Abstract
Objective
A large number of genetic loci have been associated with risk of coronary artery disease (CAD) through genome-wide association studies, however, for most loci the underlying biological mechanism is unknown. Determining the molecular pathways and cellular processes affected by these loci will provide new insights into CAD pathophysiology and may lead to new therapies. The CAD-associated variants at 10p11.23 fall in JCAD, which encodes an endothelial junction protein, however, its molecular function in endothelial cells is not known. In this study we characterize the molecular role of JCAD in endothelial cells.
Approach and Results
We show that JCAD knockdown in endothelial cells affects key phenotypes related to atherosclerosis including proliferation, migration, apoptosis, tube formation and monocyte binding. We demonstrate that JCAD interacts with LATS2 (large tumor suppressor kinase 2) and negatively regulates Hippo signaling leading to increased activity of YAP (Yes-associated protein), the transcriptional effector of the pathway. We also show by double siRNA knockdown that the phenotypes caused by JCAD knockdown require LATS2 and that JCAD is involved in transmission of RhoA-mediated signals into the Hippo pathway. In human tissues, we find that the CAD-associated lead variant, rs2487928, is associated with expression of JCAD in arteries, including atherosclerotic arteries. Gene co-expression analyses across disease-relevant tissues corroborate our phenotypic findings and support the link between JCAD and Hippo signaling.
Conclusions
Our results show that JCAD negatively regulates Hippo signaling in endothelial cells and we suggest that JCAD contributes to atherosclerosis by mediating YAP activity and contributing to endothelial dysfunction.
Keywords: Coronary artery disease, Hippo pathway, YAP/TAZ, Endothelium
Subject codes: Coronary Artery Disease, Endothelium/Vascular Type/Nitric Oxide, Cell Biology/Structural Biology, Cell Signaling/Signal Transduction
Introduction
Coronary artery disease (CAD), caused by the development of atherosclerotic plaques in the artery wall, is the leading cause of death in the world1. Plaques, which tend to localize to sites of disturbed blood flow such as bifurcations and curvature, form as a result of chronic exposure of the arterial endothelium to cardiovascular risk factors and pro-inflammatory factors2. The resulting endothelial dysfunction results in expression of adhesion molecules inducing recruitment of inflammatory cells to the site of injury alongside an increase in endothelial permeability and accumulation in lipids in the intima. Subsequently, monocytes migrate into the vessel wall where they differentiate into macrophages, ingest lipids and become foam cells2, 3. Advanced atherosclerotic plaques consist of a central core containing lipid-laden cells and extracellular deposits released from dead cells, covered by a fibrous cap consisting primarily of smooth muscle cells and connective tissue. Rupture or erosion of the fibrous cap causes thrombus formation and occlusion of the coronary arteries resulting in myocardial infarction2.
Several individual characteristics and life style factors including hypertension, elevated low density lipoprotein (LDL) cholesterol, diabetes mellitus and smoking contribute to risk of CAD. There is also a strong genetic determination4 and in the last decade an increasing number of chromosomal loci have been associated with CAD through genome-wide association studies5–10.The elucidation of underlying biological mechanisms will provide a better understanding of CAD and has the potential to lead to the development of novel therapeutics directed against processes not targeted by current treatments.
In this context, the CAD genome-wide associated variants at the 10p11.23 locus lie within JCAD (Junctional Cadherin-5 Associated Protein) gene, previously known as KIAA1462 and Junctional Protein Associated with CAD, which encodes a protein which localizes to endothelial cell (EC) junctions11 and regulates angiogenesis12, however, the molecular role of JCAD in ECs has not been examined. Two independent proteomic studies have previously identified JCAD as an interactor of members of the human Hippo signaling pathway13, 14 and during the preparation of this manuscript, Ye et al. reported that JCAD acts as an inhibitor of a core Hippo signaling protein in liver cancer15. The Hippo pathway promotes cell death and differentiation and inhibits cell proliferation16 in response to various signals including cell contact, mechanotransduction signals and GPCR mediated signaling17. A kinase cascade controls the nuclear localization of two homologous transcriptional co-activators of the pathway; YAP (Yes-associated protein) and TAZ (transcriptional coactivator with PDZ-binding motif)18. When the Hippo pathway is activated, MST1 and 2 (mammalian STE20-like protein kinase 1 and 2) phosphorylate LATS1 and 2 (large antigen tumor suppressor 1 and 2), which in turn phosphorylate YAP/TAZ resulting in them being bound by 14-3-3 proteins, excluded from the nucleus and subsequently degraded16, 18. Therefore, when the Hippo pathway is inactive, YAP/TAZ enter the nucleus and bind to the TEAD1-4 (TEA domain transcription factor 1-4) transcription factors, promoting expression of downstream genes.
