Abstract
Regulation of many physiological processes in animals, certainly those controlled by neuropeptide hormones, involves G protein-coupled receptors (GPCRs). Our work focusing on endocrine regulation of diuresis and water balance in mosquitoes and ticks started in 1997 with the kinin receptor, at the dawn of the omics era. After the genomic revolution, we began work on the endocrinology of reproduction in the red imported fire ant. We will use the template of this comparative work to summarize key points about GPCRs and signaling, and emphasize the most recent developments in the pharmacology of arthropod neuropeptide GPCRs. We will discuss omics’ contributions to the advancement of this field, and its influence on peptidomimetic design while emphasizing work on blood feeding arthropods.
Keywords: GPCRs, neuropeptides, tick, mosquito, blood feeding arthropods, High throughput screening, receptor modeling
Introduction
GPCRs, also known as seven transmembrane (7TM) receptors, are cell surface receptors and integral membrane proteins that transduce signals across cell membranes. The most comprehensive structural and functional database for GPCRs is GPCRdb [1]. GPCRs are classified in 5 major classes A-F [2], or according to the human classification GRAFS: Glutamate (Class C), Rhodopsin (Class A), Adhesion (Class B2), Frizzled (Class F) and Secretin (Class B1) [3], and additionally in vertebrates, Taste 2 and orphan GPCRs [4]. Because of the diversity in signals they transduce, these receptors regulate most physiological processes in metazoans. These signals are diverse: light, biogenic amines, neuropeptides and peptide hormones, glutamate, lipoglycoproteins, and protons. Some GPCRs have endogenous activity without the requirement for ligand binding and others have self-proteolytic and activating properties. Upon activation, GPCRs undergo a conformational change that transduces the signal across the membrane.
The GPCR designation derives from their coupling to, and activation of intracellular trimeric G proteins (Gα, Gβ and Gγ), and the G designation for the latter refers to the GTP-binding property of the GTPase subunit α (guanine nucleotide binding and GTP hydrolase). The receptor interacts with the Gα subunit through TM3, TM5, TM6 and intracellular loop 2 (ICL2) and ICL3 [5]. The Gα subunit that exchanges cGDP for cGTP, separates upon receptor activation from the two other subunits, βγ (which remain bound to each other). Downstream, both (α and βγ) modulate activity of different proteins [5]. Once activated, the GPCR signal ends when the αGTPase hydrolyzes cGTP to cGDP, and αGTPase reassociates with Gβγ [5]. Four major Gα-protein families are recognized: Gs (stimulatory, increase cAMP), Gi/o (inhibitory, decrease cAMP), Gq/11 (trigger IP3-mediated release of Ca2+ from calcium stores [5]) and G12/13 (activate RhoGTPase nucleotide exchange factors, RhoGEFs) [6]. In sum, a limited number of G protein classes link to a diverse and high number of GPCRs. All these types of Gα proteins are predicted in the Drosophila genome [7]. The Drosophila eye-specific subunit Gβe is conserved in all arthropods analyzed and most invertebrates encode two Gγ genes [8].
While the above description summarizes a generalized model for GPCR function, other signaling modalities exist, i.e. upon activation GPCRs may be phosphorylated by GPCR kinases (GRKs) and then couple to arrestins [5]; GPCRs may oligomerize [9] or heterodimerize [10]. In Drosophila GPCR desensitization by arrestin encoded by the Kurtz gene functions either at the plasma membrane (low arrestin affinity) or by internalization (high arrestin affinity) [11]. Several silkworm (Bombyx mori) GPCRs for neuropeptides are internalized by arrestin [12].
This review summarizes our opinion on the current state of GPCR research in entomology, with emphasis on arthropod vectors of diseases. Our opinion is based on our academic experience in arthropod endocrinology and neuropeptide receptors. Since the basics of GPCR signaling were summarized above, what follows is the contribution of “omics” to the advancement of GPCR deorphanization and characterization. In addition, the recent incorporation of computational structural biology to modelling of arthropod GPCRs is discussed. Until crystal structures from arthropod GPCRs are available, these models should accelerate efforts of insect physiologists and chemists to design more potent and stable receptor ligands. To end, work on insect kinins will exemplify the evolution of work in this area.
Omics aids GPCR discovery, functional characterization and deorphanization
We have summarized in Fig. 1 the general process of GPCR identification and functional characterization and we will use this framework to highlight work on specific arthropod GPCRs.
Figure 1. Omics contribution to arthropod GPCR discovery and functional analyses.
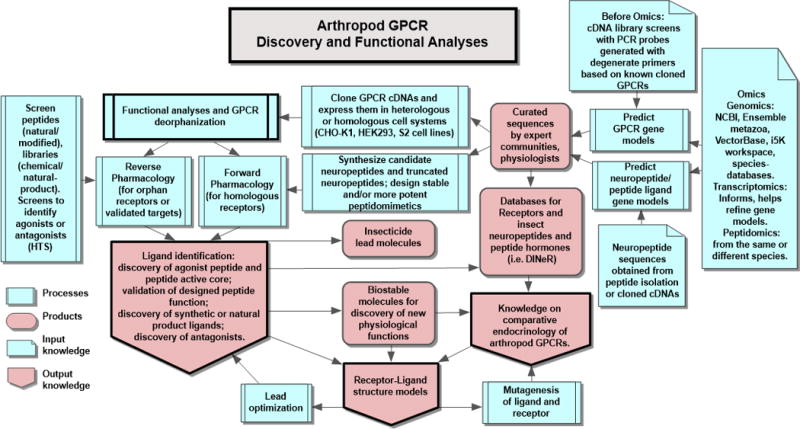
Genomes, transcriptomes, peptidomes and cDNA clones (input knowledge) aid in the curation of protein sequences for both GPCRs and peptide ligands. Curated sequences are organized in databases such as the Database for Insect Neuropeptide Research (DINeR [24]). GPCRs are expressed in cell systems for verification of function. If they are highly similar to other deorphanized receptors this constitutes a forward pharmacology approach. Alternatively, they are deorphanized through a reverse-pharmacological approach to discover either endogenous or synthetic ligands or natural small molecule active ligands by high-throughput screening (HTS). Once a ligand is a “hit” on the receptor, as agonist or antagonist, bioassays are performed, or the ligand is used as a tool to probe suspected functions in tissues (in vitro). These processes or activities result in different products, and converge as output knowledge of new ligands, receptor function, comparative endocrinology and receptor-ligand structure models. “Ligand-docking” informs ligand (= lead) optimization, and models are improved after site-directed mutagenesis experiments.
