi SY, Kim HS, Moh SH, Kim KW, Endocrinology 2015, 156, 157. [DOI] [PubMed] [Google Scholar]
- [17].Kubota K, Sumi S, Tojo H, Sumi-Inoue Y, H IC, Oi Y, Fujita H, Urata H, Nutr. Res 2011, 31, 421. [DOI] [PubMed] [Google Scholar]
- [18].Chen J, Song H, Environ. Toxicol. Pharmacol 2016, 45, 315. [DOI] [PubMed] [Google Scholar]
- [19].Kapoor MP, Sugita M, Fukuzawa Y, Okubo T, J. Nutr. Biochem 2017, 43, 1. [DOI] [PubMed] [Google Scholar]
- [20].Oliveira MR, Nabavi SF, Daglia M, Rastrelli L, Nabavi SM, Pharmacol. Res 2016, 104, 70. [DOI] [PubMed] [Google Scholar]
- [21].Verma S, Singh A, Mishra A, Environ. Toxicol. Pharmacol 2013, 35, 473. [DOI] [PubMed] [Google Scholar]
- [22].Kim HS, Quon MJ, Kim JA, Redox. Biol 2014, 2, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee PMY, Ng CF, Liu ZM, Ho WM, Lee MK, Wang F, Kan HD, He YH, Ng SSM, Wong SYS, Tse LA, Prostate Cancer Prostatic. Dis 2017, 20, 318. [DOI] [PubMed] [Google Scholar]
- [24].Khan N, Bharali DJ, Adhami VM, Siddiqui IA, Cui H, Shabana SM, Mousa SA, Mukhtar H, Carcinogenesis 2014, 35, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jacob SA, Khan TM, Lee LH, Nutr. Cancer 2017, 69, 353. [DOI] [PubMed] [Google Scholar]
- [26].Samavat H, Newman AR, Wang R, Yuan JM, Wu AH, Kurzer MS, Am. J. Clin. Nutr 2016, 104, 1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chakrawarti L, Agrawal R, Dang S, Gupta S, Gabrani R, Expert Opin. Ther. Pat 2016, 26, 907. [DOI] [PubMed] [Google Scholar]
- [28].van’t Slot G, Humpf HU, J. Agric. Food. Chem 2009, 57, 8041. [DOI] [PubMed] [Google Scholar]
- [29].Kumar NB, Pow-Sang J, Egan KM, Spiess PE, Dickinson S, Salup R, Helal M, McLarty J, Williams CR, Schreiber F, Parnes HL, Sebti S, Kazi A, Kang L, Quinn G, Smith T, Yue B, Diaz K, Chornokur G, Crocker T, Schell MJ, Cancer. Prev. Res 2015, 8, 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Guo Y, Zhi F, Chen P, Zhao K, Xiang H, Mao Q, Wang X, Zhang X, Medicine 2017, 96, e6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pi M, Kapoor K, Wu Y, Ye R, Senogles SE, Nishimoto SK, Hwang DJ, Miller DD, Narayanan R, Smith JC, Baudry J, Quarles LD, Mol. Endocrinol 2015, 29, 1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pi M, Kapoor K, Ye R, Nishimoto SK, Smith JC, Baudry J, Quarles LD, Endocrinology 2016, 157, 1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pi M, Nishimoto SK, Quarles LD, Mol. Metab 2017, 6, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ferron M, Hinoi E, Karsenty G, Ducy P, Proc. Natl. Acad. Sci. U.S.A 2008, 105, 5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ferron M, Wei J, Yoshizawa T, Del Fattore A, DePinho RA, Teti A, Ducy P, Karsenty G, Cell 2010, 142, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ferron M, McKee MD, Levine RL, Ducy P, Karsenty G, Bone 2012, 50, 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Oury F, Sumara G, Sumara O, Ferron M, Chang H, Smith CE, Hermo L, Suarez S, Roth BL, Ducy P, Karsenty G, Cell 2011, 144, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mizokami A, Yasutake Y, Gao J, Matsuda M, Takahashi I, Takeuchi H, Hirata M, PloS One 2013, 8, e57375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Oya M, Kitaguchi T, Pais R, Reimann F, Gribble F, Tsuboi T, J. Biol. Chem 2013, 288, 4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mizokami A, Yasutake Y, Higashi S, Kawakubo-Yasukochi T, Chishaki S, Takahashi I, Takeuchi H, Hirata M, Bone 2014, 69, 68. [DOI] [PubMed] [Google Scholar]
- [41].Pi M, Chen L, Huang MZ, Zhu W, Ringhofer B, Luo J, Christenson L, Li B, Zhang J, Jackson PD, Faber P, Brunden KR, Harrington JJ, Quarles LD, PLoS One 2008, 3, e3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Karsenty G, Oury F, Mol. Cell. Endocrinol 2014, 382, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pi M, Wu Y, Quarles LD, J. Bone. Miner. Res 2011, 26, 1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ye R, Pi M, Cox JV, Nishimoto SK, Quarles LD, J. Exp. Clin. Cancer Res 2017, 36, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nishimoto SK, Price PA, J. Biol. Chem 1979, 254, 437. [PubMed] [Google Scholar]
