Abstract
SPOAN syndrome is a neurodegenerative disorder mainly characterized by spastic paraplegia, optic atrophy and neuropathy (SPOAN). Affected patients are wheelchair bound after 15 years old, with progressive joint contractures and spine deformities. SPOAN patients also have sub normal vision secondary to apparently non-progressive congenital optic atrophy. A potential causative gene was mapped at 11q13 ten years ago. Here we performed next-generation sequencing in SPOAN-derived samples. While whole-exome sequencing failed to identify the causative mutation, whole-genome sequencing allowed to detect a homozygous 216-bp deletion (chr11.hg19:g.66,024,557_66,024,773del) located at the non-coding upstream region of the KLC2 gene. Expression assays performed with patient's fibroblasts and motor neurons derived from SPOAN patients showed KLC2 overexpression. Luciferase assay in constructs with 216-bp deletion confirmed the overexpression of gene reporter, varying from 48 to 74%, as compared with wild-type. Knockdown and overexpression of klc2 in Danio rerio revealed mild to severe curly-tail phenotype, which is suggestive of a neuromuscular disorder. Overexpression of a gene caused by a small deletion in the non-coding region is a novel mechanism, which to the best of our knowledge, was never reported before in a recessive condition. Although the molecular mechanism of KLC2 up-regulation still remains to be uncovered, such example adds to the importance of non-coding regions in human pathology.
Introduction
Hereditary spastic paraplegias (HSPs) are common neurodegenerative genetic disorders in which patients present progressive spasticity and lower limbs weakness. Up to date, more than 70 loci had been associated with HSPs and at least 50 genes have been identified (1). In 2005, our group identified in a geographic isolate in the backlands of Northeastern Brazil, 26 Caucasian individuals belonging to consanguineous families with an autosomal recessive (AR) complicated form of HSP, which associates spastic paraplegia, optic atrophy and neuropathy (SPOAN syndrome, OMIM #609541) (2). This condition is characterized by onset of progressive spastic paraplegia in infancy, and progressive motor and sensory axonal neuropathy in late childhood/early adolescence leading to severe motor disability. All patients are wheelchair bound after 15 years old, with progressive joint contractures and spine deformities. Patients also have sub normal vision secondary to apparently non-progressive congenital optic atrophy, dysarthria starting in the third decade of life and exacerbated acoustic startle response. Patients show no intellectual impairment. Ten years after the gene mapping, more than 70 individuals from this cluster, three unrelated affected individuals from Southern and Southeast Brazil, and a pair of Egyptian siblings were diagnosed with SPOAN. Although, all patients share the same haplotype spanning 2.3 Mb into chromosome region 11q13, Sanger sequencing of candidate genes failed to reveal the causative gene (3). Here we describe the SPOAN causative mutation, a small deletion in the non-coding region that causes gene overexpression. Gain of function in a recessive condition is a novel mechanism that, to the best of our knowledge, was never reported before.
Results
Next-generation sequencing and SPOAN mutation
Whole-exome sequencing (WES) was performed in genomic DNA from one Brazilian and one Egyptian patient diagnosed with SPOAN syndrome. We identified six homozygous variants at the critical region, but population frequency and segregation analysis excluded four variants, while the remaining two were SNPs located in non-coding region, suggesting that these two were unlikely to be associated to the clinical phenotype (Supplementary Material, Table S1). Although WES failed to reveal the SPOAN mutation, the sequencing allowed us to refine the critical interval on chromosome 11q13 to 1.77 Mb, between markers rs508548 (A>G at 65,626,289 position in CFL1) and an undescribed variant located at 67,395,410 (G>C in NUDT8). Next, using whole-genome sequencing (WGS), we identified a homozygous 216-bp deletion (chr11.hg19:g.66,024,557_66,024,773del), located at the non-coding upstream region of kinesin light chain-2 (KLC2) (Supplementary Material, Fig. S1). This variant was detected in homozygosity in all affected Brazilian individuals (n = 73), and in the Egyptian affected siblings, while it was not present in homozygosity in 111 healthy Brazilian relatives. This 216-bp deletion was also absent in 474 Brazilian healthy controls and is not described in the 1000 genomes database.
Gene expression analysis
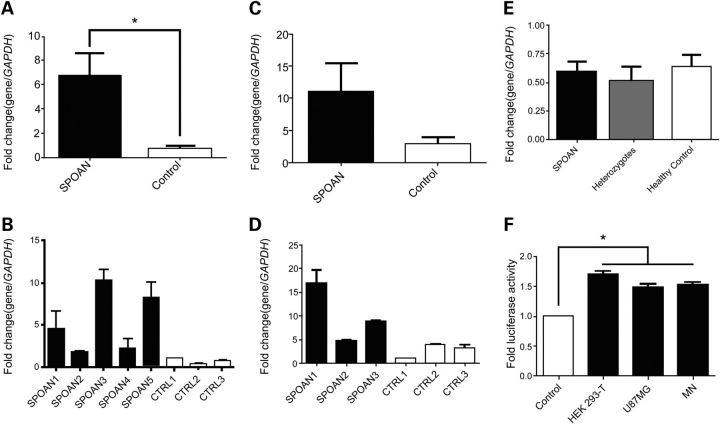
To verify if the deletion affects the expression level of genes located in SPOAN critical region, we performed expression array using cDNA from fibroblasts. Several genes (n = 23; Supplementary Material, Table S2) showed differential expression in patients compared with controls (P < 0.01). Unexpectedly, this assay revealed KLC2 overexpression. Quantitative reverse transcription PCR (RT-qPCR) performed using fibroblast cDNA samples confirmed the expression array results (Fig. 1A). We next generated induced pluripotent stem-cells (iPSC) which were differentiated into motor neurons (MN). RT-qPCR using MN samples revealed KLC2 up-regulation in SPOAN patients compared with healthy controls, confirming the over expression observed in the previous experiments (Fig. 1C). Also we investigated KLC2 expression in blood, using a larger number of cDNA samples from healthy controls, heterozygotes and affected individuals. This assay did not reveal any difference in expression levels between heterozygotes compared with SPOAN's and to healthy controls (Fig. 1E).