Identification of receptors and ligands
Kinin peptides and cognate receptors are not present in all insect species and in those where they exist, there are variations in the number of ligands and receptors. Omics and fluorescent analogues revealed in the coleopteran Tribolium castaneum kinin signaling is absent [13]. The tick Ixodes scapularis exhibits an amplification of the kinin signaling system, with 19 peptides predicted in the propeptide and four genes for cognate kinin receptors predicted, one of which we cloned (HM807526) [14]. Transcriptomics of the CNS of the triatomine Rhodnius prolixus supported the genome annotation of GPCRs for neuropeptides and neurotransmitters, with 62 GPCRs annotated [15]. Kinin has been detected in the brain of the kissing bug Triatoma infestans by nano-LC and ESI-OrbiTrap MS/MS in a comparative study supported by transcriptomics [16]. Transcriptome analyses of the southern cattle tick, Ripicephalus microplus, synganglion identified GPCRs, although the low depth of 454 sequencing did not allow to detect the kinin receptor in samples of ticks from Texas [17] which should be present [18], underscoring the often low expression of GPCRs as a challenge for detection. Without genomes available, GPCRs from the foreleg of another cattle tick, Ripicephalus australis, have been predicted from transcriptomic analyses and although no cDNAs were cloned or validated, a model was produced for a class C GABAB receptor [19]. Transcriptome analyses of the Haller’s organ in the dog tick using Illumina Hi-seq only yielded two GPCR transcripts, and expression of one was verified by qRT-PCR [20]. Both of these reports acknowledge limitations in the prediction of tick GPCRs.
Insect genome databases [21,22] facilitate the reverse genetics approach for the discovery of novel neuropeptides [23] and GPCRs. Efforts by the expert community on arthropod neuropeptides are now summarized in the Database for Insect Neuropeptide Research (DINeR) [24]. The predictions of gene products from insect genome sequences complements de novo neuropeptide identification from tissues, followed by transcriptomic studies of specific organs or tissues (Fig. 1). This approach was applied to the discovery of locust tryptopyrokinin genes [25]. The resources mentioned above are identified as input knowledge in Fig. 1 (panels on right).
Functional characterization of GPCRs
Functional characterization of arthropod GPCRs is most often achieved through recombinant expression in mammalian cells, such as Chinese Hamster Ovary cells (CHO-K1) and human embryonic kidney cells (HEK293), or embryonic insect cell lines (Drosophila Schneider S2) [26] (Fig. 1, center, Processes). Functional analyses of arthropod receptors utilizes calcium mobilization assays (using bioluminescence [27] or fluorescence), or by measurements of cAMP [28]. These are G-protein dependent assays often used in academic settings for forward pharmacology and can be low-throughput or high-throughput (Fig. 1). Other G-protein-dependent or G-protein-independent assays (β-arrestin recruitment assay, receptor trafficking and label-free assays) are commercially available [29].
Omics has provided abundant sequences for similarity searches and sequence alignments that provide information to infer the critical residues of both receptors and peptides necessary for activity. From the receptor side, this is best exemplified by the discovery that a single residue distinguishes insect periviscerokinin receptors (PVKr) from PBAN receptors [30]. Analyses of PBAN/pyrokinin (PK)/PVK receptors identified a residue in TM3 (Y125) specific for PVKr, and residue Y/T/W/L 265 in TM5, also as specific for PVKr; these residues are predicted to be involved in specific ligand recognition. We found that Y122 is present in the tick PVKr, and this was relevant to our experiments to differentiate tick pyrokinin from periviscerokinin receptors [31,32]. GPCRs for biogenic amines and most for short neuropeptides belong to the Rhodopsin-like, class A GPCRs. These GPCRs have a conserved three-amino acid residue motif, DRY or ERY, present at the cytosolic extension of TM3 region. This motif is present in tick neuropeptide GPCRs we functionally characterized for kinin, pyrokinin and periviscerokinin [18,31–33], and for serotonin receptors from tick and mosquito [34–36]. Allatostatin C in the mosquito Aedes aegypti inhibits juvenile hormone synthesis by inhibiting the citrate mitochondrial carrier [37]. In allatoregulatory peptides, a clear differentiation of function is associated with variations in this motif that are conserved across different orders: allatostatin C receptors feature the DRY motif; MIP/allatostatin B receptors, QRY; allatotropin receptors, DRW, and FGLa/allatostatin A receptors, DRF [38]. The above and other evidence [39] points to a potential role of the DRY motif in G protein interaction, but receptors also interact throughTM5, TM6 and intracellular loop 2 (ICL2) and ICL3 [5]. A limited number of G proteins display the specific patterns of amino acid residues (selective bar code) for receptor recognition, but different receptors recognize the G protein residues through distinct regions on the receptors [5,40].
If the specific ligand or ligand-type is known, then this constitutes a forward pharmacological approach (Fig. 1, left, Processes). Our work on characterizing the sNPF receptor from fire ants was complicated because all the canonical sNPF peptide ligands that end in the amino acid sequence “LRLRFa” (a= amide) failed to activate the receptor. We tested several Drosophila and mosquito sNPF peptides in both calcium mobilization and cAMP assays without success. It was not until the release of the fire ant genome and confirmation by cloning a cDNA encoding an unusual sNPYa that we succeeded: Two potential ligands predicted from the translated sequence activated the recombinant receptor by decreasing cAMP in CHO-K1 cells. Mass spectrometry of queen brain confirmed the presence of one peptide fragment [28].
Receptor deorphanization involves identifying ligands for those GPCRs that have no known activating molecule(s). This is at the core of reverse molecular pharmacology utilizing screens of candidate- or random ligands. Once a receptor is deorphanized, the physiological function of both ligand and receptor can be elucidated. A phylogenetic analysis of the Drosophila GPCRs and a list of Drosophila G proteins was recently published, where the existence of orphan receptors is acknowledged [7]. Drosophila continues to be the best studied insect and FlyAtlas 2 (www.flyatlas2.org) has recently been released [41]. This database provides tissue expression information with links to a number of fruit fly and malaria mosquito resources for each gene of interest, including GPCRs, and their associated literature. Despite the fact that some orthologs of insect GPCRs have not been identified in the Drosophila genome, the comprehensive genomic sequencing data served as the basis for novel receptor discovery in the model lepidopteran, B. mori. In silkworm, genome mining of neuropeptide GPCRs based on homology to those in Drosophila allowed the identification of contigs with partial GPCR genomic sequences. Cloning of GPCR cDNAs was followed by qRT-PCR expression analyses of tissues of interest (corpora allata and corpora cardiaca), and receptors with highly abundant transcripts were selected for recombinant expression in mammalian cells. This yielded the characterization of the first insect allatotropin receptor [42]. This is an example of reverse pharmacology (Fig. 1, left, Processes). Homologous receptors were later characterized in other insect orders [38].
