ABSTRACT
Environmental conditions inform the rate of plant growth and development. The target of rapamycin (TOR) signalling pathway is a central regulator of plant growth in response to nutrients and energy, while abscisic acid (ABA) is a main mediator of abiotic stress responses. We recently characterized Arabidopsis TIP41, a predicted TOR pathway component involved in the ABA-mediated response to abiotic stress. Here, we report the ABA sensitivity of tip41 mutants, supporting the relation between TIP41 and the hormone pathway. The analysis of predicted TIP41 functional network identified several protein phosphatases. In particular, candidate protein interactors included catalytic subunits of type 2A protein phosphatases and protein phosphatases 6, which regulate different developmental processes and responses to environmental stimuli. These results provide important information on the role of TIP41 in the cross talk between TOR and ABA pathways.
Keywords: target of rapamycin, abscisic acid, genetic network
Plants need to respond to adverse environmental conditions with a proper balance between growth and adaptive mechanisms. The target of rapamycin (TOR), an evolutionarily conserved serine/threonine protein kinase, plays a central role in signal integration, regulating growth in response to nutrients and energy availability.1 Recent studies have revealed an interplay between TOR and biotic and abiotic stress signaling. In addition, an extensive cross talk with the TOR pathway is reported for several phytohormones, such as auxin, salicylic acid, jasmonic acid and abscisic acid.2–5 Abscisic acid (ABA) is a key regulator of stress responses. Upon osmotic stress, ABA is bound by the PYR/PYL/RCAR proteins,6 resulting in a conformational change of the receptors. In this conformational state, PYLs are able to inhibit clade A PP2C protein phosphatases, leading to the activation of SnRK2 kinases, which in turn phosphorylate stress regulator and effector proteins.7–9 Although the activation of stress responses through this mechanism is well known, only recently the ABA-mediated activation of SnRK2s has been linked to TOR signalling. Wang and colleagues10 reported that activated SnRK2s inhibit RAPTOR, a major regulatory component of the TOR pathway. This inactivation leads to arrest of growth, thus shunting available energy towards the activation of stress response mechanisms. Viceversa, when favourable conditions are restored, TOR represses PYLs, to attenuate stress responses and promote growth.10
We have shown that mutants in TIP41, a predicted component of TOR signaling, are hypersensitive to ABA and NaCl, as well as to iron starvation conditions in root growth assays.11 Thus, TIP41 is an integrator of responses to nutrient availability and abiotic stress, and a further link between TOR and ABA pathways.
Here, we further characterize the involvement of TIP41 in ABA responses and identify additional candidates regulated by TIP41. We tested the sensitivity to different concentrations of ABA of plants with altered expression of TIP41 compared to wild type (Figure 1) using an ABA sensitivity assay. Sensitivity was scored in terms of presence of necrotic or bleached leaves, as previously reported.12 We used homozygous lines of two T-DNA insertion mutants (tip41_1, SALK_006384 and tip41_2, SALK_113161) and three overexpressing lines (35S::FLAG-TIP41, line #17, #18, #20). Both T-DNA lines and over-expressors were verified to have altered expression levels of TIP41, as reported in Punzo et al.11 The previously obtained RNA-seq data from tip41_1 and Col-0 exposed to ABA showed changes in the expression of genes involved in ABA response and transport (At4g37050, PATATIN-LIKE PROTEIN4; At5g28520, JACALIN-RELATED LECTIN40, At1g15520, ABC TRANSPORTER G FAMILY MEMBER40, a member of ABC transporter family).11 Consistent with our previous results,11 the hypersensitivity to abscisic acid of mutants with abolished expression of TIP41 was observed. After 11 days of exposure to 50 μM ABA, less than 60% of knockout mutants showed no signs of damage compared to 85% of wild type. When exposed to 100 μM ABA, 80% of tip41_1 and 95% of tip41_2 plants displayed damaged leaves compared to 45% of wild type. Plants overexpressing TIP41 did not show significant variation compared to the Col-0 control (Figure 1). The high sensitivity of knockout mutants to ABA in the aerial part, in addition to the previously shown sensitivity of seed germination and root growth, indicates that TIP41 regulates the ABA-mediated response to stress in multiple tissues and developmental stages. Similarly to yeast and mammals, in plants TOR is part of an extensive genetic network, which includes RAPTOR and LST8, each encoded by two different genes (RAPTOR1/RAPTOR2 and LST8-1/LST8-2), whose protein products form a complex with TOR.13,14 This core complex targets a wealth of substrates, including proteins involved in transcription, ribosome biogenesis, protein translation and autophagy.1,5,15,16
Figure 1.