Figure 1.
Effect of 216-bp deletion on KLC2 expression. (A) Relative expression of KLC2 measured by RT-qPCR performed on fibroblast cDNA isolated from SPOAN patients and healthy controls (P < 0.05; Nonparametric test [Mann–Whitney]). (B) KLC2 relative expression measured on fibroflast samples from individual patients and healthy controls. (C) Relative expression of KLC2 measured by RT-qPCR using MN. (D) KLC2 relative expression measured on MN samples from individual patients and healthy controls. (E) KLC2 relative expression measured on whole-blood cDNA samples from affected (homozygotes), heterozygotes and healthy controls. Each RT-qPCR experiment was performed in triplicate and each sample was replicated twice. (F) Expression of luciferase reporter gene controlled by the 216-bp-deleted KLC2 regulatory region relative to the expression controlled by the wild-type KLC2 regulatory region measured in HEK293 T, U87MG and MN cells. Each experiment was performed in triplicate and each cell type was replicated twice (P < 0.05; One-way ANOVA).
To investigate if the 216-bp deletion is the cause of KLC2 up-regulation, we performed luciferase gene reporter assay using three cell lines (HEK293T, U87MG and MN), which were transfected with two constructs: a KLC2 wild-type promoter and KLC2 216-bp deleted regulatory region driving the Luciferase gene. In the three cell lines, the construct with the 216-bp deletion produced a luciferase activity increment compared with wild-type promoter, varying from 48 to 74% (Fig. 1F).
Klc2 knockdown and overexpression in Danio rerio
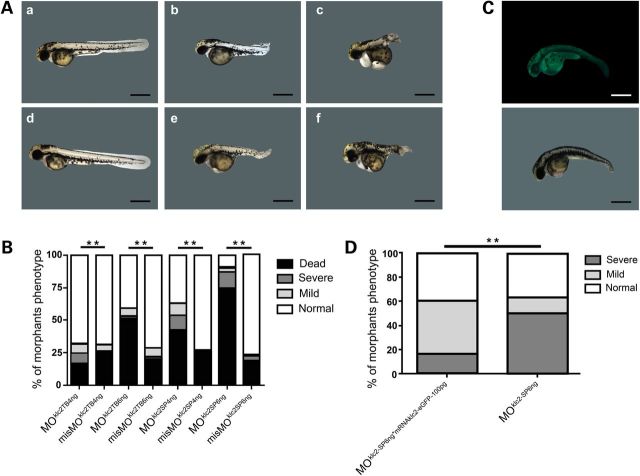
We then used Danio rerio as an animal model to study the ‘in vivo’ effect of klc2 knockdown and overexpression. Knockdown regulation was achieved by microinjecting zebrafish embryos with two different klc2 morpholinos (translation blocking morpholino [MOklc2-TB] and splice morpholino [MOklc2-SP]), each one at doses of 4 and 6 ng. Mild phenotype was defined for embryos showing curly-tail and circular swimming whereas severe phenotype for embryos with dramatically shortened and twisted tail and that were unable to swim. Both phenotypes became evident at 48-h post fertilization (hpf) (Fig. 2A). In all cases, statistically significant differences were observed between mismatch-MO and specific-MO injected embryos. For both morpholino strategies when comparing to the respective mismatch-MO controls, an increase in lethality and/or frequency of phenotypes was mainly observed in detriment of normal phenotype. Furthermore, this difference was more evident when higher amount of either MOklc2-TB or MOklc2-SP was injected (Fig. 2B). Phenotype rescue assays were performed by co-injection of 100 pg of mRNAklc2-eGFP and splice morpholino at 6 ng (Fig. 2C), and an improvement of ∼33% (P < 0.01), from severe to mild phenotype, was consistently observed (Fig. 2D).
Figure 2.
Effect of klc2 knockdown in zebrafish. (A) (a and d) Embryos microinjected with control splicing blocking morpholino: (a) misMOklc2-SP4ng (d) misMOklc2-SP6ng. (b, c, e and f) Embryos microinjected with splicing blocking morpholino: (b and c) MOklc2-SP4ng (e and f) MOklc2-SP6ng. Normal (a and d), mild curly-tail (b and e) and severe curly-tail (c–f) phenotypes were recorded at 48-hpf. (B) Frequencies of observed phenotypes among morphants. Number of microinjected embryos: MOklc2-TB4ng (292); misMOklc2-TB4ng (305); MOklc2-TB6ng (103); misMOklc2-TB6ng (249); MOklc2-SP4ng (283); misMOklc2-SP4ng (234); MOklc2-SP6ng (423); misMOklc2-SP6ng (370). P < 0.01, χ2 test. (C) Fluorescent embryo coinjected with 6 ng MOklc2-SP and 100 pg mRNAklc2-eGFP (selected by fluorescence at 24-hpf) showing mil-curly tails was recorded at 48-hpf. Scale bar 200 µm. (D) Embryos coinjected with 6 ng MOklc2-SP and 100 pg mRNAklc2-eGFP (n = 70 embryos) showed a partial rescue of morphant phenotype compared with MOklc2-SP6ng (n = 30 embryos). P < 0.01, χ2 test.
