Abstract
Bile acids are important for absorbing nutrients. Most mammals produce cholic and chenodeoxycholic bile acids. Here, we investigated genes in the bile acid synthesis pathway in four mammals that deviate from the usual mammalian bile composition. First, we show that naked-mole rats, elephants, and manatees repeatedly inactivated CYP8B1, an enzyme uniquely required for cholic acid synthesis, which explains the absence of cholic acid in these species. Second, no gene-inactivating mutations were found in any pathway gene in the rhinoceros, a species that lacks bile acids, indicating an evolutionarily recent change in its bile composition. Third, elephants and/or manatees that also lack bile acids altogether have lost additional nonessential enzymes (SLC27A5, ACOX2). Apart from uncovering genomic differences explaining deviations in bile composition, our analysis of bile acid enzymes in bile acid-lacking species suggests that essentiality prevents gene loss, while loss of pleiotropic genes is permitted if their other functions are compensated by functionally related proteins.
Keywords: bile composition, mammalian metabolism, comparative genomics, gene-inactivating mutations
Introduction
Bile is produced in the liver, often stored and concentrated in the gallbladder, and released into the small intestine, where it functions in digestion. Important components of bile are bile acids that promote the absorption of fat and fat-soluble vitamins from ingested food. Bile acids are synthesized from cholesterol by a multistep pathway involving several enzymes (fig. 1A) and represent a major route of eliminating cholesterol.
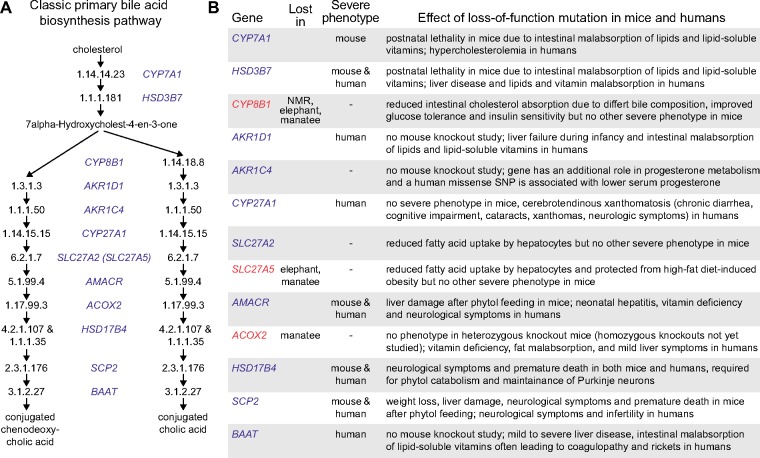
Fig. 1.
—Genes involved in the classic primary bile acid synthesis pathway. (A) Enzyme-catalyzed reactions (EC numbers, taken from KEGG map00120) and the genes encoding enzymes that catalyze these reactions. SLC27A5 is shown in parenthesis, since this enzyme is mainly required for the bile acid reconjugation and recycling but is likely less important for de novo bile acid synthesis (Hubbard et al. 2006). (B) Several of the genes involved in the bile acid synthesis pathway are lost in mammals known to deviate from the usual bile composition of placental mammals (NMR, naked-mole rat). These genes are highlighted in red font and their loss is described in this study for the first time. The right-most column lists phenotypes caused by loss-of-function mutations in mice and/or humans. The primary references describing these phenotypes are Atshaves et al. (2007), Baes et al. (2000), Carlton et al. (2003), Cheng et al. (2003), Doege et al. (2008), Duell et al. (2018), Falcon et al. (2010), Ferdinandusse et al. (2000), Ferdinandusse, Kostopoulos, et al. (2006), Ferdinandusse, Ylianttila, et al. (2006), Gonzales et al. (2004), Heinzer et al. (2003), Hubbard et al. (2006), Ishibashi et al. (1996), Johansson et al. (2011), Kaur et al. (2015), Lemonde et al. (2003), Li-Hawkins et al. (2002), Monte et al. (2017), Pullinger et al. (2002), Rosen et al. (1998), Savolainen et al. (2004), Seedorf et al. (1998), Setchell et al. (2003), Setchell et al. (2013),Shea et al. (2007), Verheijden et al. (2013), and Vilarinho et al. (2016).