- [46].Nishimoto SK, Price PA, J. Biol. Chem 1985, 260, 2832. [PubMed] [Google Scholar]
- [47].Nishimoto SK, Anal. Biochem 1990, 186, 273. [DOI] [PubMed] [Google Scholar]
- [48].Pi M, Parrill AL, Quarles LD, J. Biol. Chem 2010, 285, 39953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kuang D, Yao Y, Lam J, Tsushima RG, Hampson DR, J. Neurochem 2005, 93, 383. [DOI] [PubMed] [Google Scholar]
- [50].Pi M, Zhang L, Lei SF, Huang MZ, Zhu W, Zhang J, Shen H, Deng HW, Quarles LD, J. Bone Miner. Res 2010, 25, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pi M, Quarles LD, Prostate 2012, 72, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cao J, Huang S, Qian J, Huang J, Jin L, Su Z, Yang J, Liu J, BMC Evol. Biol 2009, 9, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Clemmensen C, Smajilovic S, Wellendorph P, Brauner-Osborne H, Br. J. Pharmacol 2014, 171, 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jacobsen SE, Norskov-Lauritsen L, Thomsen AR, Smajilovic S, Wellendorph P, Larsson NH, Lehmann A, Bhatia VK, Brauner-Osborne H, J. Pharmacol. Exp. Ther 2013, 347, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Seo KI, Choi MS, Jung UJ, Kim HJ, Yeo J, Jeon SM, Lee MK, Mol. Nutr. Food Res 2008, 52, 995. [DOI] [PubMed] [Google Scholar]
- [56].Wellendorph P, Brauner-Osborne H, Br. J. Pharmacol 2009, 156, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shahrzad S, Aoyagi K, Winter A, Koyama A, Bitsch I, J. Nutr 2001, 131, 1207. [DOI] [PubMed] [Google Scholar]
- [58].Pi M, Wu Y, Lenchik NI, Gerling I, Quarles LD, Endocrinology 2012, 153, 4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lee MJ, Maliakal P, Chen L, Meng X, Bondoc FY, Prabhu S, Lambert G, Mohr S, Yang CS, Cancer Epidemiol. Biomarkers Prev 2002, 11, 1025. [PubMed] [Google Scholar]
- [60].Figueiroa MS, Cesar Vieira JS, Leite DS, Filho RC, Ferreira F, Gouveia PS, Udrisar DP, Wanderley MI, Asian J. Androl 2009, 11, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bose M, Lambert JD, Ju J, Reuhl KR, Shapses SA, Yang CS, J. Nutr 2008, 138, 1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chen N, Bezzina R, Hinch E, Lewandowski PA, Cameron-Smith D, Mathai ML, Jois M, Sinclair AJ, Begg DP, Wark JD, Weisinger HS, Weisinger RS, Nutr. Res 2009, 29, 784. [DOI] [PubMed] [Google Scholar]
- [63].Chun L, Zhang WH, Liu JF, Acta Pharmacol. Sin 2012, 33, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kongpichitchoke T, Chiu MT, Huang TC, Hsu JL, Molecules 2016, 21, 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lovera J, Ramos A, Devier D, Garrison V, Kovner B, Reza T, Koop D, Rooney W, Foundas A, Bourdette D, J. Neurol. Sci 2015, 358, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Peredo-Silva L, Fuentes-Retamal S, Sandoval-Acuna C, Pavani M, Maya JD, Castro-Castillo V, Madrid-Rojas M, Rebolledo S, Kemmerling U, Parra E, Ferreira J, Toxicol. Appl. Pharmacol 2017, 329, 334. [DOI] [PubMed] [Google Scholar]
- [67].Cui H, Yuan J, Du X, Wang M, Yue L, Liu J, Oncol. Rep 2015, 33, 1284. [DOI] [PubMed] [Google Scholar]
- [68].Li C, Allen A, Kwagh J, Doliba NM, Qin W, Najafi H, Collins HW, Matschinsky FM, Stanley CA, Smith TJ, J. Biol. Chem 2006, 281, 10214. [DOI] [PubMed] [Google Scholar]
- [69].Kuzuhara T, Sei Y, Yamaguchi K, Suganuma M, Fujiki H, J. Biol. Chem 2006, 281, 17446. [DOI] [PubMed] [Google Scholar]
- [70].Sharma P, Shukla A, Kalani K, Dubey V, Luqman S, Srivastava SK, Khan F, Curr. Cancer Drug Targets 2017, 17, 722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.