Abstract
Cyclotides are disulfide rich macrocyclic plant peptides that are defined by their unique topology in which a head-to-tail cyclized backbone is knotted by the interlocking arrangement of three disulfide bonds. This cyclic cystine knot motif gives the cyclotides exceptional resistance to thermal, chemical or enzymatic degradation. Over 100 cyclotides have been reported and display a variety of biological activities, including a cytoprotective effect against HIV infected cells. It has been hypothesized that cyclotides from one subfamily, the Möbius subfamily, may be more appropriate than bracelet cyclotides as drug candidates given their lower toxicity to uninfected cells. He we report the anti-HIV and cytotoxic effects of three cyclotides, including two from the Möbius subfamily. We show that Möbius cyclotides have comparable inhibitory activity against HIV infection to bracelet cyclotides and that they are generally less cytotoxic to the target cells. To explore the structure activity relationships (SARs) of the 29 cyclotides tested so far for anti-HIV activity, we modeled the structures of the 21 cyclotides whose structures have not been previously solved. We show that within cyclotide subfamilies there is a correlation between hydrophobicity of certain loop regions and HIV inhibition. We also show that charged residues in these loops impact on the activity of the cyclotides, presumably by modulating membrane binding. In addition to providing new SAR data, this report is a mini-review that collates all cyclotide anti-HIV information reported so far and provides a resource for future studies on the therapeutic potential of cyclotides as natural anti-HIV agents.
Keywords: circular proteins, cyclic cystine knot, HIV
Introduction
The cyclotides are a topologically unique family of macrocyclic knotted peptides comprising over 100 peptides isolated from species of the Violaceae, Rubiaceae and Cucurbitaceae plant families.1 They are the first family of gene-expressed cyclic peptides discovered in plants, although in recent years several other families of gene-coded cyclic peptides have been discovered in bacteria, plants and animals.2–12 The cyclotides are defined by their head-to-tail cyclized peptide backbone and a knotted arrangement of the three disulfide bonds. This complex protein topology, termed the cyclic cystine knot (CCK) motif,13 gives the cyclotides remarkable resistance to thermal, chemical and enzymatic degradation.14 The rigid structure and resistance to degradation of the CCK motif has led to suggestions that cyclotides may be useful as drug leads or combinatorial peptide templates.15,16
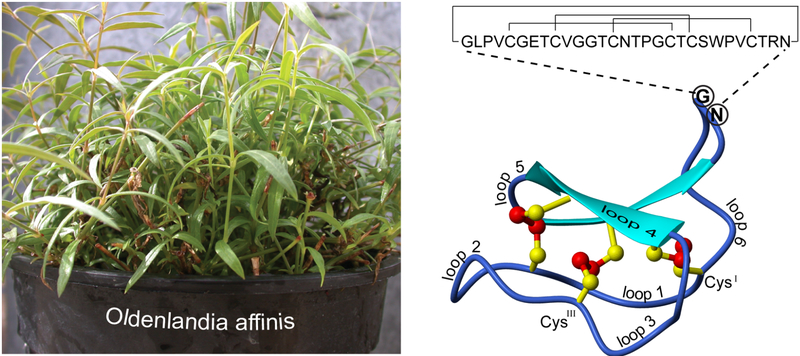
Cyclotides contain six backbone loops between the six conserved Cys residues that make the cystine knot (see Figure 1). The family members discovered so far have been classified into two main subfamilies, based on topological factors and sequence similarities. This classification was originally proposed based on the presence or absence of a cis-Pro residue in loop 5, with those containing this Pro residue referred to as Möbius cyclotides and those without referred to as bracelet cyclotides.1 Although the presence/absence of the cis-Pro residue is the defining feature of the two subfamilies, there also tends to be high sequence similarities for several loops within subfamilies but differences between subfamilies. However, the division into these subfamilies has recently been blurred with the emergence of chimeric cyclotides,17 whose sequence and structure display properties of both Möbius and bracelet families. A third minor subfamily, the trypsin inhibitor cyclotides, or cyclic knottins,2 is quite distinct from the Möbius and bracelet subfamilies and although containing the CCK motif,18,19 the two members of this subfamily have few sequence homologies with the other subfamily members.
Figure 1.
Sequence and structure of the prototypic cyclotide kalata B1. The loops and selected Cys residues on the structure are labeled. The plant, Oldenlandia affinis, from which the peptide is extracted, is shown.
The native function of cyclotides is thought to be one of plant-host defence against pests and pathogens.20,21 However, the initial discovery of the prototypic cyclotide kalata B1 was prompted by an observation of increased uterine contractions following ingestion of a tea made from the leaves of the plant Oldenlandia affinis.22 The fact that the bioactive peptide could survive boiling prompted investigations that led to characterization of the CCK motif.23 Other biological activities of cyclotides have been extensively investigated, including anti-HIV,24 cytotoxic/anti-tumor,25,26 insecticidal,21 anti-fouling,27 anti-microbial,28 and hemolytic26,29,30 activities as well as inhibition of trypsin31 and neurotensin binding.26
The anti-HIV activity of cyclotides was first reported as an outcome of a natural products-based anti-HIV screening initiative conducted by the US National Cancer Institute.6 In the initial study,24 two compounds, determined to be peptides via NMR analysis but resistant to standard peptide sequencing methods, were identified as having anti-HIV activity. These peptides inhibited the cytopathic effect of HIV infection with EC50s of approximately 70 nM, but were also found to be cytotoxic to the target cells with IC50s around 520 nM.24 Further analysis revealed that these peptides, named circulin A and circulin B, contained a head-to-tail cyclized backbone, explaining their resistance to Edman sequencing.