Bile composition is largely conserved among placental mammals with almost all species producing two main 24-carbon (C24) bile acids: cholic acid and chenodeoxycholic acid (Hagey, Vidal, et al. 2010; Hofmann et al. 2010) (supplementary table 1, Supplementary Material online). However, structural bile acid diversity exists. For example, strepsirrhini primates, horses, zebras, and solenodons produce C27 bile acids in addition to cholic acid. Another example are bears and several rodents that produce ursodeoxycholic acid in addition to cholic and chenodeoxycholic acid (Hagey, Vidal, et al. 2010; Hofmann et al. 2010). A major difference in bile composition occurs in naked-mole rats, rhinoceroses, elephants, and manatees as these four species do not produce cholic acid (Kuroki et al. 1988; Hagey, Vidal, et al. 2010; Hofmann et al. 2010) (fig. 2). Furthermore, while naked-mole rats produce the other major bile acid (chenodeoxycholic acid), the bile of rhinoceroses, elephants, and manatees completely lacks C24 bile acids and contains instead C27 bile alcohols, which are also the major bile salts in jawless fish, sharks, and lobe-finned fish (Hagey, Moller, et al. 2010; Hofmann et al. 2010). This deviation in bile composition indicates one or multiple defects in the bile acid synthesis pathway in these four mammals; however, the underlying genomic changes are not known.
Fig. 2.

—Presence of cholic and chenodeoxycholic acid in the bile of placental mammals. Bile acid data were taken from Hagey, Vidal, et al. (2010), which shows that independent lineages lack cholic acid. The bile of rhinoceroses, elephants, and manatees contains only C27 bile alcohols. Please note that all enzymes required to synthesize chenodeoxycholic acid are also required to synthesize cholic acid (fig. 1A), thus, the lack of chenodeoxycholic acid in the presence of cholic acid, as observed for mouse, microbat, and cape elephant shrew, does not indicate an enzyme defect.
Here, we sought to identify the genomic basis of bile composition differences in naked-mole rats, rhinoceroses, elephants, and manatees. Since the loss of ancestral coding genes is an important evolutionary force (Albalat and Canestro 2016), which has provided numerous insights into evolutionary processes and the genomic basis of phenotypic differences (Castro et al. 2013; Meredith et al. 2014; Gaudry et al. 2017; Hecker et al. 2017; Emerling et al. 2018; Jebb and Hiller 2018; Meyer et al. 2018; Sharma, Hecker, et al. 2018; Sharma, Lehmann, et al. 2018), we investigated whether the loss of bile acid-synthesizing enzymes could explain these differences in bile composition.
Results and Discussion
Focusing on the classic bile acid biosynthesis pathway that accounts for the majority of produced bile acids (Russell 2003) (fig. 1A), we investigated whether any of the genes in this pathway exhibit inactivating mutations that most likely result in a nonfunctional protein. To this end, we made use of a computational approach that screens a multiple alignment between the human and many other mammalian genomes (supplementary table 1, Supplementary Material online) for mutations that shift the protein’s reading frame, create in-frame stop codons, alter the conserved splice site dinucleotides, or delete parts or entire genes (Sharma and Hiller 2017; Sharma, Hecker, et al. 2018).
We first focused on the naked-mole rat. We found that the CYP8B1 gene is entirely removed by a large genomic deletion (fig. 3A). The respective genomic locus does not contain assembly gaps and searching both the genome and unassembled genomic sequencing reads of the naked-mole rat revealed no alignment to the CYP8B1 gene, indicating that this gene is truly deleted. CYP8B1 encodes a sterol 12-alpha-hydroxylase that only participates in the synthesis of cholic acid but not in the synthesis of chenodeoxycholic acid (fig. 1A) (Gafvels et al. 1999; Russell 2003). CYP8B1 knockout in mice results in the complete absence of cholic acid (Li-Hawkins et al. 2002; Kaur et al. 2015), thus, the natural knockout of CYP8B1 in naked-mole rats can explain the lack of cholic acid. All other genes in the bile acid synthesis pathway lack any gene-inactivating mutations in the naked-mole rat, which is consistent with the fact that naked-mole rats synthesize chenodeoxycholic acid, for which all other enzymes are required.