As SPOAN syndrome seems to result from KLC2 up-regulation, we mimicked this condition in zebrafish by microinjecting mRNAklc2-eGFP in specific concentrations in embryos. Fluorescent embryos displayed similar phenotype to klc2 morphants (Fig. 3A). A high lethality (over than 70%) was observed in embryos microinjected with mRNAklc2-eGFP at 200 pg at 24-hpf stage and we excluded this concentration data in phenotype analysis (Fig. 3B). We observed higher frequency of curly-tail phenotype in embryos microinjected with mRNAklc2-eGFP compared with control (mRNAeGFP), being statistical significant in embryos microinjected at 150 pg mRNA concentration (P < 0.05) (Fig. 3C).
Figure 3.
Effect of klc2 overexpression in zebrafish. (A) (a and b) Embryos microinjected with 100 pg of mRNAeGFP and (c–f) embryos microinjected with 100 pg of mRNAklc2-eGFP were selected by fluorescence at 24-hpf, and recorded at 48-hpf. (a and b) Normal Embryos, (c and d) mild curly-tail and (e and f) severe curly-tail phenotypes. Scale bars: (a and b) 200 µm; (c–f) 100 µm. (B) Lethality frequency observed in embryos at 24-hpf stage. Embryos microinjected with 200 pg mRNAklc2-eGFP showed high lethality and were excluded form phenotype analysis. (C) Frequencies of observed phenotypes among GFP-fluorescent embryos. Numbers of embryos selected by fluorescence: mRNAklc2-eGFP-100pg = 32; mRNAeGFP-100pg = 16; mRNAklc2-eGFP-150pg = 43; mRNAklc2-eGFP-150pg = 52. P < 0.05, χ2 and Fisher's exact tests.
Discussion
We previously mapped the SPOAN gene, responsible for a syndromic form of AR spastic paraplegia, at 11q13 (2,3). Based on next-generation sequencing, we were able to uncover a new causative mechanism for this condition. We observed that a small deletion in KLC2 non-coding region is responsible for the gene up-regulation and SPOAN phenotype. Additionally, BSCL2 and FLRT1, two genes previously associated with HSP and located nearby but outside the 11q13 critical region, were excluded as candidates (4,5). The Egyptian patients reported in this study as SPOAN carried the c.T2023C (stop loss) homozygous mutation in FLRT1, and were previously assigned by Novarino et al. (5) (Family 709) as SPG68. However, here we suggest that 216-bp deletion, shared by all SPOAN patients, is probably the causative mutation in both Egyptian siblings, rather than the reported FLRT1 mutation.
KLC2 codes for KLC2, a protein involved in anterograde axoplasmatic transport of organelles and macromolecules cargoes (6–10). KLC2 is a part of kinesin protein-1 complex (11), which binds to kinesins heavy chain in a stoichiometric ratio of 1:1 (12), being highly expressed in neurons. Several neurodegenerative diseases show impairment in axonal transport (13,14) and some kinesins heavy chains (KIF5A, KIF1A and KIF1C) have been associated with HSP (15–18). Animal models have also shown that disturbance of axonal transport proteins cause neurodegenerative disease and axon degeneration (10,19–21). Although the disease mechanism described here involves a homozygous deletion in a non-coding region, all these observations strongly suggest that KCL2 is the causative gene for SPOAN.
According to the RepeatMask database, KLC2 upstream region was generated by a non-LTR retrotransposon (L3/CR-1) insertion. DNA footprint and alignment of L3/CR-1 did not show conservation among distant species, but the high conservation observed among primates suggests it was inserted during the divergence of primates from other mammals. In several human populations, KLC2 surrounding region (10-kb up- and downstream) and three described SNPs surrounding the mutation location have low fixation index (FST) (Supplementary Material, Fig. S2) (rs116801155, rs190099601 and rs76627914 with FST of 0.0044, 0.0002 and 0.0427, respectively), indicating a high conservation in humans.
Surprisingly, the small deletion in its non-coding upstream region causes KLC2 overexpression, suggesting a novel molecular mechanism never report before, a gain of function in recessive condition. Intriguingly, the 216-bp deletion overlaps 9-bp of 5′-untranslated region (5′-UTR) of the largest KLC2 transcript (NM_001134775.1), which means that this mutation is located at KLC2 promoter region (upstream of the transcription start site [TSS]) and it should cause gene downregulation instead gain of function. Although this region has characteristics of a promoter (enrichment of H3Kme3, DNase I hypersensitive sites [DHS], RNA pol II binding sites, etc.), transcription factors complexes that bind at this region may act as transcriptional repressor, which could explain the gene up-regulation. Additionally, this deletion overlaps an unspliced antisense long non-coding RNA (lncRNA, AU311830.1) and regulatory elements: DHS, several transcription factors binding sites (TFBS), histone marks and DNA methylation (Supplementary Material, Fig. S1). Thus, a disruption of this non-coding and regulatory region might alter the expression level of downstream genes, which can explain SPOAN gain of function.