The anti-HIV activity associated with the circulins was not unprecedented since other naturally occurring peptides had been reported to inhibit HIV infection. For example, cyclic peptides with potent HIV inhibitory properties have been isolated from marine sources, and often contain uncommon amino acids, or amino acids with altered stereochemistry.6 Although the circulins did not contain any uncommon amino acids, they displayed a characteristic disulfide bonding pattern, namely CysI-CysIV, CysII-CysV, CysIΠ-CysVI.32 Following the complete structural elucidation of kalata B1 and the CCK motif,23 it was determined that the circulins also displayed the complex CCK motif33,34 and thus were members of the cyclotide family. The dual cytopathic HIV inhibition/cytotoxic activity observed with the circulins is a feature of many anti-HIV cyclotides.
Prior to the current study, kalata B1 and varv E were the only members of the Möbius subfamily of cyclotides to be tested for HIV inhibitory properties.35,36 Daly et al.,35 noted that as kalata B1 had a comparable HIV inhibitory activity to cyclotides from the bracelet subfamily but with a reduced toxicity to the target cells, it and other Möbius cyclotides may be more promising leads in anti-HIV therapy than their bracelet counterparts. Their study also showed that sequence variation between the subfamilies did not appear to influence the level of activity, suggesting that the overall fold of the cyclotide, rather than individual residues, was important for anti-HIV activity. Daly et al.,35 also explored the role of dynamics of the peptide backbone by testing the activity of kalata B1 acyclic permutants. These linear derivatives, despite being structurally similar to the native cyclic peptide but potentially more flexible since the N- and C-termini are free, did not have any anti-HIV activity. This reinforced the conclusion that the intact CCK fold is essential for anti-HIV activity.
In the current study, the molecular basis for anti-HIV activity of cyclotides was investigated by examining the structure activity relationships (SARs) of 29 cyclotides. Three new anti-HIV activities are reported, including one cyclotide from the bracelet subfamily and two from the Möbius subfamily. These are the first cyclotides from the plant Viola odorata to be tested for anti-HIV activity, and their contrasting hydrophobicity profiles helped to explore the relationship between cyclotide structure and function. Three dimensional structural models of 21 cyclotides were derived to supplement the available experimental structures of the eight other cyclotides studied. The investigation revealed a strong correlation between increasing regional hydrophobicity and increasing anti-HIV activity. This correlation explains the reduced anti-HIV activity and cytotoxicity of Möbius cyclotides compared to members of the bracelet subfamily.
Materials and Methods
Cycloviolacins O13, O14, and O24 were extracted and purified from the above ground parts of V. odorata according to the method of Ireland et al.37 Briefly, the procedure involved extracting the peptides from the plant tissue via chemical separation using a dichloromethane/methanol (1:1 v/v) mixture overnight at room temperature (22°C). The extract was partitioned with water and the water/methanol layer was concentrated on a rotary evaporator prior to freeze-drying. The dried product was re-dissolved in water and purified on a preparative RP (reverse phase) C18 column. The solvents used were solvent A (water/trifluoroacetic acid; 10:0.005, v/v) and solvent B (acetonitrile/water/trifluoroacetic acid; 9:1:0.005, v/v/v). Peptides were identified according to their characteristic HPLC retention times and masses (determined via mass spectrometry).
Anti-HIV activity assays
An in vitro XTT [2,3-bis-(2-methoxy-4-nitro-5-sulphophenyl)-2H-tetrazolium-5-carboxanilide]¯ based anti-HIV assay was used, as described previously,38 to examine the effect of cycloviolacins O13, O14, and O24 on virus-induced cell killing in HIV-infected cultures.
Modeling and SAR analysis
Homology models were generated using Modeller 8v1 (http://www.salilab.org/modeller), a restraint-based structure prediction program. Models of circulins C-F,39 cycloviolins A-D,40 cycloviolacins Y4 and Y5,36 and cycloviolacin O1337 were derived from a high-resolution NMR structure of the bracelet cyclotide cycloviolacin O11 (PDB ID: 1NBJ). Given the high sequence similarity between cycloviolacin Y136 and tricyclon A,41 cycloviolacin Y1 was modeled from the high-resolution NMR structure of tricyclon A (PDB ID: 1YP8). The acyclic kalata B1 permutants35 and cycloviolacin O2437 were modeled on a NMR structure of kalata B142 (PDB ID: 1NB1). The lowest energy structure from a set of 50 predicted models for each target cyclotide was evaluated and analyzed using MOLMOL.43
Results and Discussion
Extraction and purification of cycloviolacins O13, O14, and O24
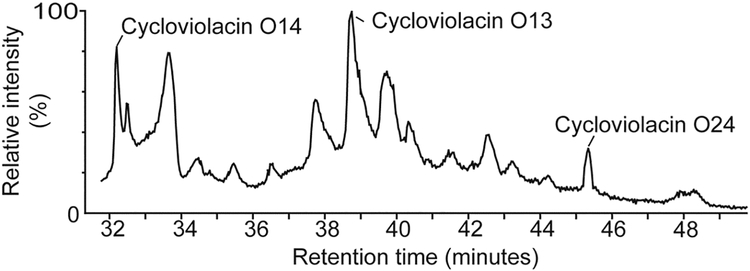
The cyclotides O13, O14 and O24 were purified from V. odorata using RP-HPLC and characterized via mass spectrometric analysis. The extractions yielded approximately 1.5 g of cyclotides (combined total mass) per kg of wet plant material, which is comparable to yields previously reported.44 Cycloviolacins O13, O14, and O24 were chosen for analysis due to their contrasting elution times (which reflect variations in hydrophobicity) and the fact that they are three of the most abundant cyclotides in V. odorata. Figure 2 shows that cycloviolacin O14 is early eluting (hydrophilic), cycloviolacin O24 is late eluting (hydrophobic), and cycloviolacin O13 has an intermediate elution time. As the suite of cyclotides expressed in V. odorata has been previously characterized,37 cyclotides O13, O14, and O24 were unambiguously identified using their characteristic retention times using RP-HPLC, monoisotopic mass using mass spectrometry, and quantitative amino acid analysis.