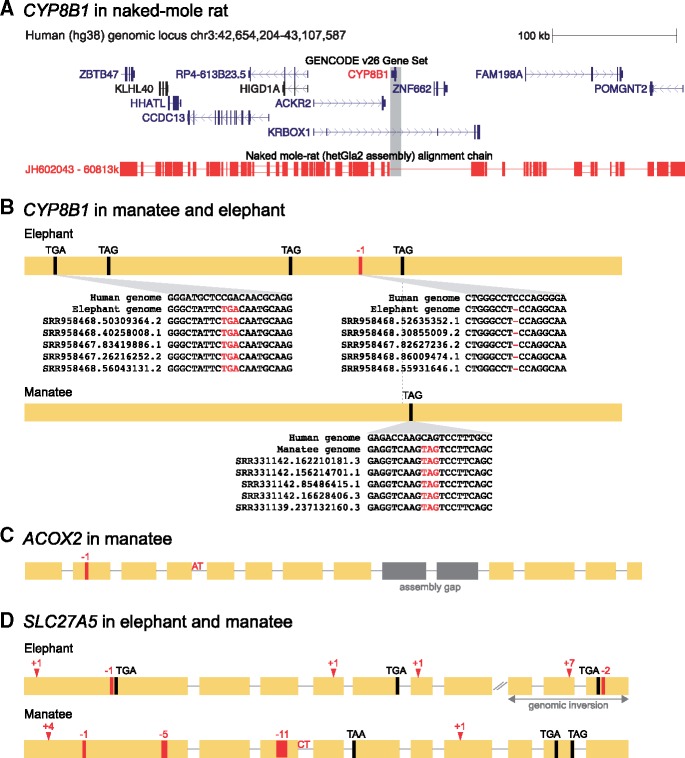
Fig. 3.
—Loss of genes involved in bile acid synthesis in mammals lacking cholic acid or C24 bile acids altogether. (A) CYP8B1 is completely deleted in the naked-mole rat genome. Screenshot of the human UCSC genome browser (Casper et al. 2018) shows the genome alignment between human and naked-mole rat (colinear alignment chain in red). Boxes in this chain represent aligning regions, single lines represent deletions, and double lines represent regions that do not align. The entire alignment chain spans >8 Mb in the human genome, showing that gene order in a larger genomic segment is conserved between both species. However, the entire region comprising CYP8B1 (highlighted in gray) is completely deleted in the naked-mole rat, as shown by the absence of any aligning blocks. In addition, the neighboring ZNF662 gene is also deleted; however, this gene is also absent in other rodents such as mouse. Consistent with a large deletion, the distance between ACKR2 and KRBOX1 is ∼70 kb in human, ∼26 kb in mouse and only ∼10 kb in the naked-mole rat. (B) Inactivating mutations in elephant and manatee CYP8B1. The yellow box represents the single coding exon of CYP8B1. Stop codon mutations are indicated, −n refers to a deletion of n bp. Three insets exemplify that all shown mutations are validated by at least 20 unassembled DNA sequencing reads (only five reads are shown for space considerations). We further found that all five mutations in the African elephant are also present in sequencing reads of the Asian elephant, a mammal that also lacks cholic acid (Hofmann et al. 2010), showing that CYP8B1 was already inactivated in the common ancestor of both elephant species. (C) Inactivating mutations in manatee ACOX2. As for (B), boxes represent coding exons of ACOX2, superimposed by inactivating mutations that are all confirmed by sequencing reads. The donor splice site of exon 4 is mutated from GT to AT. Exons 9 and 10 (gray) overlap assembly gaps and are not present in the manatee genome assembly. (D) Inactivating mutations in elephant and manatee SLC27A5, all confirmed by sequencing reads. +n refers to a frameshifting insertion of n bp. The donor splice site of exon 3 in manatee is mutated from GT to CT. In the African elephant genome assembly, the last three exons are inverted, further corroborating gene loss. The inactivating mutations in the African elephant exon 1 (−1 bp deletion and stop codon), exon 4, and exon 5 are also present in sequencing reads of the Asian elephant, indicating that SLC27A5 was already lost in their common ancestor.