Expression analysis showed an unexpected KLC2 overexpression from fibroblast and MN SPOAN samples. Because SPOAN is a recessive condition, we tried to check the KLC2 expression pattern in heterozygous samples. Whole-blood samples collected from a large number of heterozygotes did not reveal increased KLC2 expression, when compared with homozygotes and healthy controls. These results suggest a tissue-specific effect since 216-bp deletion causes KLC2 up-regulation in fibroblast and MN cell-lines, but does not in blood. Also, luciferase assay showed that reporter constructs with 216-bp deletion have increased luciferase activity when compared with the wild-type. These results support the hypothesis that the 216-bp deletion located at non-coding region is likely the responsible for the KLC2 overexpression.
Zebrafish has been an interesting animal model used in genetic studies due to its fast embryonic development and the fact it carries several human orthologues genes. The percentages of lethality and animals with curly-tail phenotype observed in morphants in this study were similar to those reported in several reports that employed zebrafish for other HSP (22–28). Microinjection of mRNAklc2-eGFP in zebrafish embryos showed a similar phenotype of klc2 morphants, which reinforces our hypothesis that klc2 is an essential gene for MN function and development. Thus, we hypothesize that imbalance of KLC2 gene expression results in neurodegenerative phenotype in humans.
Gene overexpression had been associated with several neurological disorders but none of them have AR inheritance. For example, duplication or triplication of PLP1 cause Pelizaeus-Merzbacher disease (OMIM #312080) (29–33) and PMP22 duplication causes Charcot-Marie-Tooth disease type 1A (OMIM #118220), a hereditary demyelinating neuropathy (34,35). Variants detected upstream APP region were associated with up-regulation of APP protein in Alzheimer disease and Down syndrome patients (36). Additionally, downregulation or complete disruption of protein synthesis is usually the common mechanism in HSP in which functional studies have been conducted. For instance, this is the case in X-linked [e.g. L1CAM (37)], autosomal dominant [e.g. ATL1 (38) and SPAST (39)] and AR conditions [as SPG20 (40) and FA2H (41)].
In short, several unexpected and surprising results were observed during SPOAN syndrome molecular investigation. Although the molecular mechanism of this up-regulation still remains to be uncovered, it adds another example of the importance of non-coding regions in human pathology.
Materials and Methods
Patients
Clinical information regarding SPOAN patients in the geographic cluster detected in northeastern Brazil was detailed elsewhere (2,3). Additionally, we evaluated another three Brazilian patients, with different ancestors from northeastern Brazil and two Egyptians siblings with the identified 216-bp deletion and same clinical symptoms. Blood samples were used for DNA extraction from all patients, from several obligated carriers and from unaffected siblings. Fibroblasts were obtained from dermal biopsies from five patients, one heterozygote and four Brazilian healthy controls, following informed consent under protocols approved by the Biosciences Institute, University of São Paulo (Protocol CEP 010/2003).
Molecular analysis
Previous studies conducted by our group using Sanger sequencing did not identify deleterious variants in exons of candidates genes located in the critical region for SPOAN (LRFN4, KLC2 and CCS) (3). To have a more comprehensive and detailed view over this region, WES was performed using DNA samples from two SPOAN subjects using Agilent SureSelect Human All Exon 50 Mb Kit and sequenced in Illumina HiSeq2000 (Illumina, San Diego, CA, USA). Alignment against reference GRCh37 was performed with BWA (42); genotyping with GATK (43); SNP and InDel annotation with Annovar (44) and CNV detection with the R package ExomeDepth (45). The WES coverage achieved at the candidate region was 40× and 77× in the Egyptian and Brazilian samples, respectively. The 216-bp deletion was not detectable by WES. Variants detected in the mapped linkage region were filtered by their frequency, compared with 1000 Genomes database, NHLBI GO Exome Sequencing Project (ESP), Exome Aggregation Consortium (ExAC) and with sequences obtained from 1484 Brazilian controls.
Whole-genome sequencing was performed in DNA from a third affected patient (a distant cousin from Brazilian series) using Illumina TruSeq DNA kit. Alignment against reference GRCh37 was performed with BWA (42); genotyping with GATK (43); SNP and InDel annotation with SnpEff (46) and CNV detection using R package ExomeDepth (45) restricted to exon regions (using bedfile template of the Agilent V4Plus kit), Pindel and additional manual screening in the target linkage region. The achieved coverage at the candidate region was 26×. Variants were filtered by comparison with 1000 Genomes. SPOAN mutation (chr11.hg19:g.66,024,557_66,024,773del) was checked for co-segregation in affected and family health controls (also checked in 474 unrelated health controls) by PCR followed by agarose gel electrophoresis using primer ID 1 (Supplementary Material, Table S3).
Induced pluripotent stem-Cells (iPSC)
Retrovirus vectors containing the Oct4, c-Myc, Klf4 and Sox2 human cDNAs were obtained from Muotri's group and the protocol is described elsewhere (47). Embryoid bodies (EBs) were formed by mechanical dissociation of cell clusters (pre-treated with dorsomorphin, 1 nM, for 2 days) and plating onto low-adherence dishes in NB media (DMEM/F12 plus 0.5X N2 and 0.5X B27 supplements) plus dorsomorphin for 2 days and in the next 5 days in NB media plus FGF and EGF. After that, mature EBs were dissociated with accutase for 5 min at 37°C and plated in matrigel in NB media plus FGF 20 ng/ml and EGF 20 ng/ml. Rosettes were visible for collection after 7 days and were then dissociated with accutase (Chemicon, EMD Millipore, Darmstadt, Germany) and plated onto poly-ornitine/laminin-coated dishes (Sigma) with NB media plus FGF and EGF. Homogeneous populations of neural progenitor cells (NPCs) were achieved after 1–2 passages with accutase in the same condition. To improve cell differentiation, brain-derived neurotrophic factor (20 ng/ml), glial cell-derived neurotrophic factor (20 ng/ml), insulin-like growth factor-1 (20 ng/ml), Ri (5uM) and SHH (100 ng/ml; neuronal maturation medium) were added to neuronal cultures for 5 weeks. NPCs were differentiated in MN following a protocol modified from study described elsewhere (48).