Figure 2.
Partial HPLC trace of an extract from Viola odorata. The peaks corresponding to the cyclotides cycloviolacin O13, O14, and O24 are indicated.
Anti-HIV activities of cycloviolacins O13, O14 and O24
The anti-HIV activities of O13, O14 and O24 are presented in Table 1. These cyclotides inhibited the cytopathic effects of HIV-1 infection in cultured human T-lymphoblast (CEM-SS) cells with antiviral cytoprotective concentrations (EC50) of 320, 440 and 308 nM, while the cytotoxic concentration (IC50) was >6.4, 4.8, and 6.2 μM respectively. Although other cyclotides have more potent anti-HIV activities, O13, O14 and O24 are three of the least cytotoxic.
Table 1.
Sequences, properties and anti-HIV activities of cyclotides
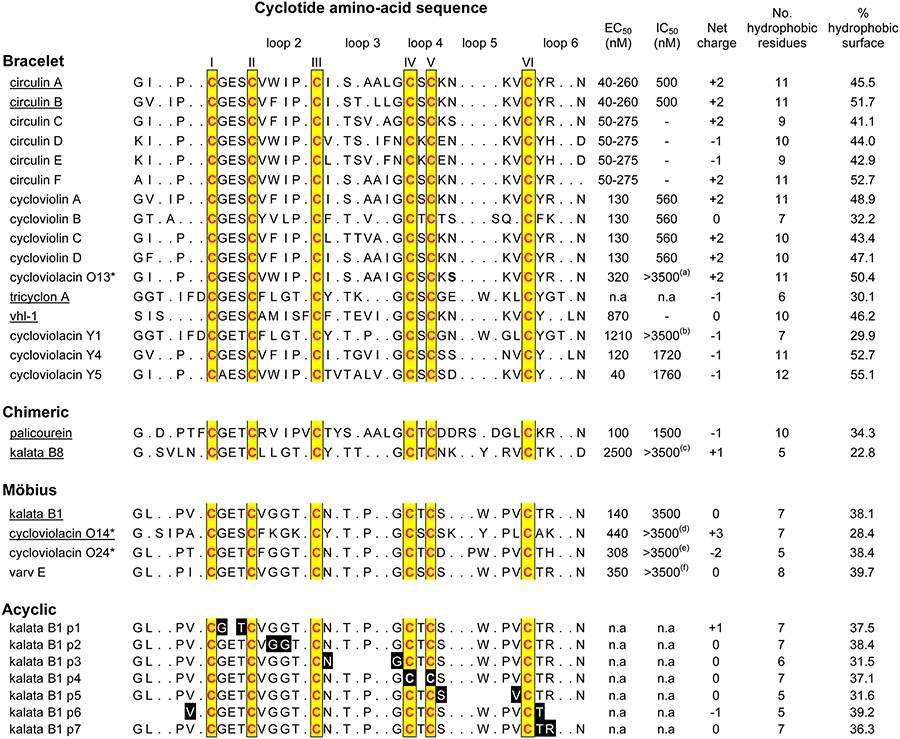
|
The amino acid residues are presented in the single-letter style. The termini of the acyclic permutants (kalata B1 p1, p2 etc) are shaded black with the amino acids in white. EC50 refers to the cytopathic inhibitory activity and IC50 refers to the cytotoxicity to the target cells. n.a. refers to no activity. The cyclotides tested in this study are marked with an *. (a) 6400, (b) >4470, (c) >11,000, (d) 4800, (e) 6170, (f) 3980. Underlined cyclotides have had their structures resolved with NMR. circulin A (PDB ID: 1BH4), circulin B (PDB ID: 2ERI), tricyclon A (PDB ID: 1YP8), vhl-1 (PDB ID: 1ZA8), palicourein (PDB ID: 1R1F), kalata B1 (PDB ID: 1NB1), kalata B8 (PDB ID: 2B38), cycloviolacin O14 (PDB ID: 2GJ0). Hydrophobic residues include A, I, L, M, F, P, W, and V.
Structure analysis and implications for anti-HIV activity
Although the six Cys residues are absolutely conserved in all cyclotides and are responsible for the preservation of the CCK motif, from Table 1 it is clear that cyclotides exhibit extensive variations in the composition and size of their loops between the Cys residues. Loop 1 shows the least variation, with the Glu residue in this loop absolutely conserved across the cyclotides tested for anti-HIV activity. Loop 4 is similarly highly conserved, always comprising just a single residue, namely Ser, Thr or Lys. Both of these loops form part of the embedded ring of the cystine knot motif and hence likely play a structural role in defining the knot. By contrast, loops 2, 3, 5 and 6 show extensive variations in their composition and size, particularly between subfamilies. The most highly conserved element within them is an Asn (or occasionally an Asp) residue in loop 6 which appears to play a functional role in the biosynthetic cyclisation.21,45,46 This variation in the residues in loops 2, 3, 5 and 6, superimposed on an otherwise conserved framework makes cyclotides an excellent template in which to examine structure-activity relationships.