For the rhinoceros that produces neither cholic nor chenodeoxycholic acid, we surprisingly found no gene-inactivating mutation in any of the genes encoding enzymes in the pathway (fig. 1A). The lack of any clear gene-inactivation mutations would be consistent with an evolutionarily recent loss of bile acid synthesis in the white rhinoceros, the species with a sequenced genome. Supporting a putative recent change, some individuals of the related Sumatran and Indian rhinoceros still produce a mixture of C24 bile acids and C27 bile alcohols (Hagey LR, personal communication). A previous study showed that the C27 bile alcohols of rhinoceroses are derived from an intermediate product of the multifunctional CYP27A1 enzyme that catalyzes the 27-hydroxylation reaction but fails to subsequently oxidize the side chain (Hagey, Vidal, et al. 2010). Interestingly, we found significant evidence that the rhinoceros CYP27A1 evolves under relaxed selection to preserve the encoded protein. Using RELAX, a method to estimate whether selection was relaxed or intensified in selected branches of the phylogeny (Wertheim et al. 2015), we obtained a relaxation factor estimate of 0.25 (values <1 indicate relaxed selection) with a P value of 0.005. This raises the possibility that amino acid mutations affecting CYP27A1 activity or regulatory mutations affecting its expression are responsible for the putative recent loss of bile acid synthesis in the white rhinoceros; however, the exact causal mutation(s) remain to be identified.
For the African elephant and manatee that also lack C24 bile acids altogether, our analysis detected gene-inactivating mutations in CYP8B1 (fig. 3B), which explains the absence of cholic acid and the lack of 12α-hydroxyl groups in any of the manatee bile alcohols (Kuroki et al. 1988). To rule out potential artifacts that can mimic gene loss (Hecker et al. 2017; Sharma, Lehmann, et al. 2018), we manually validated the inactivation of CYP8B1. Similar to the naked-mole rat (fig. 3A), gene order in elephant and manatee is conserved for >2.5-Mb upstream and downstream of CYP8B1, showing that the remnants of this gene are located in the ancestral locus. Using unassembled genomic sequencing reads, we found support by at least 20 sequencing reads for all five inactivating mutations in the elephant and for the stop codon mutation in the manatee that would lead to a protein with a severely truncated cytochrome P450 domain (fig. 3B). The respective ancestral alleles that are not gene-inactivating are not supported by a single read, which excludes the possibility that the inactivating mutations observed in the genome are sequencing or assembly errors. Finally, consistent with CYP8B1 loss, RELAX analysis shows that the remnants of the CYP8B1 coding sequence evolve under relaxed or no selection (elephant: relaxation factor 0.09, P < 0.0001; manatee: relaxation factor 0.34, P = 0.0015). Thus, patterns of relaxed selection together with gene-inactivating mutations that are validated by sequencing reads show that CYP8B1 is truly inactivated in the elephant and manatee.
The bile alcohols of elephant and manatee are, as for the rhinoceros, derived from an intermediate CYP27A1 product, showing that the bile acid synthesis pathway stops at this reaction (Kuroki et al. 1988; Hagey, Vidal, et al. 2010; Hofmann et al. 2010). This indicates that the enzyme-encoding genes upstream of CYP27A1 (CYP7A1, HSD3B7, AKR1D1, AKR1C4; see fig. 1A) should lack inactivating mutations. Our analysis shows that this is indeed the case, which confirms and extends previous findings (Hagey, Vidal, et al. 2010). CYP27A1 does not evolve under relaxed selection in elephant or manatee (RELAX: P > 0.33). However, analyzing the other enzyme-encoding genes downstream of CYP27A1, we first found that the manatee (but not elephant) has inactivating mutations in ACOX2 (fig. 3C), which are also supported by >20 sequencing reads and a moderate but not significant pattern of relaxed selection (relaxation factor 0.22, P = 0.11). ACOX2 encodes a peroxisomal enzyme that oxidizes CoA esters of bile acid intermediates and branched fatty acids. Mutations in human ACOX2 lead to the absence of bile acids, but individuals with ACOX2 deficiency have only mild clinical symptoms (elevated serum transaminase; fig. 1B) (Monte et al. 2017; Ferdinandusse et al. 2018), showing that ACOX2 is not an essential enzyme. Second, we detected and validated the loss of SLC27A5 in the African elephant and manatee (fig. 3D). This liver-specific enzyme is required for bile acid reconjugation and recycling (Hubbard et al. 2006), which would be consistent with a reduced necessity to recycle bile acids in mammals that do not produce them.