Human RNA extraction and cDNA synthesis
RNA extraction from fibroblasts (n = 5 affected; n = 1 heterozygote; n = 4 healthy controls) and MN (n = 3 affected; n = 1 heterozygote; n = 3 healthy controls) was performed with TRIZOL® reagent (Invitrogen) and Norgen Biotek RNA/DNA/Protein Purification Kit (Norgen Biotek Corp., Ontario, Canada); RNA from whole-blood (n = 7 affected; n = 7 heterozygotes; n = 6 family healthy controls +1 unrelated healthy control from the same region) were extracted using PAXgene Blood RNA Kit (Qiagen); RNA was reverse-transcribed with oligo(dT) primers using SuperScript™ III First-strand Synthesis System (Life Technologies).
Expression array
Fibroblast cDNA samples were submitted to array expression assay using GeneChip® Scanner 3000 7G System (Affymetrix, Santa Clara, CA, USA). The results of expression array were normalized by Robust Multi-array Average (49) and statistical method (test-T) was performed using CLCbio Genomics Workbench, adjusted by Bonferroni and false discovery rate (FDR). Data weresubmitted to GEO (accession number: GSE67527).
Quantitative reverse transcription PCR (RT-qPCR)
KLC2 primers for RT-qPCR were detailed in Supplementary Material, Table S3 (primer ID 2). RT-qPCR was normalized to GAPDH and was performed using LightCycler® 480 (Roche Diagnostics). KLC2 expression data were calculated using 2−ΔΔCT method (50). Mann–Whitney test (Nonparametric) was performed using GraphPad Prism version 5.00 (San Diego, CA, USA). Each experiment was performed in triplicate and each sample was replicated twice.
TaqMan Gene Expression Assay probes: MNX1/HB9 (Hs00907365_m1), CHAT (Hs00252848_m1) and ISL1 (Hs00158126_m1) were used to validate the neurons derived cells from iPSC as MN (Applied Biosystems, USA). RT-qPCR was normalized to Human ACTB (β-actin; Hs01060665_g1). RT-qPCR was performed using the Applied Biosystems® 7500 Fast Real-time PCR System.
Immunofluorescence and MN validation
For immunofluorescence evaluation of MN, cells were fixed with 4% paraformaldehyde, followed by permeabilization and blocking with 0.05% (v/v) Triton X in PBS containing 5% (v/v) donkey serum. Primary antibodies were incubated overnight at 4°C. Samples were washed three times before secondary antibodies incubation (Alexa Fluor Dyes, Life Technologies). Dapi was added in the last 20 min of secondary antibody incubation. Primary antibody concentrations were: a-NeuN mouse monoclonal 1:500 (Millipore); a-Hb9 mouse polyclonal 1:500 (DSHB) and a-Islet 1 rabbit polyclonal 1:1000 (BD Bio-science). Images were obtained through Axio Observer.A1 immunofluorescence microscope (Zeiss). cDNA obtained from fibroblasts, NPC and MN were used for MN validation using TaqMan probes described above. RT-qPCR using fibroblast samples did not show expression of MN probes. RT-qPCR of MN samples showed expression of MNX1/HB9 probe, which was not amplified in NPC samples. MN samples showed higher significant (P < 0.05) expression of CHAT compared with NPC (Supplementary Material, Fig. S3B). We confirmed the presence of 216-bp deletion in DNA extracted from MN patient samples (Supplementary Material, Fig. S3C).
Gene reporter assay
The full-length (3,313-bp) and deleted 216-bp (3,097-bp) KLC2 upstream region was synthesized (Genone) and cloned into promoterless firefly luciferase vector pGL4 (Promega). pShuttle/RL was used for transfection normalization, which expresses the reporter gene Renilla luciferase (51). Assays using HEK239T, U87MG and MN about 1 × 104 cells were plated in 96-well dishes in triplicate for each point. In HEK239T and U87MG a total of 200 ng of plasmids (180 ng pGL4 and 20 ng pShuttle/RL) were used for transfection using Lipofectamine 2000 Transfection Reagent (Invitrogen). In MN we used 480 ng pGL4 and 20 ng pShuttle/RL. Two days after DNA transfection, the luciferase activities were measured in Glomax luminometer (Promega) with the Dual-Glo Luciferase Assay System (Promega) according to manufacturer's instructions. One-way ANOVA was performed using GraphPad Prism version 5.00 (San Diego, CA, USA).
Zebrafish animal model
Adult zebrafish were maintained at 28°C on a 14 h light/10 h dark cycle and the embryos were obtained by natural mating. Zebrafish presents only one klc2 gene in its genome (ZFIN ID: ZDB-GENE-030131-2670), which turns appropriate the use Danio rerio as animal model in this study. The use of Danio rerio in this study was approved by the Committee on the Ethics of Animal Experiments of Pharmacology and Biochemistry Sciences department of National University of Rosario, Argentina (No. 429/2014).