Another important structural feature in the cyclotides is the propensity for amino acid side chains that are not associated with the cystine knot to protrude outwards to the molecular surface, since the molecular core is occupied by the disulfide bonds of the cystine knot. These solvent exposed amino acids include a number of residues that form a hydrophobic patch that is a characteristic of the surface of cyclotide structures.16 This surface-exposed hydrophobic patch contributes to some of the key biophysical properties of the cyclotide family, including their late elution on HPLC. Furthermore, the hydrophobic surface has been implicated in various activities of cyclotides, including antibacterial,28 insecticidal,20 anti-HIV,36 hemolytic activities.37,47 In the case of hemolytic activities the hydrophobic surface areas and hemolytic activities of cycloviolacin H4, tricyclon A, kalata B1 and circulin A were compared and there was a strong correlation between increasing hydrophobic surface area and increasing hemolytic activity.47 This correlation is consistent with the fact that melittin, which is the principal toxic component in the venom of the European honeybee and the standard control reagent for hemolytic activity assays, achieves its hemolytic activity via its hydrophobic residues disrupting the membrane.48
For the case of anti-HIV activity, a recent study of cyclotides from the Chinese medicinal herb V. yedeonsis extended the correlation between hydrophobicity and biological activity.36 That study found that for a limited series of five cyclotides tested there was a precise correlation between hydrophobicity and anti-HIV activity. The aim of the current study was to see if this correlation could be broadened to a wider range of cyclotides.
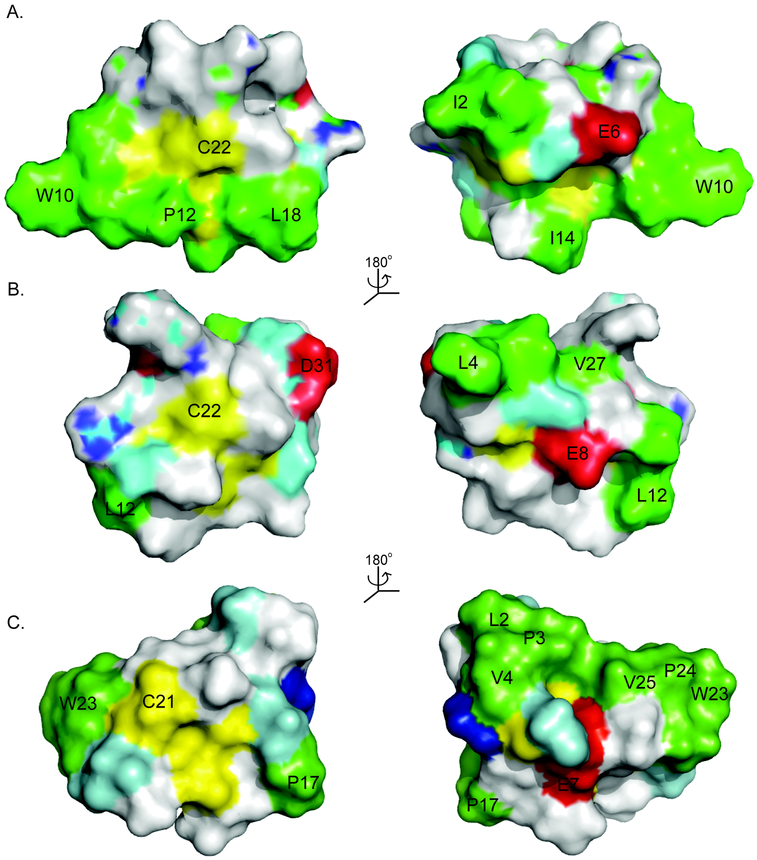
As illustrated in Table 1, an increase in the number of hydrophobic residues does correlate broadly with an increase in anti-HIV activity, with the average number of hydrophobic residues for the bracelet cyclotides being 9.8 compared to 7.5 and 6.8 for the chimeric and Möbius cyclotides respectively. Figure 3 compares the surface hydrophobicity of circulin A33 (PDB ID: 1BH4), kalata B817 (PDB ID: 2B38), and kalata B1 (PDB ID: 1NB1), representative cyclotides from the bracelet, chimeric and Möbius subfamilies. It is clear from this Figure and Table 1 that the surface characteristics differ between the subfamilies, with bracelet cyclotides appearing to be generally more hydrophobic, with hydrophobic residues on both faces of the molecule, and more potent anti-HIV agents than their Möbius, or chimeric, counterparts. The data also broadly suggest a correlation between increasing hydrophobicity and increasing cytotoxicity, with the average toxicity of the (more hydrophobic) bracelet cyclotides being greater than the (less hydrophobic) Möbius cyclotides.
Figure 3.
The surface structures of (A) circulin A, (B) kalata B8, and (C) kalata B1 are presented as representatives of the bracelet, chimera and Möbius subfamilies to show the distribution of hydrophobic residues. The hydrophobic residues are colored in green, the Cys residues in yellow, the polar residues in cyan, the positively charged in blue, and the negatively charged in red. Selected residues are labeled with a single-letter amino acid code to help orientation. In general the bracelet cyclotides are more hydrophobic than cyclotides from the other subfamilies having solvent exposed hydrophobic residues on both faces of the molecule. Möbius cyclotides have hydrophobic residues on one side of the molecule whereas chimeric cyclotides have few solvent exposed hydrophobic residues. The locations of the charged residues also differ.
However the correlation between hydrophobicity and anti-HIV activity is not as precise as seen for the limited number of cyclotides tested in V. yedeonsis, suggesting that more than just the total number of hydrophobic residues is needed to explain the anti-HIV activity of cyclotides. To further explore the features responsible for anti-HIV activity, the distribution of hydrophobic residues within cyclotides was investigated. To do this, the 3D structures of all the cyclotides tested for anti-HIV activity were required. As structures for only eight of these cyclotides had been solved, the structures of the remaining 21 cyclotides were calculated using homology modeling. Modeled cyclotide structures are generally very similar to experimentally determined structures given the high degree of structure conservation owing to the interlocking nature of the CCK motif. From the modeled and experimental structures the proportions of surface-exposed hydrophobic residues were calculated and are shown in Table 1. Analysis of these data suggests that the correlation between hydrophobicity and anti-HIV activity is stronger within the subfamilies than across subfamilies.