Given the observed loss of CYP8B1, ACOX2, and SLC27A5 in at least one of the four mammals that deviate from the usual bile composition, we asked how specific these gene losses are. Analyzing all other 58 placental mammals contained in our multiple genome alignment (supplementary table 1, Supplementary Material online) (Sharma and Hiller 2017), we found no inactivating mutation in any of these three genes, with the exception of SLC27A5 that is also lost in the cape golden mole (supplementary fig. 1, Supplementary Material online), a mammal whose bile composition has not been characterized yet. The average nonsynonymous/synonymous (Ka/Ks) ratios of 0.25, 0.27, and 0.32 further indicate that the CYP8B1, SLC27A5, and ACOX2 genes evolve under purifying selection in other placental mammals.
Overall, our analysis shows that the majority of genes encoding bile acid-synthesizing enzymes lack inactivating mutations in mammals that lack bile acids. The enzymes up to CYP27A1 are required to produce bile alcohols in rhinoceroses, elephants, and manatees, which explains the absence of inactivating mutations in the respective genes. Loss of enzymes downstream of CYP27A1 is likely restricted by essentiality of the respective genes (fig. 1B). For example, loss-of-function mutations in AMACR, HSD17B4, SCP2, or BAAT lead to severe phenotypes in human and/or mouse, ranging from vitamin deficiency to liver damage and premature death (fig. 1B). Apart from the synthesis of bile acids, AMACR, HSD17B4, and SCP2 are also required for the catabolism of plant-derived phytol. The lack of any of these genes causes accumulation of phytol intermediates, liver damage, and neuropathy in human and in mouse (Baes et al. 2000; Savolainen et al. 2004; Ferdinandusse, Kostopoulos, et al. 2006; Atshaves et al. 2007), suggesting these three genes are also important for obligate herbivores such as naked-mole rats, rhinoceroses, elephants, and manatees. Furthermore, HSD17B4 has essential roles in peroxisomal beta-oxidation and in maintaining Purkinje neurons in the brain (Baes et al. 2000; Verheijden et al. 2013).
In contrast to essentiality, pleiotropy of genes does not seem to be major factor preventing the natural loss of bile acid-synthesizing enzymes. For example, ACOX2 is involved in both bile acid synthesis and phytol catabolism. However, while being pleiotropic, ACOX2 is not an essential enzyme, since ACOX3 can compensate a defect in oxidizing phytol intermediates and other branched-chain fatty acids in case of ACOX2 deficiency (Ferdinandusse et al. 2018). Similarly, SLC27A5 conjugates bile acids and contributes to fatty acid uptake by the liver. However, knockout mice have no severe phenotypes, likely because other fatty acid transport proteins provide sufficient fatty acid uptake capacity (Hubbard et al. 2006; Doege et al. 2008), suggesting that SLC27A5 is pleiotropic but not essential. Thus, pleiotropic genes can get lost in evolution if one of their functions (here bile acid synthesis) becomes dispensable and loss of their other functions is compensated by functionally related proteins.
In summary, we detected the recurrent loss of CYP8B1 in three mammals that do not produce cholic acid, which provides a molecular explanation for the loss of cholic acid production in these species. Differences in bile composition in elephants and manatees are further associated with the loss of additional genes. Overall, this highlights the potential of comparative genome analysis to link gene inactivation to metabolic differences between species. Given that more and more genome sequences will be available in future, comparative genome analysis can obtain additional insights into how changes in bile acid-synthesizing enzymes relate to evolutionary changes in bile composition. More generally, our analysis suggests that gene essentiality rather than pleiotropy restricts the loss of bile acid-synthesizing enzymes in bile acid-lacking mammals.