YAP/TAZ are key regulators of EC function and are essential for angiogenesis and vascular homeotsasis19, 20. Notably, two recent studies have produced compelling evidence that YAP/TAZ activity is a critical mediator of endothelial dysfunction and atherogenesis at sites of disturbed blood flow and in response to inflammatory cytokines20,21. Unidirectional blood flow, which is important for normal vascular function and is atheroprotective, was shown to signal through Integrin-β3 and inhibit the small GTPase RhoA, suppressing YAP/TAZ activity. Atherogenic disturbed flow induced YAP/TAZ activity and mediated a proinflammatory response, promoting endothelial adhesion molecule expression and monocyte adhesion21, 22. In Apoe-/- mice, inhibition of YAP by injection of morpholino oligonucleotides21 or EC-specific CRISPR/Cas YAP knockdown22 reduced plaque size in partial carotid ligation disturbed flow models. EC-specific YAP overexpression increased atherosclerosis22. Interestingly, YAP/TAZ, via RhoA, have been identified as mediators of the athero-protective and anti-inflammatory effects of statins23, 24. Statin treatment of ECs caused inhibition of YAP/TAZ activity and blocked EC atherosclerotic phenotypes, suggesting that Hippo signaling and YAP/TAZ might be viable targets for new therapeutics21, 22.
Here, we present our investigation of JCAD and its function in ECs. We demonstrate that JCAD is required for normal EC function, interacts with LATS2 and promotes YAP/TAZ activity by acting as a negative regulator of Hippo signaling in ECs. We show that JCAD is required for regulation of Hippo signaling by RhoA and that the phenotypic effects of JCAD require LATS2. We use translational network analysis of gene expression in disease relevant human tissues to corroborate our findings.
Materials and Methods
All data that support the findings of this study are either available within the article, the supplemental material or are available from the corresponding author upon reasonable request.
Cell culture, siRNA and drug treatments
HUVECs were obtained from Promocell. Three independent lines were used for all experiments. HUVECs were cultured in Endothelial Cell Growth Medium MV (Promocell). To knock-down JCAD, 2 unique siRNAs (Qiagen, SI04210136, SI04344704) were reverse transfected into the cells using RNAiMAX transfection reagent (Life Technologies, 10514953), LATS2 knockdown was achieved using a Dharmacon Smart pool of 4 siRNAs (GE Life Sciences, L-003865-00-0005). HAECs were cultured in EGM-2 media (Lonza, CC-3156) and JCAD knockdown achieved with a Dharmacon Smart pool of 4 siRNAs (GE Life Sciences, L-026476-02-0005), also transfected using RNAiMAX.Thp-1s were cultured in RPMI (Corning, 10-040-CVR) supplemented with 10% FBS. HEK293A cells were cultured in DMEM with 10% FBS. Drug treatments were carried out in full Endothelial media, for 1 hour treatment time, TRAP6 (Sigma-Aldrich, T1573) was used at 1 µM and Y-27632 (Stemcell Technologies, 72304) at 10 µM.
Proliferation, apoptosis, migration and tube formation assays
For proliferation assays, HUVECs were seeded in 96-well plates at 2x103 cells/well, with 8 replicates per sample. The sulforhodamine-B assay was used. HAECs were seeded at 5x103 cells/well, fixed with 4% paraformaldehyde after 72 hours, DAPI stained and imaged using an Operetta imaging system (Perkin-Elmer, HH12000000). Apoptosis was measured by staining with Annexin-V-FITC (Biolegend, 640906) and measured with a Cyan ADP Flow Cytometer (Beckman Coulter, no catalog number) using standard methods. Cell migration was measured using a wound-healing assay. Cells were grown in a 24-well plate until confluent, then a scratch created across the whole well using a pipette tip. A Nikon Eclipse Ti microscope equipped with an LED light source for epifluorescence and a Nikon Perfect Focus System at the Advanced Imaging Facility of the University of Leicester was used to record images every hour. The microscope has an environmental chamber with temperature control and CO2 supply. An Andor iXonEM+ EMCCD DU 885 camera is attached to the microscope for image collection using NIS-Elements software (Nikon Instruments Europe). Tube formation was carried out in Ibidi angiogenesis slides. Matrigel (Corning, 354277) was plated into the well, allowed to polymerise for 1 hour, then 104 cells were added to each well. After 6 hours, images were taken of each well at 4x magnification using an EVOS Fluorescent Microscope (Fisher Scientific) or Nikon Eclipse TE2000-U. The FIJI distribution of ImageJ25 with the plugin Angiogenesis Analyzer was used to measure the total tubule length in each image.