With the different surfaces characteristics between Möbius and bracelet cyclotides it is reasonable to suggest that the way in which they exhibit their biological activity may also differ, possibly explaining the lack of precise SAR correlations across subfamilies. It has been suggested that the mechanism of action of cyclotides is, like melittin, through membrane disruption.21,25,28,49 A recent study investigated the orientation of kalata B1 in DPC micelles and found that it binds to the micelle surface via two hydrophobic loops, loops 5 and 6.50 The study also showed that the charged residues in loops 1 and 6 play a role in membrane binding by interacting with the polar head groups of lipids. There is no experimental structure of a bracelet cyclotide bound to membranes, but an excellent protocol51 for predicting membrane binding of peptides has been reported and was applied to cyclotides.
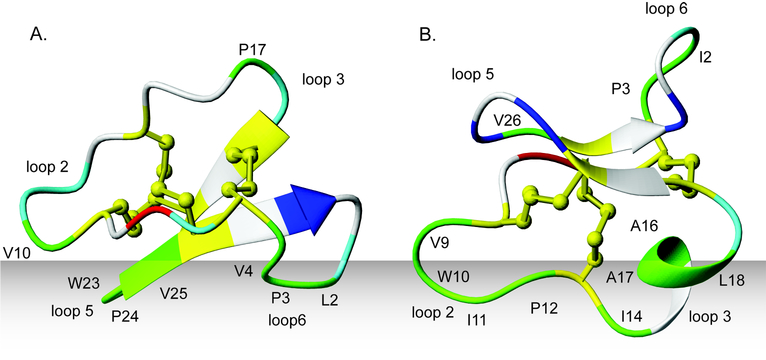
Figure 4 illustrates the predicted orientations of circulin A, a bracelet cyclotide, and kalata B1, a Möbius cyclotide, in a lipid bilayer, as predicted by the Orientations of Proteins in Membranes (OPM) database.51 The predicted orientation of kalata B1 in a lipid bilayer is in excellent agreement with the experimentally determined orientation in DPC micelles as studied by NMR,50 providing confidence that the predicted orientation for the bracelet cyclotides is also reasonable. It appears that while Möbius cyclotides interact via loops 5 and 6, bracelet cyclotides interact with the membrane via loops 2 and 3. Whether cyclotides act independently or aggregate to form multimers to disrupt the membrane is uncertain, but their ability to selfassociate has previously been suggested to be dependent on the combination of a solvent-exposed bipolar patch in loop 6 and a hydrophobic patch in loop 2.25,52
Figure 4.
Proposed mode of interaction of cyclotides (A) kalata B1 and (B) circulin A with cell membranes. The hydrophobic residues are colored in green, the Cys residues in yellow, the polar residues in cyan, the positively charged in blue, and the negatively charged in red. Selected loops and residues have been labeled to help orientation. Cyclotides most likely bind to membrane through the hydrophobic patch on their surface. The location of the hydrophobic patch on Möbius cyclotides is in loops 5 and 6 and bracelet cyclotides in loops 2 and 3. The orientations of kalata B1 and circulin A are as predicted by the OPM database.51 The predicted orientation of kalata B1 is in excellent agreement with the experimentally determined orientation in DPC micelles.50
Excluding the acyclic kalata B1 permutants, whose lack of activity is a result of destabilization due to linearization of the peptide backbone and disruption to the hydrogen bonding network,35,53 the three cyclotides with the weakest anti-HIV activity are tricyclon A,17 kalata B8,17 and cycloviolacin Y1.36 These cyclotides are from the bracelet subfamily and have on average four fewer hydrophobic residues, equating to 20% less hydrophobic surface area, than the remaining cyclotides from this subfamily. But although the observed decreased anti-HIV activity of tricyclon A, kalata B8 and cycloviolacin Y1 may be a consequence of their reduced surface hydrophobicity, the reduction in hydrophobicity seems to be localized to particular regions. Table 2 summarizes the loop characteristics of all cyclotides tested for anti-HIV activity, including the relative proportions of hydrophobic, positive and negative residues in each loop. The reduced surface hydrophobicity of tricyclon A, kalata B8 and cycloviolacin Y1 occurs predominately in loops 2 and 3, the loops that appear to be involved in membrane binding for bracelet cyclotides. The reduced hydrophobicity of these loops may account for the lower anti-HIV activity of these cyclotides, as they may be less able to bind to and disrupt the cell membranes. This reinforces the notion that it is the location of the hydrophobic residues that is important for eliciting the anti-HIV activity.
Table 2.
Surface hydrophobic and charged properties (as a percentage of total surface area) of cyclotides tested for anti-HIV activity.