Materials and Methods
We used the human Ensembl v91 gene annotation as a reference and screened the aligned gene loci of other mammals for in-frame stop codon mutations, frameshifting insertions or deletions, mutations that disrupt the conserved donor (GT/GC) or acceptor (AG) dinucleotide, and deletions of coding exons (Sharma, Hecker, et al. 2018). Large deletions were only considered if the corresponding locus did not overlap assembly gaps in the genome assembly of the other mammal. Our approach makes use of CESAR (Sharma et al. 2016, 2017) to exclude false inactivating mutations that are due to alignment ambiguities. To validate inactivating mutations, we extracted the genomic sequence around the mutation with a 25-bp flank and used megablast (parameters match score −1, mismatch scores −2, gap costs linear, expectation value threshold 10) to search unassembled sequencing reads stored in the TRACE and Sequence Read Archives (supplementary table 2, Supplementary Material online). To test if the ancestral noninactivating allele is supported by reads, we reversed the mutation by replacing stop codons with the conserved sense codon and inserting the single base pair that is deleted in elephants. To test for relaxed selection, we used RELAX (Wertheim et al. 2015) with the partition descriptive method that fits three Ka/Ks classes to the test and the reference branches and also estimates an overall Ka/Ks value. RELAX estimates a relaxation/intensification parameter k that either indicates relaxed selection (k < 1) or intensified selection (k > 1). We included either the rhinoceros, the elephant, or the manatee sequence as the foreground and specified the branches to all other mammals as background.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the genomics community for sequencing and assembling the genomes and the UCSC genome browser group for providing software and genome annotations. We also thank Lee Hagey for valuable discussions, and the Computer Service Facilities of the MPI-CBG and MPI-PKS for their support. This work was supported by the Max Planck Society and the German Research Foundation (HI 1423/3-1).
Literature Cited
- Albalat R, Canestro C.. 2016. Evolution by gene loss. Nat Rev Genet. 17(7):379–391. [DOI] [PubMed] [Google Scholar]
- Atshaves BP, et al. 2007. Effect of SCP-x gene ablation on branched-chain fatty acid metabolism. Am J Physiol Gastrointest Liver Physiol. 292(3):G939–G951. [DOI] [PubMed] [Google Scholar]
- Baes M, et al. 2000. Inactivation of the peroxisomal multifunctional protein-2 in mice impedes the degradation of not only 2-methyl-branched fatty acids and bile acid intermediates but also of very long chain fatty acids. J Biol Chem. 275(21):16329–16336. [DOI] [PubMed] [Google Scholar]
- Carlton VE, et al. 2003. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet. 34(1):91–96. [DOI] [PubMed] [Google Scholar]
- Casper J, et al. 2018. 2018. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 46(D1):D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro LF, et al. 2013. Recurrent gene loss correlates with the evolution of stomach phenotypes in gnathostome history. Proc Biol Sci. 281(1775):20132669.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JB, et al. 2003. Molecular genetics of 3beta-hydroxy-Delta5-C27-steroid oxidoreductase deficiency in 16 patients with loss of bile acid synthesis and liver disease. J Clin Endocrinol Metab. 88(4):1833–1841. [DOI] [PubMed] [Google Scholar]
- Doege H, et al. 2008. Silencing of hepatic fatty acid transporter protein 5 in vivo reverses diet-induced non-alcoholic fatty liver disease and improves hyperglycemia. J Biol Chem. 283(32):22186–22192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duell PB, et al. 2018. Diagnosis, treatment and clinical outcomes in 43 cases with cerebrotendinous xanthomatosis. J Clin Lipidol. 12(5):1169–1178. [DOI] [PubMed] [Google Scholar]