GPCRs as successful targets in the druggable genome
Arthropod GPCRs are of great interest as potential new targets for pesticide development [43–45]. Human GPCRs are the targets of about 34% of marketed therapeutic drugs [46]. Further, 50% of the marketed drugs targeting peptidergic GPCRs are small molecules [47]. Chemical validation (Fig. 2) of a potential target by a small synthetic molecule is critical to generate initial commercial interest. Current drug discovery approaches for the identification of new compounds that activate or inhibit GPCRs integrate molecular informatics, structural biology, combinatorial library design, and high-throughput screening (HTS) (Fig. 1, left). HTS involves assay development, miniaturization, and automation. For assays relying on G protein coupling, it is a common practice to manipulate the signaling pathway via use of chimeric, mutated, and/or a promiscuous G protein to allow non-Gq-coupled receptors to couple to PLC-IP3-Ca2+ pathway [48]. Most work on functional characterization of arthropod GPCRs or HTS report results after testing one signaling modality (i.e. calcium or cAMP) or using Gα16 for universal signaling through the calcium cascade. This is a limitation in advancing knowledge of arthropod GPCRs because the same GPCR may signal through more than one pathway. Other GPCR technologies for HTS are described elsewhere [29,49].
Figure 2. Key elements of GPCR target validation.
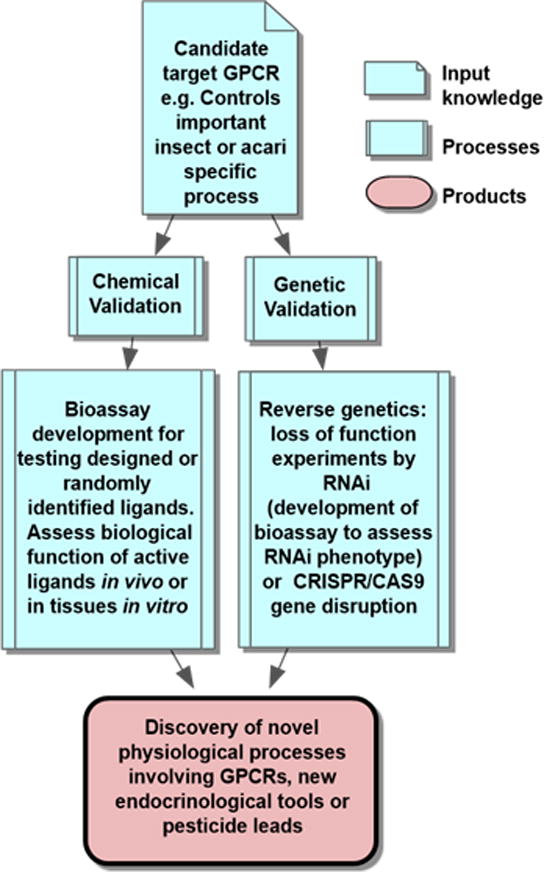
For chemical validation of a GPCR as a candidate target, the designed or identified ligand that is active on the recombinant receptor is applied in vivo and in vitro in tissues. Bioactivity must be determined, either as mortality or by another adverse biological effect derived from its action as antagonist or superagonist (i.e. paralysis). For genetic validation, “loss of function” experiments most typically, or “gain of function” experiments, must confirm the disruption of receptor function has a measurable effect. Through these processes, we validated stable potent peptidomimetics for, and discovered novel functions of the kinin receptor [73].
Perhaps the major obstacle in the exploitation of the GPCR superfamily for insect management is that only one group of GPCRs, the tick octopamine and tyramine/octopamine receptors (biogenic amines GPCRs) are believed to be successful targets of the commercial formamidine acaricide, amitraz [50]. Gross et al. characterized the previously named octopamine receptor as a tyramine/octopamine receptor (TAR1) which is also activated by the amitraz metabolite BTS27271, but only in the presence of tyramine [51]. The presence of mutations associated with resistance to amitraz in another tick octopamine receptor (β-adrenergic-like) gene [52], points to the biogenic amine receptors, β-AOR and perhapsTAR1, as the likely targets of formamidines in ticks. In the lepidopteran Bombyx mori, α- and β-like adrenergic octopamine receptors responded differentially to the amitraz metabolite N2-(2,4-Dimethylphenyl)-N1-methyformamidine (DPMF), being DPMF more potent (at pMolar concentrations) on recombinant β-AOR in a cAMP synthesis assay [53]. Altogether, these results point to insect and tick β-AORs as targets for amitraz metabolites. In an academic setting, dopamine receptors have been targeted for these HTS approaches in both tick Ix. scapularis [54] and mosquitoes [43]. D1-like receptor antagonists were toxic to mosquito larvae [55]. Several vertebrate biogenic amine receptor crystal structures are available to facilitate homologous arthropod receptor modelling [1].
Omics informs computational biology in absence of crystal structures: structural modeling and design of receptor-ligand interactions
The elucidation of GPCR crystal structures has advanced drug design for specificity and potency for medical pharmacology [56,57]. The structural characterization of GPCRs has been possible by baculovirus-mediated expression in insects cells [58]. While there are no published crystal structures for arthropod GPCRs [1], the first homology model for red pigment-concentrating hormone receptor from the water flea crustacean is available [59,60]. While advances have been done in the crystallization of GPCRs, this is still a highly technically challenging field [61]. A strategy to produce abundant allatostatin C receptor protein from silkworm involved expressing both receptor and combinations of eight endogenous G proteins in the baculovirus system. Receptor co-expression in cells with a Gα4β3γ trimer produced higher protein yields, perhaps by stabilizing the receptor [62]. Selecting the optimal G proteins for co-expression may help in producing abundant GPCR protein for other receptors [62].