| loop 1 | loop 2 | loop 3 | loop 4 | loop 5 | loop 6 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hyd | Pos | Neg | Hyd | Pos | Neg | Hyd | Pos | Neg | Hyd | Pos | Neg | Hyd | Pos | Neg | Hyd | Pos | Neg | EC50 | IC50 | |
| Bracelet | ||||||||||||||||||||
| cycloviolacin Y5 | 5 | 0 | 2 | 22 | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 6 | 10 | 0 | 0 | 40 | 1760 |
| Circulin A | 0 | 0 | 4 | 23 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 1 | 10 | 0 | 8 | 6 | 0 | 40–260 | 500 |
| Circulin B | 0 | 0 | 1 | 20 | 0 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 2 | 8 | 0 | 11 | 4 | 0 | 40–260 | 500 |
| Circulin C | 0 | 0 | 3 | 21 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 2 | 12 | 0 | 7 | 5 | 0 | 50–275 | - |
| Circulin D | 0 | 0 | 2 | 20 | 0 | 0 | 14 | 0 | 0 | 0 | 5 | 0 | 1 | 5 | 4 | 8 | 6 | 4 | 50–275 | - |
| Circulin E | 0 | 0 | 2 | 19 | 0 | 0 | 15 | 0 | 0 | 0 | 6 | 0 | 1 | 5 | 3 | 8 | 7 | 4 | 50–275 | - |
| Circulin F | 0 | 0 | 3 | 21 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 2 | 10 | 0 | 14 | 7 | 0 | 50–275 | - |
| palicourein | 0 | 0 | 2 | 16 | 5 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 3 | 6 | 8 | 6 | 6 | 3 | 100 | 1500 |
| cycloviolin A | 0 | 0 | 2 | 19 | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 2 | 11 | 0 | 11 | 7 | 0 | 130 | 560 |
| cycloviolin B | 0 | 0 | 5 | 14 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 5 | 0 | 130 | 560 |
| cycloviolin C | 0 | 0 | 2 | 20 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 2 | 10 | 0 | 8 | 6 | 0 | 130 | 560 |
| cycloviolin D | 0 | 0 | 2 | 20 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 2 | 11 | 0 | 10 | 7 | 0 | 130 | 560 |
| cycloviolacin Y4 | 0 | 0 | 2 | 22 | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 12 | 0 | 0 | 120 | 1720 |
| cycloviolacin O13 | 0 | 0 | 1 | 23 | 0 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 1 | 10 | 0 | 8 | 6 | 0 | 320 | >3500 |
| vhl-1 | 0 | 0 | 2 | 12 | 0 | 0 | 18 | 0 | 2 | 0 | 0 | 0 | 2 | 12 | 0 | 8 | 0 | 0 | 870 | - |
| cycloviolacin Y1 | 0 | 0 | 1 | 14 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 8 | 0 | 4 | 1210 | >3500 |
| kalata B8 | 0 | 0 | 3 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 14 | 0 | 11 | 4 | 4 | >3500 | >3500 |
| tricyclon A | 0 | 0 | 1 | 13 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 10 | 4 | 7 | 8 | 0 | 4 | n.a | n.a |
| Möbius | ||||||||||||||||||||
| kalata B1 | 0 | 0 | 1 | 5 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 13 | 4 | 0 | 140 | 3500 |
| cycloviolacin O14 | 0 | 0 | 1 | 6 | 17 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 6 | 5 | 0 | 11 | 3 | 0 | 440 | >3500 |
| cycloviolacin O24 | 0 | 0 | 1 | 7 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 17 | 0 | 3 | 8 | 0 | 0 | 308 | >3500 |
| varv e | 0 | 0 | 1 | 5 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 14 | 4 | 0 | 350 | >3500 |
| Acyclic | ||||||||||||||||||||
| kalata B1 p1 | 0 | 0 | 0 | 4 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 12 | 4 | 0 | n.a | n.a |
| kalata B1 p2 | 0 | 0 | 2 | 4 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 13 | 4 | 0 | n.a | n.a |
| kalata B1 p3 | 0 | 0 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 12 | 6 | 0 | n.a | n.a |
| kalata B1 p4 | 0 | 0 | 1 | 5 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 12 | 5 | 0 | n.a | n.a |
| kalata B1 p5 | 0 | 0 | 2 | 7 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 13 | 4 | 0 | n.a | n.a |
| kalata B1 p6 | 0 | 0 | 2 | 5 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 17 | 0 | 0 | 9 | 0 | 0 | n.a | n.a |
| kalata B1 p7 | 0 | 0 | 2 | 4 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 12 | 10 | 0 | n.a | n.a |
Cyclotides are arranged in descending order of anti-HIV activity. The surface characteristics, hydrophobicity, positive and negative charge, for each loop of each cyclotide are given as a percentage of the total surface area for the given cyclotide. For instance, 4.7% of the total surface area of cycloviolacin Y5 is hydrophobic and in loop 1.
The net (total) charge of an individual cyclotide appears to have no direct impact on its HIV inhibitory properties, as the net charges range from +3 to −2 with no obvious correlation with activity (Table 1). For example, despite circulins C-F having similar activity, their net charges range from −1 to +2. Likewise tricyclon A and circulin D have similar charges but their activities range from no activity to an EC50 of 50–270 nM respectively. But, as with hydrophobicity, it appears that the location of charge/s on cyclotide surfaces affects anti-HIV activity (Table 2). For example, for bracelet cyclotides whose hydrophobic and membrane binding region is in loops 2 and 3, the presence of a negative charge in this region, as in the case of vhl-1, may inhibit the interaction with the negatively charged cell membrane thereby decreasing the anti-HIV activity. By contrast, whereas a negative charge may inhibit the binding of a cyclotide to the cell membrane, a strategically located positive charge may act to enhance the binding. Of the four Möbius cyclotides tested for anti-HIV activity, three have a positive charge in loops 5 or 6. As shown in Figures 3 and 4, Möbius cyclotides interact with membranes through a hydrophobic interaction with loops 5 and 6. By improving the binding of this surface region with the membrane this positive charge may enhance the anti-HIV activity of Möbius cyclotides without the need for additional hydrophobic residues. Overall, it is clear that the location of charged residues, rather than the net charge, modulates the anti-HIV activity of cyclotides.
The cytotoxicity of bracelet cyclotides also seems to be influenced by the location of charges. A negatively charged surface exposed residue in loops 5 or 6 seems to reduce cytotoxicity. Cycloviolacin Y5, the most anti-HIV cyclotide tested to date, is one of the least cytotoxic bracelet cyclotides and has the unusual feature of a negative charge in loop 5. This negative charge may interfere with the ability of the cycloviolacin Y5 to interact with and disrupt the negatively charged cell membrane.