Zebrafish RNA extraction and cDNA synthesis
Total RNA was extracted from whole embryos at different embryonic stages (6, 24, 48 and 72-hpf). RNA extraction was performed using TRIZOL® reagent (Invitrogen), following the manufacturer's protocol. First-strand cDNA was synthetized using SuperScript Reverse Transcriptase (Invitrogen) with a specific primer (primer ID 3) for Danio rerio klc2 gene transcript (Ensembl ENSDARG00000075485). The complete klc2 CDS was amplified by PCR using primers ID 4, forward including EcoRI and reverse including SacI restriction sites.
Plasmids and DNA constructs
The complete CDS sequence from klc2 (mRNAklc2) was cloned using EcoRI and SacI sites into an engineered version of pCS2+MT as described elsewhere (52). This plasmid was used to transcribe mRNAklc2-eGFP coding for KLC2 fused to eGFP. Plasmid without klc2 insert was used to transcribe mRNAeGFP as a control. For mRNAklc2-eGFP and mRNAeGFP transcription, plasmids were linearized by NotI and the SP6 promoter was used for in vitro transcription using mMESSAGE mMACHINE® Kit (Ambion, Applied Biosystems). The mRNAklc2-eGFP was used to perform the overexpression assay and for rescue of morphant's phenotype.
Knockdown and overexpression assays
Microinjection of morpholino oligonucleotides (MO) in the yolk of embryos at one- to two-cell stage were performed in specific concentrations (4 and 6 ng). Translation blocking morpholino (MOklc2-TB) sequence was 5′-GGTGGACATCACCCACTGACACACA-3′ (misMOklc2-TB was 5′-GGaGcACATgACCCAgTcACACACA-3′) and splicing blocking morpholino (MOklc2-SP) sequence was 5′-CGTGTGTGTTTCACCTGTGCTTCCC-3′ (misMOklc2-SP was 5′-CGTcTcTGTTTgACCTcTcCTTCCC-3′). MOklc2-SP target exon 2 of klc2 gene. The rescue of phenotype was performed by co-injecting 6 ng MOklc2-SP and 100 pg mRNAklc2-eGFP in the yolk of embryos staged at one- to two-cells. Chi-square and Fisher's exact tests were performed using GraphPad Prism version 5.00 (San Diego, CA, USA).
Overxpression of klc2 gene in zebrafish was performed by microinjecting mRNAklc2-eGFP at specific concentrations (100, 150 and 200 pg), as described in previous study (53). Same concentrations of mRNAeGFP were microinjected in zebrafish embryos to be used as controls. Both microinjected embryos (mRNAklc2-eGFP and mRNAeGFP) were selected by fluorescence at 24-hpf stage and evaluated at 48-hpf under MVX10 Olympus Microscope, and recorded with MVXTV1XC Olympus digital camera. Chi-square test was performed using GraphPad Prism version 5.00 (San Diego, CA, USA).
Funding
This work was supported by Propesq/UEPB, PPSUS/FAPESQ/PB, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Instituto Nacional de Ciência e Tecnologia (INCT), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP)/Centro de Pesquisa, Inovação e Difusão (CEPID) and National Counsel of Technological and Scientific Development (CNPq).
Supplementary Material
Acknowledgements
We are grateful to H. Miranda for her help in conducting MN differentiation. We also thank R. Moura for his contribution in bioinformatics analysis.
Conflict of Interest statement. None declared.
References
- 1. Lo Giudice T., Lombardi F., Santorelli F.M., Kawarai T., Orlacchio A. (2014) Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Exp. Neurol. Nov., 261, 518–539. [DOI] [PubMed] [Google Scholar]
- 2. Macedo-Souza L., Kok F., Santos S., Amorim S.C., Starling A., Nishimura A., Lezirovitz K., Lino A.M., Zatz M. (2005) Spastic paraplegia, optic atrophy, and neuropathy is linked to chromosome 11q13. Ann. Neurol., 57, 730–737. [DOI] [PubMed] [Google Scholar]
- 3. Macedo-Souza L., Kok F., Santos S., Licinio L., Lezirovitz K., Cavaçana N., Bueno C., Amorim S., Pessoa A., Graciani Z. et al. (2009) Spastic paraplegia, optic atrophy, and neuropathy: new observations, locus refinement, and exclusion of candidate genes. Ann. Hum. Genet., 73, 382–387. [DOI] [PubMed] [Google Scholar]
- 4. Windpassinger C., Auer-Grumbach M., Irobi J., Patel H., Petek E., Hörl G., Malli R., Reed J.A., Dierick I., Verpoorten N. et al. (2004) Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat. Genet., 36, 271–276. [DOI] [PubMed] [Google Scholar]
- 5. Novarino G., Fenstermaker A.G., Zaki M.S., Hofree M., Silhavy J.L., Heiberg A.D., Abdellateef M., Rosti B., Scott E., Mansour L. et al. (2014) Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science, 31, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vale R.D. (2003) The molecular motor toolbox for intracellular transport. Cell, 112, 467–480. [DOI] [PubMed] [Google Scholar]
- 7. Hirokawa N., Noda Y., Tanaka Y., Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol., 10, 682–696. [DOI] [PubMed] [Google Scholar]
- 8. Visscher K., Schnitzer M.J., Block S.M. (1999) Single kinesin molecules studied with a molecular force clamp. Nature, 400, 184–189. [DOI] [PubMed] [Google Scholar]
- 9. Verhey K.J., Hammond J.W. (2009) Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol., 10, 765–777. [DOI] [PubMed] [Google Scholar]
- 10. Hirokawa N., Niwa S., Tanaka Y. (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron, 68, 610–638. [DOI] [PubMed] [Google Scholar]