The lack of crystal structures for insect or tick GPCRs is a major obstacle to conducting in silico screens on them and for the rational design of active ligands targeting them (Fig. 1, below, center). However, an alternative approach is to predict the structure by computational modeling. Sahbaz et al. made in silico structure models of an allatostatin receptor from a stick insect Carausius morosus, and computationally docked a putative peptide ligand into the receptor [63]. They concluded the N-terminus and extracellular loops (ECL2 and ECL3) form the binding pocket. From these modeled receptor-ligand interactions they proposed binding motifs on the receptor extracellular loops and performed receptor site-directed alanine-mutagenesis accordingly. By measuring binding forces between mutant receptors and the ligand through atomic force microscopy, they experimentally validated the importance of a 5-residue motif in ECL3 in the receptor-ligand interaction [63]. Similar computational protocols including sequence and phylogeny analyses, protein structure prediction, protein-ligand docking, and sometimes ligand redesign were used in other studies for arthropod GPCRs [19,64–66]. In the absence of crystal structures these studies point to the power of computational biology, especially when combined with structure-activity relationship (SAR) experiments (including receptor mutagenesis), in studying GPCR-ligand binding and for designing ligands. Meanwhile, obstacles remain in modeling the often-flexible receptor ECLs for structure prediction or ligand docking.
Aided by the accurate peptide structure prediction and the ability of massive peptide production, the trend of small proteins as drugs has started in pharmacological drug design [67]. The European Union funded project nEUROSTRESSPEP focuses on discovery of novel biocontrol agents for insect pests from neuroendocrinology that would be harmless to pollinators. The project aims at testing in field trials rationally designed and selective peptides against arthropod pests by 2020 (http://www.neurostresspep.eu/home).
The arthropod kinin signaling system as a model of neuropeptide GPCR validation towards pesticide discovery
Invertebrate kinins, also known as “leucokinins” are multifunctional neuropeptides [68]. They are part of the neuropeptide “variable set” in insect genomes, not present in all insects [69]. We have studied the kinin signaling system utilizing chemical and reverse genetics tools for validation, as summarized in Fig. 2. Several kinin functions have emerged since their discovery as myotropins: are diuretic in dipterans [70], control pre-ecdysis movements during development, centrally regulate the size of a meal, modulate desiccation and cold tolerance [71] and olfaction. In Ae. aegypti three kinin peptides are encoded in a single mRNA and all three activate the receptor [72]. We recently discovered that the kinin receptor is expressed in labella and tarsi and modulates sugar taste perception in Ae. aegypti females, results that were unexpected considering the known involvement of the kinin receptor in mosquito diuresis [73]. Kinin signaling is crucial for tracheal clearance and air filling prior to ecdysis in Drosophila [74].
The selection of kinin peptides to initiate a validation pipeline of neuropeptide GPCRs as targets for arthropod control fulfilled the a-priori requirement of being a small peptide, i.e. the kinin core analog is 5 amino-acid residues long. This is an important criterion because small peptides may have lesser interactions with the receptor and thus, it should be easier to design or randomly screen libraries to find a small synthetic molecule [75] as ligand that may have commercial potential as lead compounds (pronounced [lēd]) (Fig. 1). The “Lipinski rule of five” which defines small molecules for therapeutic purposes (oral drugs <500 Da) [75], is often violated for many agrochemicals and antibiotics.
The cloning of the R. microplus kinin receptor represented the first neuropeptide GPCR cloned from the Acari and the first kinin arthropod receptor known [18]. For this, we used degenerate primers similar to the lymnokinin receptor from a snail (Fig. 1). Sequence similarity searches allowed us to predict the gene CG10626 encoded the Drosophila kinin receptor [18,76]. The development of CHO-K1 cell lines stably expressing the cattle tick kinin receptor and the Ae. aegypti kinin receptor allowed us to confirm receptor function in calcium bioluminescence assays [33,72] (Fig. 1), and perform comparative structure-activity relationship studies of core kinin analogs and alanine scans to determine critical residues for activity [77]. These studies verified that F at position 1 and W at position 4 of the core kinin pentapeptide of sequence FFSWGa are critical for activity. With this information other analogues were designed by R. Nachman. We validated a potent biostable kinin peptidomimetic “K-Aib-1” (also named 1728; contains α-aminoisobutyric acid) in functional assays with recombinant kinin receptors from the tick and Ae. aegypti and showed it had equal or greater potency than endogenous kinins [78]. Further, this analogue showed antifeedant activity and caused mortality in aphids [79]. Subsequently, Chinese scientists have evaluated a shorter lead Aib analog [80] related to K-Aib-1 (and derivatives thereof) and found that it exhibited two-fold greater potency, underscoring the aphicidal potential for these lead compounds [81]. The K-Aib-1 analog reduced the blood meal size in R. prolixus, which by failing to fully engorge also exhibited a reduced rate of normal ecdysis [82]. Parallel to chemical validation of diuretic peptides in Ae. aegypti females, we began validation of the kinin receptor [70] by RNAi, a reverse genetics approach (Fig. 2). Silencing of the kinin receptor reduced fluid excretion by 50% after a blood meal [70]. We discovered a novel function of the kinin receptor by chemical and genetic validation (Fig. 2). When we provided the K-Aib-1 analog in a sugar solution to starved Ae. aegypti females, contact of tarsi and labella with the mixture elicited their aversive behavior (ET50 ~ 6s). Females walked-, jumped- or flew-away after contacting the mixture [73] and they did not ingest it. The elicited aversive behaviors point to the possibility of exploiting GPCR-modulated sensory perception in mosquitoes by developing environmental or human protection compounds that are deterrent before the female feeds.
Conclusions
The practical exploitation of the insect and Acari (ticks and mites) “GPCRome” for pest control is still in its infancy. Genomics, proteomics and peptidomics should provide needed information on tick GPCRs and ligands for advancing endocrinology. The possible role of TAR1 as a target site for amitraz advances efforts in molecular acaricide resistance monitoring in tick populations. Loss-of-function studies of GPCRs by generating stable mutant strains with CRISPR-Cas9 technology for both receptor and ligand should accelerate discovery in insects, as shown for Ae. aegypti [83]. Further functional analyses of diverse sequences will provide a clearer picture of constraints, commonalities and behaviors of these receptors between vertebrates and invertebrates.
Highlights.
Arthropod GPCRs are under-exploited targets for pest management.
Omics contributes to functional characterization of GPCR-ligand pairs.
Insect GPCR structural models will improve small molecule design.
Kinin peptidomimetics reduced feeding in insect pests.
Acknowledgments
P.V.P. research program is under the United States Department of Agriculture- National Institute of Food and Agriculture Hatch project TEX0-1-9206. Tick research is supported by USDA-NIFA-Agriculture and Food Research Initiative award 2016-67015-24918 to P.V.P., K. Temeyer and J. Coats. Research on fire ants was supported by National Science Foundation- Integrative Organismal Systems award 1257837 to P.V.P and C. Tamborindeguy. Mosquito research is funded by a Texas A&M AgriLife Research seed grant in Vector Biology to P.V.P. and Y.S., the grant that supports C. X., a Ph.D. student in the Entomology graduate program at Texas A&M University.