Conclusions and Future Directions
This paper reports new anti-HIV activity data for three cyclotides from V. odorata. The SARs of these and 26 other cyclotides previously tested for anti-HV activity was investigated. Building on previous work that examined the global structure of cyclotides and how this affects anti-HIV activity, this current study examined the distribution of hydrophobic and charged residues. It was found that in addition to an intact cyclotide backbone being essential for anti-HIV activity and there being a broad correlation between increasing hydrophobicity and anti-HIV activity, cyclotides from the bracelet and Möbius subfamilies are affected differently by the location of surfaces patches of hydrophobic and charged residues. These differences relate to their ability to interact and disrupt membranes. For bracelet cyclotides, the anti-HIV activity is dependent on the presence of hydrophobic residues in loops 2 and 3, the loops that most likely interact with cell membranes. It also appears that the cytotoxicity of bracelet cyclotides can be reduced by the presence of a negative charge in loop 5. For Möbius cyclotides a concentration of hydrophobic residues in loops 5 and 6 seems essential for anti-HIV activity and that the addition of a positive residue in either loop can improve the anti-HIV activity.
With both chemical28,54,55 and “intein-based”56 approaches now available for the synthesis of cyclotides, the opportunity for a structured approach to anti-HIV drug design is available. Using the knowledge from this study, future studies could be aimed at improving the therapeutic index of cyclotides, that is enhance their anti-HIV activity and reduce their cytotoxicity to uninfected cells, by designing novel sequences with an emphasis on location of hydrophobicity and charge.
References
- 1.Craik DJ; Daly NL; Bond T; Waine C J Mol Biol 1999, 294, 1327–1336. [DOI] [PubMed] [Google Scholar]
- 2.Chiche L; Heitz A; Gelly JC; Gracy J; Chau PT; Ha PT; Hernandez JF; Le-Nguyen D Curr Protein Pept Sci 2004, 5, 341–349. [DOI] [PubMed] [Google Scholar]
- 3.Craik DJ Science 2006, 311, 1563–1564. [DOI] [PubMed] [Google Scholar]
- 4.Craik DJ; Daly NL; Mulvenna J; Plan MR; Trabi M Curr Prot Pept Sci 2004, 5, 297–315. [DOI] [PubMed] [Google Scholar]
- 5.Goransson U; Svangard E; Claeson P; Bohlin L Curr Protein Pept Sci 2004, 5, 317329. [DOI] [PubMed] [Google Scholar]
- 6.Gustafson KR; McKee TC; Bokesch HR Curr Protein Pept Sci 2004, 5, 331–340. [DOI] [PubMed] [Google Scholar]
- 7.Kawai Y; Kemperman R; Kok J; Saito T Curr Protein Pept Sci 2004, 5, 393–398. [DOI] [PubMed] [Google Scholar]
- 8.Korsinczky ML; Schirra HJ; Craik DJ Curr Protein Pept Sci 2004, 5, 351–364. [DOI] [PubMed] [Google Scholar]
- 9.Maqueda M; Galvez A; Bueno MM; Sanchez-Barrena MJ; Gonzalez C; Albert A; Rico M; Valdivia E Curr Protein Pept Sci 2004, 5, 399–416. [DOI] [PubMed] [Google Scholar]
- 10.Selsted ME Curr Protein Pept Sci 2004, 5, 365–371. [DOI] [PubMed] [Google Scholar]
- 11.Trabi M; Craik DJ Trends Biochem Sci 2002, 27, 132–138. [DOI] [PubMed] [Google Scholar]
- 12.Trabi M; Schirra HJ; Craik DJ Biochemistry 2001, 40, 4211–4221. [DOI] [PubMed] [Google Scholar]
- 13.Craik DJ; Daly NL; Waine C Toxicon 2001, 39, 43–60. [DOI] [PubMed] [Google Scholar]
- 14.Colgrave ML; Craik DJ Biochemistry 2004, 43, 5965–5975. [DOI] [PubMed] [Google Scholar]
- 15.Craik DJ; Čemažar M; Daly NL Curr Opin Drug Discov Devel 2006, 9, 251–260. [PubMed] [Google Scholar]
- 16.Craik DJ; Čemažar M; Wang CK; Daly NL Biopolymers 2006, 84, 250–266. [DOI] [PubMed] [Google Scholar]
- 17.Daly NL; Clark RJ; Plan MR; Craik DJ Biochem J 2006, 393, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felizmenio-Quimio ME; Daly NL; Craik DJ J Biol Chem 2001, 276, 2287522882. [DOI] [PubMed] [Google Scholar]
- 19.Heitz A; Hernandez JF; Gagnon J; Hong TT; Pham TT; Nguyen TM; Le-Nguyen D; Chiche L Biochemistry 2001, 40, 7973–7983. [DOI] [PubMed] [Google Scholar]
- 20.Jennings CV; Rosengren KJ; Daly NL; Plan M; Stevens J; Scanlon MJ; Waine C; Norman DG; Anderson MA; Craik DJ Biochemistry 2005, 44, 851860. [DOI] [PubMed] [Google Scholar]
- 21.Jennings C; West J; Waine C; Craik D; Anderson M Proc Natl Acad Sci U S A 2001, 98, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gran L Lloydia 1973, 36, 207–208.4744557 [Google Scholar]