- Emerling CA, Delsuc F, Nachman MW.. 2018. Chitinase genes (CHIAs) provide genomic footprints of a post-Cretaceous dietary radiation in placental mammals. Sci Adv. 4(5):eaar6478.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon A, et al. 2010. FATP2 is a hepatic fatty acid transporter and peroxisomal very long-chain acyl-CoA synthetase. Am J Physiol Endocrinol Metab. 299(3):E384–E393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandusse S, et al. 2000. Mutations in the gene encoding peroxisomal alpha-methylacyl-CoA racemase cause adult-onset sensory motor neuropathy. Nat Genet. 24(2):188–191. [DOI] [PubMed] [Google Scholar]
- Ferdinandusse S, Kostopoulos P, et al. 2006. Mutations in the gene encoding peroxisomal sterol carrier protein X (SCPx) cause leukencephalopathy with dystonia and motor neuropathy. Am J Hum Genet. 78(6):1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandusse S, Ylianttila MS, et al. 2006. Mutational spectrum of D-bifunctional protein deficiency and structure-based genotype-phenotype analysis. Am J Hum Genet. 78(1):112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandusse S, et al. 2018. A novel case of ACOX2 deficiency leads to recognition of a third human peroxisomal acyl-CoA oxidase. Biochim Biophys Acta. 1864(3):952–958. [DOI] [PubMed] [Google Scholar]
- Gafvels M, et al. 1999. Structure and chromosomal assignment of the sterol 12alpha-hydroxylase gene (CYP8B1) in human and mouse: eukaryotic cytochrome P-450 gene devoid of introns. Genomics 56:184–196. [DOI] [PubMed] [Google Scholar]
- Gaudry MJ, et al. 2017. Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades. Sci Adv. 3(7):e1602878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales E, et al. 2004. SRD5B1 (AKR1D1) gene analysis in delta(4)-3-oxosteroid 5beta-reductase deficiency: evidence for primary genetic defect. J Hepatol. 40(4):716–718. [DOI] [PubMed] [Google Scholar]
- Hagey LR, Moller PR, Hofmann AF, Krasowski MD.. 2010. Diversity of bile salts in fish and amphibians: evolution of a complex biochemical pathway. Physiol Biochem Zool. 83(2):308–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagey LR, Vidal N, Hofmann AF, Krasowski MD.. 2010. Evolutionary diversity of bile salts in reptiles and mammals, including analysis of ancient human and extinct giant ground sloth coprolites. BMC Evol Biol. 10(1):133.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker N, Sharma V, Hiller M.. 2017. Transition to an aquatic habitat permitted the repeated loss of the pleiotropic KLK8 gene in mammals. Genome Biol Evol. 9(11):3179–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzer AK, et al. 2003. A very long-chain acyl-CoA synthetase-deficient mouse and its relevance to X-linked adrenoleukodystrophy. Hum Mol Genet. 12(10):1145–1154. [DOI] [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR, Krasowski MD.. 2010. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res. 51(2):226–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard B, et al. 2006. Mice deleted for fatty acid transport protein 5 have defective bile acid conjugation and are protected from obesity. Gastroenterology 130:1259–1269. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Schwarz M, Frykman PK, Herz J, Russell DW.. 1996. Disruption of cholesterol 7alpha-hydroxylase gene in mice. I. Postnatal lethality reversed by bile acid and vitamin supplementation. J Biol Chem. 271(30):18017–18023. [DOI] [PubMed] [Google Scholar]
- Jebb D, Hiller M.. 2018. Recurrent loss of HMGCS2 shows that ketogenesis is not essential for the evolution of large mammalian brains. Elife 7:e38906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson AG, Nikamo P, Schalling M, Landen M.. 2011. AKR1C4 gene variant associated with low euthymic serum progesterone and a history of mood irritability in males with bipolar disorder. J Affect Disord. 133(1–2):346–351. [DOI] [PubMed] [Google Scholar]
- Kaur A, et al. 2015. Loss of Cyp8b1 improves glucose homeostasis by increasing GLP-1. Diabetes 64(4):1168–1179. [DOI] [PubMed] [Google Scholar]
- Kuroki S, et al. 1988. Bile salts of the West Indian manatee, Trichechus manatus latirostris: novel bile alcohol sulfates and absence of bile acids. J Lipid Res. 29(4):509–522. [PubMed] [Google Scholar]