Footnotes
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
References
- 1••.Pándy-Szekeres G, Munk C, Tsonkov TM, Mordalski S, Harpsøe K, Hauser AS, Bojarski AJ, Gloriam DE. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Research. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. GPCRdb is the most comprehensive database for structural models of GPCRs, ligands, and corresponding G-proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nature Reviews Molecular Cell Biology. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 3.Venter JC. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.Isberg V, de Graaf C, Bortolato A, Cherezov V, Katritch V, Marshall FH, Mordalski S, Pin J-P, Stevens RC, Vriend G, et al. Generic GPCR residue numbers – aligning topology maps while minding the gaps. Trends in Pharmacological Sciences. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Hilger D, Masureel M, Kobilka BK. Structure and dynamics of GPCR signaling complexes. Nature Structural & Molecular Biology. 2018;25:4–12. doi: 10.1038/s41594-017-0011-7. This paper reviews the structural relationships of GPCRs with key molecules regulating signal transduction, G proteins and arrestins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siehler S. Regulation of RhoGEF proteins by G(12/13)-coupled receptors. British Journal of Pharmacology. 2009;158:41–49. doi: 10.1111/j.1476-5381.2009.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanlon CD, Andrew DJ. Outside-in signaling – a brief review of GPCR signaling with a focus on the Drosophila GPCR family. Journal of Cell Science. 2015;128:3533–3542. doi: 10.1242/jcs.175158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan A, Mustafa A, Almén MS, Fredriksson R, Williams MJ, Schiöth HB. Evolutionary hierarchy of vertebrate-like heterotrimeric G protein families. Molecular Phylogenetics and Evolution. 2015;91:27–40. doi: 10.1016/j.ympev.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Pediani JD, Ward RJ, Marsango S, Milligan G. Spatial intensity distribution analysis: Studies of G protein-coupled receptor oligomerisation. Trends in Pharmacological Sciences. 2018;39:175–186. doi: 10.1016/j.tips.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borroto-Escuela OD, Brito I, Romero-Fernandez W, Di Palma M, Oflijan J, Skieterska K, Duchou J, Van Craenenbroeck K, Suárez-Boomgaard D, Rivera A, et al. The G protein-coupled receptor heterodimer network (GPCR-HetNet) and its hub components. International Journal of Molecular Sciences. 2014;15 doi: 10.3390/ijms15058570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson EC, Tift FW, McCauley A, Liu L, Roman G. Functional characterization of kurtz, a Drosophila non-visual arrestin, reveals conservation of GPCR desensitization mechanisms. Insect Biochemistry and Molecular Biology. 2008;38:1016–1022. doi: 10.1016/j.ibmb.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Shen Z, Jiang X, Yang H, Huang H, Jin L, Chen Y, Shi L, Zhou N. Agonist-Activated Bombyx Corazonin Receptor Is Internalized via an Arrestin-Dependent and Clathrin-Independent Pathway. Biochemistry. 2016;55:3874–3887. doi: 10.1021/acs.biochem.6b00250. [DOI] [PubMed] [Google Scholar]
- 13.Halberg KA, Terhzaz S, Cabrero P, Davies SA, Dow JA. Tracing the evolutionary origins of insect renal function. Nature Communications. 2015;6:6800. doi: 10.1038/ncomms7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, Sattelle DB, De La Fuente J, Ribeiro JM, Megy K. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nature Communications. 2016;7:10507. doi: 10.1038/ncomms10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ons S, Lavore A, Sterkel M, Wulff JP, Sierra I, Martínez-Barnetche J, Rodriguez MH, Rivera-Pomar R. Identification of G protein coupled receptors for opsines and neurohormones in Rhodnius prolixus Genomic and transcriptomic analysis. Insect Biochemistry and Molecular Biology. 2016;69:34–50. doi: 10.1016/j.ibmb.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Traverso L, Sierra I, Sterkel M, Francini F, Ons S. Neuropeptidomics in Triatoma infestans. Comparative transcriptomic analysis among triatomines. Journal of Physiology-Paris. 2016;110:83–98. doi: 10.1016/j.jphysparis.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Guerrero FD, Kellogg A, Ogrey AN, Heekin AM, Barrero R, Bellgard MI, Dowd SE, Leung M-Y. Prediction of G protein-coupled receptor encoding sequences from the synganglion transcriptome of the cattle tick, Rhipicephalus microplus. Ticks and Tick-borne Diseases. 2016;7:670–677. doi: 10.1016/j.ttbdis.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes S, He H, Chen A, Lvie G, Pietrantonio P. Cloning and transcriptional expression of a leucokinin‐like peptide receptor from the Southern cattle tick, Boophilus microplus (Acari: Ixodidae) Insect Molecular Biology. 2000;9:457–465. doi: 10.1046/j.1365-2583.2000.00208.x. [DOI] [PubMed] [Google Scholar]
- 19.Munoz S, Guerrero FD, Kellogg A, Heekin AM, Leung MY. Bioinformatic prediction of G protein-coupled receptor encoding sequences from the transcriptome of the foreleg, including the Haller’s organ, of the cattle tick, Rhipicephalus australis. PLoS One. 2017;12:e0172326. doi: 10.1371/journal.pone.0172326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carr AL, D Mitchell R, III, Dhammi A, Bissinger BW, Sonenshine DE, Roe RM. Tick Haller’s organ, a new paradigm for arthropod olfaction: How ticks differ from insects. International Journal of Molecular Sciences. 2017;18:1563. doi: 10.3390/ijms18071563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chilana P, Sharma A, Rai A. Insect genomic resources: status, availability and future. Current Science. 2012:571–580. [Google Scholar]
- 22.Robinson GE, Hackett KJ, Purcell-Miramontes M, Brown SJ, Evans JD, Goldsmith MR, Lawson D, Okamuro J, Robertson HM, Schneider DJ. Creating a buzz about insect genomes. Science. 2011;331:1386–1386. doi: 10.1126/science.331.6023.1386. [DOI] [PubMed] [Google Scholar]