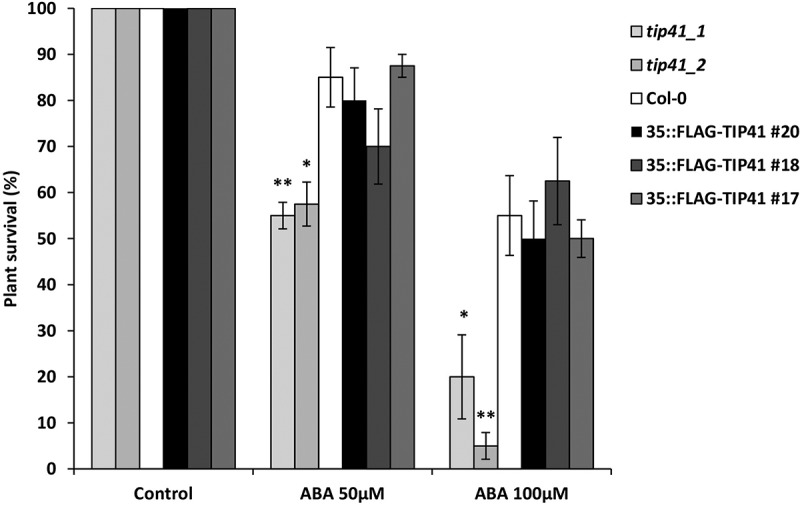
ABA sensitivity test of TIP41 knockout mutants (tip41_1, tip41_2) and three 35S::FLAG-TIP41 over-expressing lines (#20,#18,#17) compared to wild type (Col-0). 16-day-old seedlings germinated for 5 days on germination media and then exposed to ABA (0, 50 or 100 μM) for 11 days. Sensitivity was scored in terms of presence of necrotic or bleached leaves. Data are means ± SE (n = 40) of four biological replicates. The asterisk denotes a significant difference compared to wild type according to Student’s t-test (*P ≤ 0.05, **P ≤ 0.01).
For a preliminary insight on the functional network involving TIP41, we used the GeneMANIA prediction software,17 which constructs gene functional networks based on studies of co-expression, co-localization, physical protein interaction and homology to other systems, such as human, yeast and fly (https://genemania.org) (Figure 2). The network, visualized by Cytoscape (v3.3.0), showed that TIP41 may be associated with several proteins. Although the interaction between TIP41 and related genes will have to be validated through ad hoc experiments, based on the specific identified genes, the predicted interaction between RAPTOR1, RAPTOR2 and TIP41 further supports the role of TIP41 in the TOR pathway. The co-localization of TIP41 and TAP46, a downstream component of TOR pathway and regulator of Type 2A protein phosphatases (PP2A), suggests that these two proteins may have related functions. The TAP46 regulation of PP2A was reported.18,19 In particular, Hu and colleagues19 reported that tap46 knockdown mutants showed increased PP2A activity and decreased ABA sensitivity during germination. The hypersensitivity to ABA that here we observed in plant lacking TIP41, and during germination as reported in Punzo et al.,11 may suggest a different role of TIP41 in the regulation of PP2A. Future experiments will address the impact of TIP41 on PP2A activity. Several phosphatase-associated genes, such as At1g30470, At1g07990, At3g45190, At2g28360, in addition to protein phosphatases 4, (PPX1, PPX2), and the two genes encoding the catalytic subunits of the serine/threonine protein phosphatases 6 (FYPP3, FYPP1) were also predicted to be in TIP41 network based on protein interactions observed in other organisms. FYPPs act as antagonists of SnRK2s to regulate ABI5 phosphorylation and ABA responses.20 Interestingly, the 5 proteins encoding the C subunits of PP2A21 (PP2A-C1, At1g59830; PP2A-C2, At1g10430; PP2A-C3, At3g58500; PP2A-C4, At2g42500; PP2A-C5, At169960) have predicted interaction with TIP41. PP2A-C2 and PP2A-C5 are also co-expressed with TIP41 (Figure 2). This bioinformatic prediction is confirmed by previously reported results of physical interaction between PP2A-C1 and TIP41.11 In addition to the catalytic C subunit, the PP2A holoenzymes are composed of a scaffolding (A) and a regulatory (B) subunit. The presence of multiple genes encoding each subunit leads to a large number of possible PP2A trimer combinations with different substrate specificities. Indeed, PP2As play key roles in several signaling pathways, such as auxin transport,22 ABA and ethylene signaling,23-25 as well as growth and development regulation.26,27 Based on mutant or over-expressing lines phenotypes, individual plant PP2A subunits have been linked to responses to abiotic stresses, including salt and photo-oxidative stresses.