- Lemonde HA, et al. 2003. Mutations in SRD5B1 (AKR1D1), the gene encoding delta(4)-3-oxosteroid 5beta-reductase, in hepatitis and liver failure in infancy. Gut 52(10):1494–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Hawkins J, et al. 2002. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest. 110(8):1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Zhang G, Gilbert MT, Jarvis ED, Springer MS.. 2014. Evidence for a single loss of mineralized teeth in the common avian ancestor. Science 346(6215):1254390.. [DOI] [PubMed] [Google Scholar]
- Meyer WK, et al. 2018. Ancient convergent losses of Paraoxonase 1 yield potential risks for modern marine mammals. Science 361(6402):591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte MJ, et al. 2017. ACOX2 deficiency: an inborn error of bile acid synthesis identified in an adolescent with persistent hypertransaminasemia. J Hepatol. 66(3):581–588. [DOI] [PubMed] [Google Scholar]
- Pullinger CR, et al. 2002. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J Clin Invest. 110(1):109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen H, et al. 1998. Markedly reduced bile acid synthesis but maintained levels of cholesterol and vitamin D metabolites in mice with disrupted sterol 27-hydroxylase gene. J Biol Chem. 273(24):14805–14812. [DOI] [PubMed] [Google Scholar]
- Russell DW. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 72(1):137–174. [DOI] [PubMed] [Google Scholar]
- Savolainen K, et al. 2004. A mouse model for alpha-methylacyl-CoA racemase deficiency: adjustment of bile acid synthesis and intolerance to dietary methyl-branched lipids. Hum Mol Genet. 13(9):955–965. [DOI] [PubMed] [Google Scholar]
- Seedorf U, et al. 1998. Defective peroxisomal catabolism of branched fatty acyl coenzyme A in mice lacking the sterol carrier protein-2/sterol carrier protein-x gene function. Genes Dev. 12(8):1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setchell KD, et al. 2003. Liver disease caused by failure to racemize trihydroxycholestanoic acid: gene mutation and effect of bile acid therapy. Gastroenterology 124(1):217–232. [DOI] [PubMed] [Google Scholar]
- Setchell KD, et al. 2013. Genetic defects in bile acid conjugation cause fat-soluble vitamin deficiency. Gastroenterology 144(5):945–955. e946; quiz e914–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Elghafari A, Hiller M.. 2016. Coding exon-structure aware realigner (CESAR) utilizes genome alignments for accurate comparative gene annotation. Nucleic Acids Res. 44(11):e103.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Hecker N, et al. 2018. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat Commun. 9:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Hiller M.. 2017. Increased alignment sensitivity improves the usage of genome alignments for comparative gene annotation. Nucleic Acids Res. 45(14):8369–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Lehmann T, Stuckas H, Funke L, Hiller M.. 2018. Loss of RXFP2 and INSL3 genes in Afrotheria shows that testicular descent is the ancestral condition in placental mammals. PLoS Biol. 16(6):e2005293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Schwede P, Hiller M.. 2017. CESAR 2.0 substantially improves speed and accuracy of comparative gene annotation. Bioinformatics 33(24):3985–3987. [DOI] [PubMed] [Google Scholar]
- Shea HC, Head DD, Setchell KD, Russell DW.. 2007. Analysis of HSD3B7 knockout mice reveals that a 3alpha-hydroxyl stereochemistry is required for bile acid function. Proc Natl Acad Sci U S A. 104(28):11526–11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijden S, et al. 2013. Peroxisomal multifunctional protein-2 deficiency causes neuroinflammation and degeneration of Purkinje cells independent of very long chain fatty acid accumulation. Neurobiol Dis. 58:258–269. [DOI] [PubMed] [Google Scholar]
- Vilarinho S, et al. 2016. ACOX2 deficiency: a disorder of bile acid synthesis with transaminase elevation, liver fibrosis, ataxia, and cognitive impairment. Proc Natl Acad Sci U S A. 113(40):11289–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32(3):820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.