- 23.De Haes W, Van Sinay E, Detienne G, Temmerman L, Schoofs L, Boonen K. Functional neuropeptidomics in invertebrates. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2015;1854:812–826. doi: 10.1016/j.bbapap.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 24.Yeoh JGC, Pandit AA, Zandawala M, Nässel DR, Davies S-A, Dow JAT. DINeR: database for insect neuropeptide research. Insect Biochemistry and Molecular Biology. 2017;86:9–19. doi: 10.1016/j.ibmb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Redeker J, Bläser M, Neupert S, Predel R. Identification and distribution of products from novel tryptopyrokinin genes in the locust, Locusta migratoria. Biochemical and Biophysical Research Communications. 2017;486:70–75. doi: 10.1016/j.bbrc.2017.02.135. [DOI] [PubMed] [Google Scholar]
- 26.Towers PR, Sattelle DB. A Drosophila melanogaster cell line (S2) facilitates post-genome functional analysis of receptors and ion channels. BioEssays. 2002;24:1066–1073. doi: 10.1002/bies.10178. [DOI] [PubMed] [Google Scholar]
- 27.Lu HL, Kersch CN, Taneja-Bageshwar S, Pietrantonio PV. A calcium bioluminescence assay for functional analysis of mosquito (Aedes aegypti) and tick (Rhipicephalus microplus) G protein-coupled receptors. Jove Immunology and Infection. 2011;50 doi: 10.3791/2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bajracharya P, Lu H-L, Pietrantonio PV. The red imported fire ant (Solenopsis invicta Buren) kept Y not F: predicted sNPY endogenous ligands deorphanize the short NPF (sNPF) receptor. PloS One. 2014;9:e109590. doi: 10.1371/journal.pone.0109590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, Xie X. Tools for GPCR drug discovery. Acta Pharmacologica Sinica. 2012;33:372. doi: 10.1038/aps.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurenka R, Nusawardani T. The pyrokinin/pheromone biosynthesis‐activating neuropeptide (PBAN) family of peptides and their receptors in Insecta: evolutionary trace indicates potential receptor ligand‐binding domains. Insect Molecular Biology. 2011;20:323–334. doi: 10.1111/j.1365-2583.2010.01065.x. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Bajracharya P, Castillo P, Nachman RJ, Pietrantonio PV. Molecular and functional characterization of the first tick CAP2b (periviscerokinin) receptor from Rhipicephalus (Boophilus) microplus (Acari: Ixodidae) General and Comparative Endocrinology. 2013;194:142–151. doi: 10.1016/j.ygcen.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Nachman RJ, Pietrantonio PV. Molecular and pharmacological characterization of the Chelicerata pyrokinin receptor from the southern cattle tick, Rhipicephalus (Boophilus) microplus. Insect Biochemistry and Molecular Biology. 2015;60:13–23. doi: 10.1016/j.ibmb.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 33.Holmes S, Barhoumi R, Nachman R, Pietrantonio P. Functional analysis of a G protein‐coupled receptor from the Southern cattle tick Boophilus microplus (Acari: Ixodidae) identifies it as the first arthropod myokinin receptor. Insect Molecular Biology. 2003;12:27–38. doi: 10.1046/j.1365-2583.2003.00384.x. [DOI] [PubMed] [Google Scholar]
- 34.Pietrantonio P, Jagge C, McDowell C. Cloning and expression analysis of a 5HT7‐like serotonin receptor cDNA from mosquito Aedes aegypti female excretory and respiratory systems. Insect Molecular Biology. 2001;10:357–369. doi: 10.1046/j.0962-1075.2001.00274.x. [DOI] [PubMed] [Google Scholar]
- 35.Chen A, Holmes S, Pietrantonio P. Molecular cloning and functional expression of a serotonin receptor from the Southern cattle tick, Boophilus microplus (Acari: Ixodidae) Insect Molecular Biology. 2004;13:45–54. doi: 10.1111/j.1365-2583.2004.00457.x. [DOI] [PubMed] [Google Scholar]
- 36.Lee DW, Pietrantonio P. In vitro expression and pharmacology of the 5HT7‐like receptor present in the mosquito Aedes aegypti tracheolar cells and hindgut‐associated nerves. Insect Molecular Biology. 2003;12:561–569. doi: 10.1046/j.1365-2583.2003.00441.x. [DOI] [PubMed] [Google Scholar]
- 37.Nouzova M, Rivera-Perez C, Noriega FG. Allatostatin-C reversibly blocks the transport of citrate out of the mitochondria and inhibits juvenile hormone synthesis in mosquitoes. Insect Biochemistry and Molecular Biology. 2015;57:20–26. doi: 10.1016/j.ibmb.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verlinden H, Gijbels M, Lismont E, Lenaerts C, Broeck JV, Marchal E. The pleiotropic allatoregulatory neuropeptides and their receptors: A mini-review. Journal of Insect Physiology. 2015;80:2–14. doi: 10.1016/j.jinsphys.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Meyer JM, Ejendal KFK, Watts VJ, Hill CA. Molecular and pharmacological characterization of two D1-like dopamine receptors in the Lyme disease vector, Ixodes scapularis. Insect Biochemistry and Molecular Biology. 2011;41:563–571. doi: 10.1016/j.ibmb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Flock T, Hauser AS, Lund N, Gloriam DE, Balaji S, Babu MM. Selectivity determinants of GPCR–G-protein binding. Nature. 2017;545:317. doi: 10.1038/nature22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Leader DP, Krause SA, Pandit A, Davies SA, Dow JAT. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Research. 2017;46:D809–D815. doi: 10.1093/nar/gkx976. FlyAtlas 2 provides tissue expression information with links to a number of fruit fly and malaria mosquito resources for each gene of interest (including GPCRs) and associated literature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamanaka N, Yamamoto S, Žitňan D, Watanabe K, Kawada T, Satake H, Kaneko Y, Hiruma K, Tanaka Y, Shinoda T. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLOS One. 2008;3:e3048. doi: 10.1371/journal.pone.0003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Sharan S, Hill CA. Potential of GPCR-targeting insecticides for control of arthropod vectors. In: Gross AD, Ozoe Y, Coats JR, editors. Advances in Agrochemicals: Ion Channels and G Protein-Coupled Receptors (GPCRs) as Targets for Pest Control. Vol. 1265. American Chemical Society; 2017. pp. 55–84. (ACS Symposium Series). This review summarizes the current status of the field in finding small molecules as lead compounds targeting GPCRs towards pesticide development. It points out the opportunities and challenges in this emerging field, focusing on GPCRs for biogenic amines such as octopamine, dopamine and serotonin. [Google Scholar]