Figure 2.
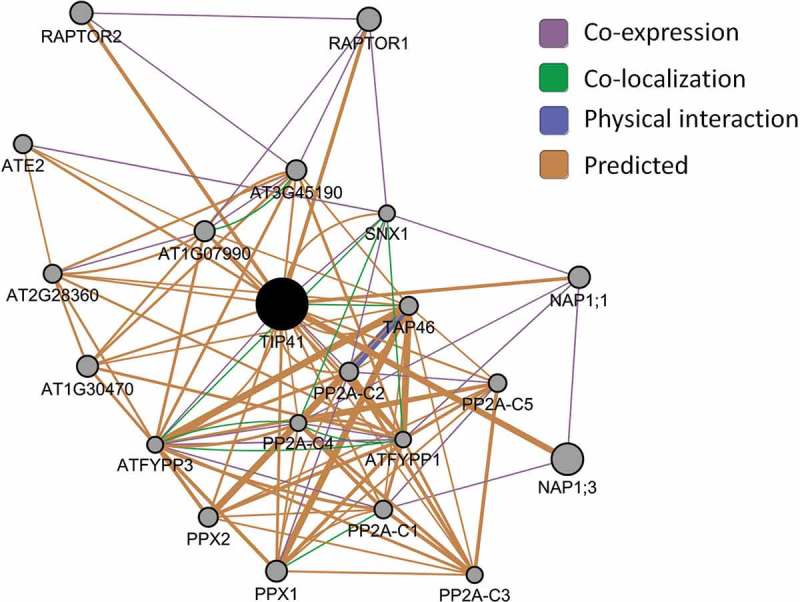
Arabidopsis TIP41 putative network of functionally related genes generated through GeneMANIA prediction software. Co-expression (violet lines), co-localization (green), physical (blue) and predicted (orange) interactions are shown. The network was constructed using GeneMANIA cytoscape app (v 3.3.0).
While we have confirmed the interaction between TIP41 and PP2A-C1, other PP2A-Cs may also interact with TIP41 and subsequently regulate stress responses through PP2A holoenzyme (Figure 3). Altogether, the predicted network identifies a set of testable candidate interactors of TIP41, which will be verified for physical and genetic interaction to clarify the putative role of TIP41 in the activation of stress response mechanisms in a possible crosslinked network relating the TOR and ABA pathways.
Figure 3.