- 11. Pernigo S., Lamprecht A., Steiner R.A., Dodding M.P. (2013) Structural basis for kinesin-1: cargo recognition. Science, 340, 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rahman A., Friedman D.S., Goldstein L.S.B. (1998) Two kinesin light chain genes in mice: identification and characterization of the encoded proteins. J. Biol. Chem., 273, 15395–15403. [DOI] [PubMed] [Google Scholar]
- 13. Perlson E., Maday S., Fu M.M., Moughamian A.J., Holzbaur E.L. (2010) Retrograde axonal transport: pathways to cell death? Trends Neurosci., 33, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Millecamps S., Julien J.P. (2013) Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci., 14, 161–176. [DOI] [PubMed] [Google Scholar]
- 15. Reid E., Kloos M., Ashley-Koch A., Hughes L., Bevan S., Svenson I.K., Graham F.L., Gaskell P.C., Dearlove A., Pericak-Vance M.A. et al. (2002) A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10). Am. J. Hum. Genet., 71, 1189–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ebbing B., Mann K., Starosta A., Jaud J., Schöls L., Schüle R., Woehlke G. (2008) Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum. Mol. Genet., 17, 1245–1252. [DOI] [PubMed] [Google Scholar]
- 17. Caballero Oteyza A., Battaloğlu E., Ocek L., Lindig T., Reichbauer J., Rebelo A.P., Gonzalez M.A., Zorlu Y., Ozes B., Timmann D. et al. (2014) Motor protein mutations cause a new form of hereditary spastic paraplegia. Neurology, 3, 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J., Srour M., Kim D., Hamdan F.F., Lim S.H., Brunel-Guitton C., Décarie J.C., Rossignol E., Mitchell G.A., Schreiber A. et al. (2015) De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy and cerebellar atrophy. Hum. Mutat., 36, 69–78. [DOI] [PubMed] [Google Scholar]
- 19. Munch C., Rosenbohm A., Sperfeld A.D., Uttner I., Reske S., Krause B.J., Sedlmeier R., Meyer T., Hanemann C.O., Stumm G. et al. (2005) Heterozygous R1101K mutation of the DCTN1 gene in a family with ALS and FTD. Ann. Neurol., 58, 777. [DOI] [PubMed] [Google Scholar]
- 20. Falzone T.L., Stokin G.B., Lillo C., Rodrigues E.M., Westerman E.L., Williams D.S., Goldstein L.S. (2009) Axonal stress kinase activation and tau misbehavior induced by kinesin-1 transport defects. J. Neurosci., 29, 5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Füger P., Sreekumar V., Schüle R., Kern J.V., Stanchev D.T., Schneider C.D., Karle K.N., Daub K.J., Siegert V.K., Flötenmeyer M. et al. (2012) Spastic paraplegia mutation N256S in the neuronal microtubule motor KIF5A disrupts axonal transport in a Drosophila HSP model. PLoS Genet., 8, e1003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Valdmanis P.N., Meijer I.A., Reynolds A., Lei A., MacLeod P., Schlesinger D., Zatz M., Reid E., Dion P.A., Drapeau P. et al. (2007) Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am. J. Hum. Genet., 80, 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin P., Li J., Liu Q., Mao F., Li J., Qiu R., Hu H., Song Y., Yang Y., Gao G. et al. (2008) A missense mutation in SLC33A1, which encodes the acetyl-CoA transporter, causes autosomal-dominant spastic paraplegia (SPG42). Am. J. Hum. Genet., 83, 752–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clemen C.S., Tangavelou K., Strucksberg K.H., Just S., Gaertner L., Regus-Leidig H., Stumpf M., Reimann J., Coras R., Morgan R.O. et al. (2010) Strumpellin is a novel valosin-containing protein binding partner linking hereditary spastic paraplegia to protein aggregation diseases. Brain, 133, 2920–2941. [DOI] [PubMed] [Google Scholar]
- 25. Fassier C., Hutt J.A., Scholpp S., Lumsden A., Giros B., Nothias F., Schneider-Maunoury S., Houart C., Hazan J. (2010) Zebrafish atlastin controls motility and spinal motor axon architecture via inhibition of the BMP pathway. Nat. Neurosci., 13, 1380–1387. [DOI] [PubMed] [Google Scholar]
- 26. Martin E., Yanicostas C., Rastetter A., Naini S.M., Maouedj A., Kabashi E., Rivaud-Péchoux S., Brice A., Stevanin G., Soussi-Yanicostas N. (2012) Spatacsin and spastizin act in the same pathway required for proper spinal motor neuron axon outgrowth in zebrafish. Neurobiol. Dis., 48, 299–308. [DOI] [PubMed] [Google Scholar]
- 27. Martin E., Schüle R., Smets K., Rastetter A., Boukhris A., Loureiro J.L., Gonzalez M.A., Mundwiller E., Deconinck T., Wessner M. et al. (2013) Loss of function of glucocerebrosidase GBA2 is responsible for motor neuron defects in hereditary spastic paraplegia. Am. J. Hum. Genet., 92, 238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song Y., Wang M., Mao F., Shao M., Zhao B., Song Z., Shao C., Gong Y. (2013) Knockdown of Pnpla6 protein results in motor neuron defects in zebrafish. Dis. Model Mech., 6, 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cremers F.P.M., Pfeiffer R.A., van de Pol T.J., Hofker M.H., Kruse T.A., Wieringa B., Ropers H.H. (1987) An interstitial duplication of the X chromosome in a male allows physical fine mapping of probes from the Xq13-q22 region. Hum. Genet., 77, 23–27. [DOI] [PubMed] [Google Scholar]
- 30. Hodes M.E., Pratt V.M., Dlouhy S.R. (1993) Genetics of Pelizaeus-Merzbacher disease. Dev. Neurosci., 15, 383–394. [DOI] [PubMed] [Google Scholar]