- 44.Audsley N, Down RE. G protein coupled receptors as targets for next generation pesticides. Insect Biochemistry and Molecular Biology. 2015;67:27–37. doi: 10.1016/j.ibmb.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 45.Ohta H, Ozoe Y. Chapter Two - Molecular signalling, pharmacology, and physiology of octopamine and tyramine receptors as potential insect pest control targets. In: Cohen E, editor. Advances in Insect Physiology. Vol. 46 Academic Press; 2014. pp. 73–166. [Google Scholar]
- 46.Hauser AS, Chavali S, Masuho I, Jahn LJ, Martemyanov KA, Gloriam DE, Babu MM. Pharmacogenomics of GPCR drug targets. Cell. 2018;172:41–54.e19. doi: 10.1016/j.cell.2017.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu F, Song G, de Graaf C, Stevens RC. Structure and function of peptide-binding G protein-coupled receptors. Journal of Molecular Biology. 2017;429:2726–2745. doi: 10.1016/j.jmb.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 48.Hansen KB, Bräuner-Osborne H. FLIPR® assays of intracellular calcium in GPCR drug discovery. In: Leifert WR, editor. G Protein-Coupled Receptors in Drug Discovery. Springer; 2009. pp. 269–278. [DOI] [PubMed] [Google Scholar]
- 49.Miyano K, Sudo Y, Yokoyama A, Hisaoka-Nakashima K, Morioka N, Takebayashi M, Nakata Y, Higami Y, Uezono Y. History of the G protein–coupled receptor (GPCR) assays from traditional to a atate-of-the-art biosensor assay. Journal of Pharmacological Sciences. 2014;126:302–309. doi: 10.1254/jphs.14R13CP. [DOI] [PubMed] [Google Scholar]
- 50.Gross AD, Ozoe Y, Coats JR, editors. Advances in Agrochemicals: Ion Channels and G Protein-Coupled Receptors (GPCRs) as Targets for Pest Control. American Chemical Society; 2017. [Google Scholar]
- 51.Gross AD, Temeyer KB, Day TA, Pérez de León AA, Kimber MJ, Coats JR. Pharmacological characterization of a tyramine receptor from the southern cattle tick, Rhipicephalus (Boophilus) microplus. Insect Biochemistry and Molecular Biology. 2015;63:47–53. doi: 10.1016/j.ibmb.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Corley SW, Jonsson NN, Piper EK, Cutullé C, Stear MJ, Seddon JM. Mutation in the RmβAOR gene is associated with amitraz resistance in the cattle tick Rhipicephalus microplus. Proceedings of the National Academy of Sciences. 2013;110:16772–16777. doi: 10.1073/pnas.1309072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomo K, Takeshi H, Tomohiro O, Haruka T, Hiroshi T, Genyan L, Hiroto O, Fumiyo O, Yoshihisa O. Amitraz and its metabolite differentially activate α‐ and β‐adrenergic‐like octopamine receptors. Pest Management Science. 2017;73:984–990. doi: 10.1002/ps.4412. [DOI] [PubMed] [Google Scholar]
- 54.Ejendal KFK, Meyer JM, Brust TF, Avramova LV, Hill CA, Watts VJ. Discovery of antagonists of tick dopamine receptors via chemical library screening and comparative pharmacological analyses. Insect Biochemistry and Molecular Biology. 2012;42:846–853. doi: 10.1016/j.ibmb.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 55.Nuss AB, Ejendal KF, Doyle TB, Meyer JM, Lang EG, Watts VJ, Hill CA. Dopamine receptor antagonists as new mode-of-action insecticide leads for control of Aedes and Culex mosquito vectors. PLoS Neglected Tropical Diseases. 2015;9:e0003515. doi: 10.1371/journal.pntd.0003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nature Reviews Drug Discovery. 2017;16:829. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Alexander SPH, Christopoulos A, Davenport AP, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD, Pawson AJ, Sharman JL, et al. The Concise guide to pharmacology 2017/18: G protein-coupled receptors. British Journal of Pharmacology. 2017;174:S17–S129. doi: 10.1111/bph.13878. This reference has active links to other online pharmacological resources for GPCRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saarenpää T, Jaakola V-P, Goldman A. Chapter Nine - Baculovirus-mediated expression of GPCRs in insect cells. In: Shukla AK, editor. Methods in Enzymology. Vol. 556. Academic Press; 2015. pp. 185–218. [DOI] [PubMed] [Google Scholar]
- 59.Jackson GE, Pavadai E, Gäde G, Timol Z, Andersen NH. Data for the homology modelling of the red pigment-concentrating hormone receptor (Dappu-RPCHR) of the crustacean Daphnia pulex, and docking of its cognate agonist (Dappu-RPCH) Data in Brief. 2017;15:941–947. doi: 10.1016/j.dib.2017.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson GE, Pavadai E, Gäde G, Timol Z, Andersen NH. Interaction of the red pigment-concentrating hormone of the crustacean Daphnia pulex, with its cognate receptor, Dappu-RPCHR: A nuclear magnetic resonance and modeling study. International Journal of Biological Macromolecules. 2018;106:969–978. doi: 10.1016/j.ijbiomac.2017.08.103. [DOI] [PubMed] [Google Scholar]
- 61.Ghosh E, Kumari P, Jaiman D, Shukla AK. Methodological advances: the unsung heroes of the GPCR structural revolution. Nature Reviews Molecular Cell Biology. 2015;16:69. doi: 10.1038/nrm3933. [DOI] [PubMed] [Google Scholar]
- 62.Chen J, Xu J, Hino M, Yamashita M, Hirata K, Patil AA, Tatsuke T, Mon H, Banno Y, Kusakabe T, et al. Co-expression of silkworm allatostatin-C receptor BNGR-A1 with its cognate G protein subunits enhances the GPCR display on the budding baculovirus. Journal of Asia-Pacific Entomology. 2016;19:753–760. [Google Scholar]
- 63••.Sahbaz BD, Sezerman OU, Torun H, Iyison NB. Ligand binding pocket of a novel allatostatin receptor type C of stick insect, Carausius morosus. Scientific Reports. 2017;7:41266. doi: 10.1038/srep41266. The study points to the power of computational biology in identifying critical residues in GPCR in the absence of available crystal structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim J-h, Kim S-K, Lee J-H, Kim Y-J, Goddard WA, III, Kim Y-C. Homology modeling and molecular docking studies of Drosophila and Aedes sex peptide receptors. Journal of Molecular Graphics and Modelling. 2016;66:115–122. doi: 10.1016/j.jmgm.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 65.Leander M, Bass C, Marchetti K, Maynard BF, Wulff JP, Ons S, Nichols R. Cardiac contractility structure-activity relationship and ligand-receptor interactions; the discovery of unique and novel molecular switches in myosuppressin signaling. PloS one. 2015;10:e0120492. doi: 10.1371/journal.pone.0120492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu N, Zotti MJ, Scheys F, Braz ASK, Penna PHC, Nachman RJ, Smagghe G. Flexibility and extracellular opening determine the interaction between ligands and insect sulfakinin receptors. Scientific Reports. 2015;5:12627. doi: 10.1038/srep12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kruger RP. The coming peptide tidal wave. Cell. 2017;171:497. doi: 10.1016/j.cell.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 68.Coast GM, Schooley DA. Diuretic/Antidiuretic. In: Kastin AJ, editor. Handbook of Biologically Active Peptides. Second. Elsevier; 2013. pp. 229–236. [Google Scholar]