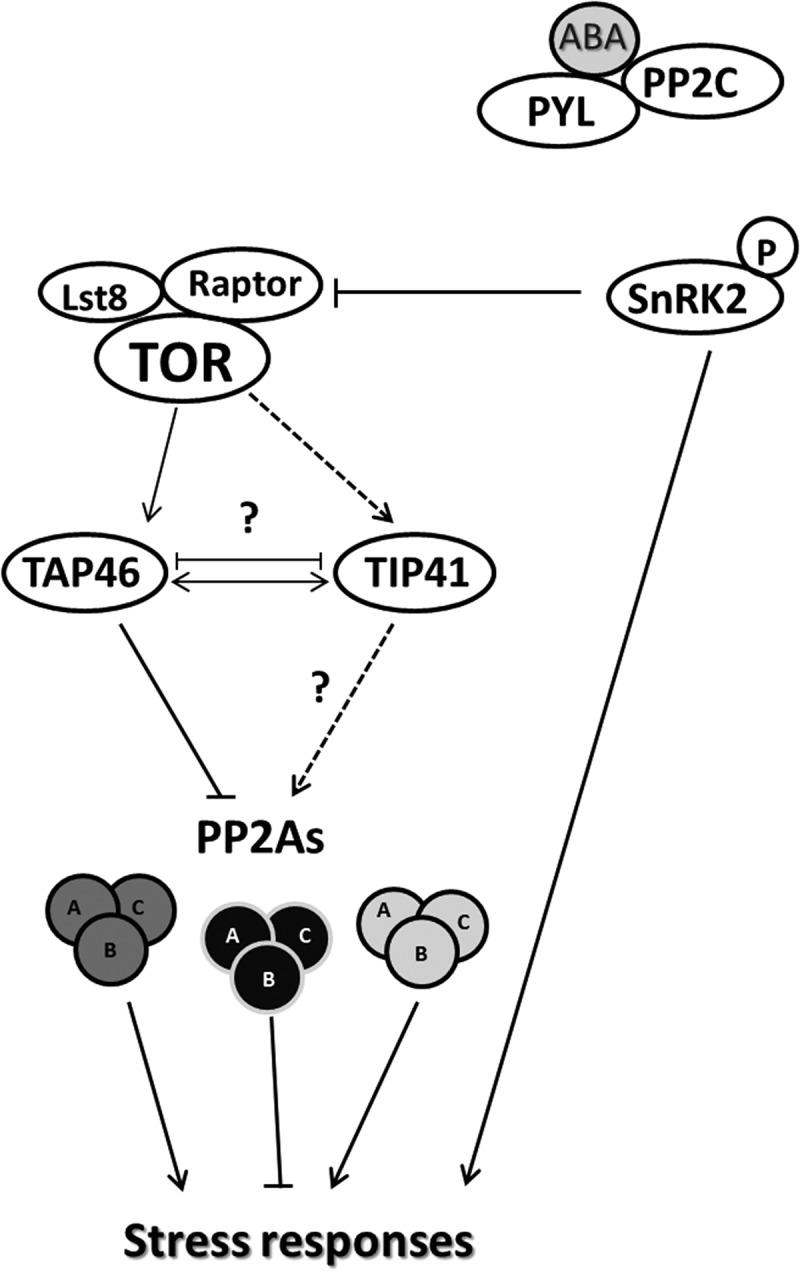
Model of TIP41 regulation of PP2As in the ABA signaling pathway. Interaction network between ABA-activated SnRK2-type protein kinases and TOR complex as reported in Wang et al.10 is shown. In presence of ABA, the activated SnRK2s phosphorylate RAPTOR, a component of the TOR complex, to reduce plant growth by inhibiting TOR activity. The TOR kinase phosphorylates TAP46, a regulator of PP2A, which, based on reported phenotypes, may have an opposite function as TIP41.11,25,26 PP2As are composed of three subunits (A, B and C) with a large number of possible dimer or trimer combinations (reported in the figure with different colors) with different cellular functions. TIP41 interacts with and may regulate PP2As affecting different stress responses, based on PP2A subunit combinations. Full arrows indicate established positive (pointed) and negative (barred) protein interactions. Dashed arrows indicate hypothetical regulatory functions.
Funding Statement
This work was supported by the Ministero dell’Istruzione, dell’Università e della Ricerca [PON02_003_3082360].
Acknowledgments
This work was supported by the Italian Ministry of University and Research under Grant PON02_00395_3082360, (GenoPOM-PRO). Paola Punzo received a fellowship by Regione Campania, POR Campania FSE 2014/2020 Program “Sviluppo di processi innovativi e di prodotti di qualità per il benessere dei consumatori e la valorizzazione del comparto agroalimentare campano”.