- 31. Inoue K., Osaka H., Sugiyama N., Kawanishi C., Onishi H., Nezu A., Kimura K., Yamada Y., Kosaka K. (1996) A duplicated PLP gene causing Pelizaeus-Merzbacher disease detected by comparative multiplex PCR. Am. J. Hum. Genet., 59, 32–39. [PMC free article] [PubMed] [Google Scholar]
- 32. Mimault C., Giraud G., Courtois V., Cailloux F., Boire J.Y., Dastugue B., Boespflug-Tanguy O. (1999) Clinical European network on brain dysmyelinating disease. Proteolipoprotein gene analysis in 82 patients with sporadic Pelizaeus-Merzbacher disease: duplications, the major cause of the disease, originate more frequently in male germ cells, but point mutations do not. Am. J. Hum. Genet., 65, 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carvalho C.M.B., Ramocki M.B., Pehlivan D., Franco L.M., Gonzaga-Jauregui C., Fang P., McCall A., Pivnick E.K., Hines-Dowell S., Seaver L.H. et al. (2011) Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat. Genet., 43, 1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hertz J.M., Borglum A.D., Brandt C.A., Flint T., Bisgaard C. (1994) Charcot-Marie-Tooth disease type 1A: the parental origin of a de novo 17p11.2-p12 duplication. Clin. Genet., 46, 291–294. [DOI] [PubMed] [Google Scholar]
- 35. Sorour E., Thompson P., MacMillan J., Upadhyaya M. (1995) Inheritance of CMT1A duplication from a mosaic father. J. Med. Genet., 32, 483–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Theuns J., Brouwers N., Engelborghs S., Sleegers K., Bogaerts V., Corsmit E., De Pooter T., van Duijn C.M., De Deyn P.P., Van Broeckhoven C. (2006) Promoter mutations that increase amyloid precursor-protein expression are associated with Alzheimer disease. Am. J. Hum. Genet., 78, 936–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nagaraj K., Kristiansen L.V., Skrzynski A., Castiella C., Garcia-Alonso L., Hortsch M. (2009) Pathogenic human L1-CAM mutations reduce the adhesion-dependent activation of EGFR. Hum. Mol. Genet., 18, 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meijer I.A., Dion P., Laurent S., Dupré N., Brais B., Levert A., Puymirat J., Rioux M.F., Sylvain M., Zhu P.P. et al. (2007) Characterization of a novel SPG3A deletion in a French-Canadian family. Ann. Neurol., 61, 599–603. [DOI] [PubMed] [Google Scholar]
- 39. Havlicek S., Kohl Z., Mishra H.K., Prots I., Eberhardt E., Denguir N., Wend H., Plötz S., Boyer L., Marchetto M.C. et al. (2014) Gene dosage-dependent rescue of HSP neurite defects in SPG4 patients’ neurons. Hum. Mol. Genet., 15, 2527–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bakowska J.C., Wang H., Xin B., Sumner C.J., Blackstone C. (2008) Lack of spartin protein in Troyer syndrome: a loss-of-function disease mechanism? Arch. Neurol., 65, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kruer M.C., Paisán-Ruiz C., Boddaert N., Yoon M.Y., Hama H., Gregory A., Malandrini A., Woltjer R.L., Munnich A., Gobin S. et al. (2010) Defective FA2H leads to a novel form of neurodegeneration with brain iron accumulation (NBIA). Ann. Neurol., 68, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li H., Durbin R. (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics, 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang K., Li M., Hakonarson H. (2010) ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res., 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Plagnol V., Curtis J., Epstein M., Mok K.Y., Stebbings E., Grigoriadou S., Wood N.W., Hambleton S., Burns S.O., Thrasher A.J. et al. (2012) A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics, 28, 2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. (2010) A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell, 12, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mitne-Neto M., Machado-Costa M., Marchetto M.C., Bengtson M.H., Joazeiro C.A., Tsuda H., Bellen H.J., Silva H.C., Oliveira A.S., Lazar M. et al. (2011) Downregulation of VAPB expression in motor neurons derived from induced pluripotent stem cells of ALS8 patients. Hum. Mol. Genet., 15, 3642–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264. [DOI] [PubMed] [Google Scholar]
- 50. Schmittgen T.D., Livak K.J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc., 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 51. Soltys D.T., Rocha C.R., Lerner L.K., de Souza T.A., Munford V., Cabral F., Nardo T., Stefanini M., Sarasin A., Cabral-Neto J.B. et al. (2013) Novel XPG (ERCC5) mutations affect DNA repair and cell survival after ultraviolet but not oxidative stress. Hum. Mutat., 34, 481–489. [DOI] [PubMed] [Google Scholar]
- 52. Favaro F.P., Alvizi L., Zechi-Ceide R.M., Bertola D., Felix T.M., de Souza J., Raskin S., Twigg S.R., Weiner A.M., Armas P. et al. (2014) A noncoding expansion in EIF4A3 causes Richieri-Costa-Pereira syndrome, a craniofacial disorder associated with limb defects. Am. J. Hum. Genet., 94, 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Niederriter A., Davis E.E., Golzio C., Oh E.C., Tsai I.C., Katsanis N. (2013) In vivo modeling of the morbid human genome using Danio rerio. J. Vis. Exp., 24, e50338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.