- 23.Saether O; Craik DJ; Campbell ID; Sletten K; Juul J; Norman DG Biochemistry 1995, 34, 4147–4158. [DOI] [PubMed] [Google Scholar]
- 24.Gustafson KR; Sowder II RC; Henderson LE; Parsons IC; Kashman Y; Cardellina II JH; McMahon JB; Buckheit RW Jr; Pannell LK; Boyd MR J Am Chem Soc 1994, 116, 9337–9338. [Google Scholar]
- 25.Lindholm P; Göransson U; Johansson S; Claeson P; Gulbo J; Larsson R; Bohlin L; Backlund A Mol Cancer Ther 2002, 1, 365–369. [PubMed] [Google Scholar]
- 26.Witherup KM; Bogusky MJ; Anderson PS; Ramjit H; Ransom RW; Wood T; Sardana M J Nat Prod 1994, 57, 1619–1625. [DOI] [PubMed] [Google Scholar]
- 27.Göransson U; Sjogren M; Svangard E; Claeson P; Bohlin L J Nat Prod 2004, 67, 1287–1290. [DOI] [PubMed] [Google Scholar]
- 28.Tam JP; Lu YA; Yang JL; Chiu KW Proc Natl Acad Sci U S A 1999, 96, 89138918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barry DG; Daly NL; Clark RJ; Sando L; Craik DJ Biochemistry 2003, 42, 6688–6695. [DOI] [PubMed] [Google Scholar]
- 30.Schöpke T; Hasan Agha MI; Kraft R; Otto A; Hiller K Sci Pharm 1993, 61, 145153. [Google Scholar]
- 31.Hernandez JF; Gagnon J; Chiche L; Nguyen TM; Andrieu JP; Heitz A; Trinh Hong T; Pham TT; Le Nguyen D Biochemistry 2000, 39, 5722–5730. [DOI] [PubMed] [Google Scholar]
- 32.Derua R; Gustafson KR; Pannell LK Biochem Biophys Res Commun 1996, 228, 632–638. [DOI] [PubMed] [Google Scholar]
- 33.Daly NL; Koltay A; Gustafson K,R; Boyd MR; Casas-Finet JR; Craik DJ J Mol Biol 1999, 285, 333–345. [DOI] [PubMed] [Google Scholar]
- 34.Koltay A; Daly NL; Gustafson KR; Craik DJ Int J Pept Res Therap 2005, 11, 99–106. [Google Scholar]
- 35.Daly NL; Gustafson KR; Craik DJ FEBS Lett 2004, 574, 69–72. [DOI] [PubMed] [Google Scholar]
- 36.Wang CKL; Colgrave ML; Gustafson KR; Ireland DC; Goransson U; Craik DJ J Nat Prod, Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ireland DC; Colgrave ML; Craik DJ Biochem J 2006, 400, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gulakowski RJ; McMahon JB; Staley PG; Moran RA; Boyd MR J Virol Methods 1991, 33, 87–100. [DOI] [PubMed] [Google Scholar]
- 39.Gustafson KR; Walton LK; Sowder II RC; Johnson DG; Pannell LK; Cardellina II JH; Boyd MR J Nat Prod 2000, 63, 176–178. [DOI] [PubMed] [Google Scholar]
- 40.Hallock YF; Sowder II RC; Pannell LK; Hughes CB; Johnson DG; Gulakowski R; Cardellina II JH; Boyd MR J Org Chem 2000, 65, 124–128. [DOI] [PubMed] [Google Scholar]
- 41.Mulvenna JP; Sando L; Craik DJ Structure 2005, 13, 691–701. [DOI] [PubMed] [Google Scholar]
- 42.Rosengren KJ; Daly NL; Plan MR; Waine C; Craik DJ J Biol Chem 2003, 278, 8606–8616. [DOI] [PubMed] [Google Scholar]
- 43.Koradi R; Billeter M; Wuthrich K J Mol Graphics 1996, 14, 51–55. [DOI] [PubMed] [Google Scholar]
- 44.Gran L Lloydia 1973, 36, 174–178. [PubMed] [Google Scholar]
- 45.Dutton JL; Renda RF; Waine C; Clark RJ; Daly NL; Jennings CV; Anderson MA; Craik DJ J Biol Chem 2004, 279, 46858–46867. [DOI] [PubMed] [Google Scholar]
- 46.Ireland DC; Colgrave ML; Nguyencong P; Daly NL; Craik DJ J Mol Biol 2006, 357, 1522–1535. [DOI] [PubMed] [Google Scholar]
- 47.Chen B; Colgrave ML; Wang C; Craik DJ J Nat Prod 2006, 69, 23–28. [DOI] [PubMed] [Google Scholar]
- 48.Raghuraman H; Chattopadhyay A Biopolymers 2006, 83, 111–121. [DOI] [PubMed] [Google Scholar]
- 49.Kamimori H; Hall K; Craik DJ; Aguilar MI Anal Biochem 2005, 337, 149–153. [DOI] [PubMed] [Google Scholar]
- 50.Shenkarev ZO; Nadezhdin KD; Sobol VA; Sobol AG; Skjeldal L; Arseniev AS Febs J 2006, 273, 2658–2672. [DOI] [PubMed] [Google Scholar]
- 51.Lomize MA; Lomize AL; Pogozheva ID; Mosberg HI Bioinformatics 2006, 22, 623–625. [DOI] [PubMed] [Google Scholar]
- 52.Nourse A; Trabi M; Daly NL; Craik DJ J Biol Chem 2004, 279, 562–570. [DOI] [PubMed] [Google Scholar]
- 53.Simonsen SM; Daly NL; Craik DJ FEBS Lett 2004, 577, 399–402. [DOI] [PubMed] [Google Scholar]
- 54.Daly NL; Love S; Alewood PF; Craik DJ Biochemistry 1999, 38, 10606–10614. [DOI] [PubMed] [Google Scholar]
- 55.Gunasekera S; Daly NL; Anderson MA; Craik DJ IUBMB Life 2006, 58, 515524. [DOI] [PubMed] [Google Scholar]
- 56.Kimura RH; Tran AT; Camarero JA Angew Chem Int Ed Engl 2006, 45, 973–976. [DOI] [PubMed] [Google Scholar]