References
- 1.Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C.. TOR signaling and nutrient sensing. Annu Rev Plant Biol. 2016;67:261–285. doi: 10.1146/annurev-arplant-043014-114648. [DOI] [PubMed] [Google Scholar]
- 2.Dong P, Xiong F, Que Y, Wang K, Yu L, Li Z, Maozhi R. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front Plant Sci. 2015;6:677. doi: 10.3389/fpls.2015.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song Y, Zhao G, Zhang X, Li L, Xiong F, Zhuo F, Zhang C, Yang Z, Datla R, Ren M, et al. The crosstalk between Target of Rapamycin (TOR) and Jasmonic Acid (JA) signaling existing in Arabidopsis and cotton. Sci Rep. 2017;7:45830. doi: 10.1038/srep45830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kravchenko A, Citerne S, Jéhanno I, Bersimbaev RI, Veit B, Meyer C, Leprince AS. Mutations in the Arabidopsis Lst8 and Raptor genes encoding partners of the TOR complex, or inhibition of TOR activity decrease abscisic acid (ABA) synthesis. Biochem Biophys Res Commun. 2015;467(4):992–997. doi: 10.1016/j.bbrc.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 5.Schepetilnikov M, Ryabova LA. Recent discoveries on the role of TOR (target of rapamycin) signaling in translation in plants. Plant Physiol. 2018;176(2):1095–1105. doi: 10.1104/pp.17.01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 7.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 9.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1012–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang PC, Zhao Y, Li Z, Hsu CC, Liu X, Fu L, Hou YJ, Du Y, Xie S, Zhang C, et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell. 2017;69:100–112. doi: 10.1016/j.molcel.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Punzo P, Ruggiero A, Possenti M, Nurcato R, Costa A, Morelli G, Grillo S, Batelli G. The PP 2A‐interactor TIP 41 modulates ABA responses in Arabidopsis thaliana. Plant J. 2018;94(6):991–1009. doi: 10.1111/tpj.13913. [DOI] [PubMed] [Google Scholar]
- 12.Ambrosone A, Batelli G, Nurcato R, Aurilia V, Punzo P, Bangarusamy D, Ruberti I, Sassi M, Leone A, Costa A, et al. The Arabidopsis AtRGGA RNA binding protein regulates tolerance to salt and drought stress. Plant Physiol. 2015;168:292–306. doi: 10.1104/pp.114.255802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreau M, Azzopardi M, Clément G, Dobrenel T, Marchive C, Renne C, Martin-Magniette ML, Taconnat L, Renou JP, Robaglia C, et al. Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell. 2012;24:463–481. doi: 10.1105/tpc.112.098962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong Y, Sheen J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem. 2012;287:2836–2842. doi: 10.1074/jbc.M111.300749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonseca BD, Smith EM, Yelle N, Alain T, Bushell M, Pause A (2014) The ever-evolving role of mTOR in translation In: Bouchard M, Tee AR, Balda MS, Matter K, editors. Seminars in cell & developmental biology (Vol. 36, p. 102–112). Academic Press. doi: 10.1016/j.semcdb.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;Web Server issue:W214–20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn CS, Han JA, Lee HS, Lee S, Pai HS. The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell. 2011;23(1):185–209. doi: 10.1105/tpc.110.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu R, Zhu Y, Shen G, Zhang H. TAP46 plays a positive role in the ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis. Plant Physiol. 2014;164:721–734. doi: 10.1104/pp.113.233684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai M, Xue Q, Mccray T, Margavage K, Chen F, Lee JH, Nezames CD, Guo L, Terzaghi W, Wan J, et al. The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell. 2013;25(2):517–534. doi: 10.1105/tpc.112.105767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farkas I, Dombradi V, Miskei M, Szabados L, Koncz C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007;12(4):169–176. doi: 10.1016/j.tplants.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130(6):1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 23.Saito N, Munemasa S, Nakamura Y, Shimoishi Y, Mori IC, Murata Y. Roles of RCN1, regulatory A subunit of protein phosphatase 2A, in methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid. Plant Cell Physiol. 2008;49(9):1396–1401. doi: 10.1093/pcp/pcn106. [DOI] [PubMed] [Google Scholar]
- 24.Charpentier M, Sun J, Wen J, Mysore KS, Oldroyd GE. Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiol. 2014;166(4):2077–2090. doi: 10.1104/pp.114.246371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skottke KR, Yoon GM, Kieber JJ, DeLong A. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 2011;7(4):e1001370. doi: 10.1371/journal.pgen.1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou HW, Nussbaumer C, Chao Y, DeLong A. Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell. 2004;16(3):709–722. doi: 10.1105/tpc.018994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blakeslee JJ, Zhou HW, Heath JT, Skottke KR, Barrios JA, Liu SY, DeLong A. Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol. 2008;146:539–553. doi: 10.1104/pp.107.110361. [DOI] [PMC free article] [PubMed] [Google Scholar]