- 69.Hauser F, Neupert S, Williamson M, Predel R, Tanaka Y, Grimmelikhuijzen CJP. Genomics and peptidomics of neuropeptides and protein hormones present in the parasitic wasp Nasonia vitripennis. Journal of Proteome Research. 2010;9:5296–5310. doi: 10.1021/pr100570j. [DOI] [PubMed] [Google Scholar]
- 70.Kersch CN, Pietrantonio PV. Mosquito Aedes aegypti (L.) leucokinin receptor is critical for in vivo fluid excretion post blood feeding. FEBS letters. 2011;585:3507–3512. doi: 10.1016/j.febslet.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 71.Cannell E, Dornan AJ, Halberg KA, Terhzaz S, Dow JA, Davies S-A. The corticotropin-releasing factor-like diuretic hormone 44 (DH44) and kinin neuropeptides modulate desiccation and starvation tolerance in Drosophila melanogaster. Peptides. 2016;80:96–107. doi: 10.1016/j.peptides.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pietrantonio P, Jagge C, Taneja‐Bageshwar S, Nachman R, Barhoumi R. The mosquito Aedes aegypti (L.) leucokinin receptor is a multiligand receptor for the three Aedes kinins. Insect Molecular Biology. 2005;14:55–67. doi: 10.1111/j.1365-2583.2004.00531.x. [DOI] [PubMed] [Google Scholar]
- 73••.Kwon H, Agha MA, Smith RC, Nachman RJ, Marion-Poll F, Pietrantonio PV. Leucokinin mimetic elicits aversive behavior in mosquito Aedes aegypti (L.) and inhibits the sugar taste neuron. Proceedings of the National Academy of Sciences. 2016;113:6880–6885. doi: 10.1073/pnas.1520404113. The first report of the kinin receptor, a diuretic peptide GPCR, in sensory structures and regulating sugar taste perception. A biostable kinin mimetic, analog 1728 (K-Aib-1) is a feeding deterrent and promotes behavioral avoidance. The approach combines feeding assays, behavioral recordings, receptor cloning, immunolocalization, electrophysiology and RNAi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim D-H, Kim Y-J, Adams ME. Endocrine regulation of airway clearance in Drosophila. Proceedings of the National Academy of Sciences. 2018:201717257. doi: 10.1073/pnas.1717257115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lipinski CA. Rule of five in 2015 and beyond: Target and ligand structural limitations, ligand chemistry structure and drug discovery project decisions. Adv Drug Deliv Rev. 2016;101:34–41. doi: 10.1016/j.addr.2016.04.029. [DOI] [PubMed] [Google Scholar]
- 76.Radford JC, Davies SA, Dow JA. Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. Journal of Biological Chemistry. 2002;277:38810–38817. doi: 10.1074/jbc.M203694200. [DOI] [PubMed] [Google Scholar]
- 77•.Taneja‐Bageshwar S, Strey A, Zubrzak P, Pietrantonio PV, Nachman RJ. Comparative structure‐activity analysis of insect kinin core analogs on recombinant kinin receptors from southern cattle tick Boophilus microplus (Acari: Ixodidae) and mosquito Aedes aegypti (Diptera: Culicidae) Archives of Insect Biochemistry and Physiology. 2006;62:128–140. doi: 10.1002/arch.20129. [DOI] [PubMed] [Google Scholar]
- 78.Taneja-Bageshwar S, Strey A, Isaac RE, Coast GM, Zubrzak P, Pietrantonio PV, Nachman RJ. Biostable agonists that match or exceed activity of native insect kinins on recombinant arthropod GPCRs. General and Comparative Endocrinology. 2009;162:122–128. doi: 10.1016/j.ygcen.2008.10.013. Kinin peptidomimetics incorporating aminoisobutyric acid were systematically tested on mosquito Ae. aegypti and tick R. micropus kinin receptors stably expressed in CHO-K1 cell lines, allowing for comparisions of activity and establishing the high potency of analog 1728 (K-Aib-1). [DOI] [PubMed] [Google Scholar]
- 79•.Smagghe G, Mahdian K, Zubrzak P, Nachman RJ. Antifeedant activity and high mortality in the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidae) induced by biostable insect kinin analogs. Peptides. 2010;31:498–505. doi: 10.1016/j.peptides.2009.07.001. Designed kinin analog 1728 (K-Aib-1) exhibits high aphicidal activity. [DOI] [PubMed] [Google Scholar]
- 80.Nachman RJ, Isaac RE, Coast GM, Holman GM. Aib-containing analogues of the insect kinin neuropeptide family demonstrate resistance to an insect angiotensin-converting enzyme and potent diuretic Activity. Peptides. 1997;18:53–57. doi: 10.1016/s0196-9781(96)00233-1. [DOI] [PubMed] [Google Scholar]
- 81.Zhang C, Qu Y, Wu X, Song D, Ling Y, Yang X. Eco-friendly insecticide discovery via peptidomimetics: Design, synthesis, and aphicidal activity of novel insect kinin analogues. Journal of Agricultural and Food Chemistry. 2015;63:4527–4532. doi: 10.1021/acs.jafc.5b01225. [DOI] [PubMed] [Google Scholar]
- 82•.Lange AB, Nachman RJ, Kaczmarek K, Zabrocki J. Biostable insect kinin analogs reduce blood meal and disrupt ecdysis in the blood-gorging Chagas’ disease vector, Rhodnius prolixus. Peptides. 2016;80:108–113. doi: 10.1016/j.peptides.2016.01.012. Kinin analog 1728 is effective in reducing blood meal size and interfering with normal ecdysis by preventing full blood-goring in another Hemipteran. [DOI] [PubMed] [Google Scholar]
- 83••.Duvall LB, Basrur NS, Molina H, McMeniman CJ, Vosshall LB. A peptide signaling system that rapidly enforces paternity in the Aedes aegypti mosquito. Current Biology. 2017;27:3734–3742.e3735. doi: 10.1016/j.cub.2017.10.074. This group employed CRISPR-Cas9 to generate both receptor and neuropeptide mutant strains of Ae. aegypti for functional assays in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]