JCAD-GFP cloning, expression and immunoprecipitation
The full-length open reading frame (ORF) of JCAD in the pF1K vector was obtained from Promega. The full ORF was PCR amplified from this, and subcloned into the pLEICS-29 vector by the Protein Expression Laboratory at the University of Leicester. JCAD-GFP or empty pLEICS-29 expressing GFP alone were transfected into HUVECs using Lipofectamine-LTX transfection reagent (Life Technologies, A12621). Cells were then trypsinised, lysed in protein lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA and 0.5% NP-40) with Roche cOmplete protease inhibitor cocktail (Sigma-Aldrich, 4693159001) and Roche PhosSTOP phosphatase inhibitors (Sigma-Aldrich, 4906845001). Immunoprecipitation was carried out using GFP-Trap A beads (Chromotek, gta-10) according to the manufacturer’s instructions.
Western blotting
Cells were lysed in protein lysis buffer (10 mM Tris-HCl ph 7.5, 150 mM NaCl, 0.5 mM EDTA and 0.5% NP-40). Western blotting was carried out using the NuPAGE electrophoresis system (Life Technologies, NP0335BOX) according to standard procedures. Primary antibodies used were Anti-LATS2 at 1 µg/ml (NEB), Anti-phospho-YAP at 1 µg/ml (NEB, 13008S), anti-YAP at 1 µg/ml (NEB, 4912S) and anti-β-actin at 0.56 µg/ml (abcam, ab2676). Secondary anti-mouse IgG (NEB, 7076S) and anti-rabbit IgG (NEB, 7074S) were used at 0.067 µg/ml
Immunofluoresence and microscopy
Cells were grown on Ibidi µ-slide chamber slides until the right level of confluence was obtained. Cells were then fixed with 4% paraformaldehyde, permeabilised with Triton-X-100, then stained according to standard procedures. Anti-YAP was used at 10 µg/ml (NEB, 4912S). Anti-JCAD was used at 4 µg/ml (Sigma-Aldrich, HPA017956). Images were obtained using an inverted Leica TCS SP5 confocal microscope with an x63 oil immersion objective at the Advanced Imaging Facility of the University of Leicester.
Gene expression analysis
RNA was extracted from cells using an RNeasy miniprep kit (Sigma-Aldrich, 74104), reverse transcribed with Superscript III (Fisher Scientific, 18080093) and qPCR carried out using Sensimix (Bioline, QT650-05) on a Rotorgene Q qPCR machine (Qiagen, 9001550).
Monocyte adhesion assay
THP-1 monocytes were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were pelleted, resuspended to a concentration of 1 x 105 cells/ml in full medium. Cells were labelled with 5mM Calcein-AM (Fisher Scientific, C3099) for 30 minutes. The HUVEC monolayers were treated with TNFα (R&D Systems, 210-TA-005) for 30 minutes as appropriate, washed with RPMI-1640, then 1x104 THP-1 cells added and incubated for 30 minutes. The THP-1s were then removed and the THP-1-bound monolayer washed 4 times with RPMI-1640. The final wash was removed, then the cells fixed with 4% paraformaldehyde. Images were taken with an EVOS Fluorescent Microscope (Fisher Scientific) and the number of bound cells counted manually in ImageJ.
ICAM-1 Flow cytometry
Cells were stained in staining buffer (PBS, pH 7.2, 0.5% bovine serum albumin, 2mM EDTA) with 1:10 diluted anti-CD54-APC (Miltenyi Biotec, 130-103-840) for 20 minutes in the dark at 4°C. Cells were then analysed using Cyan ADP Flow Cytometer (Beckman Coulter) using standard methods.
Network analysis
The RNA-seq expression profiles in 44 tissues (n≥50; subcutaneous adipose, artery, etc.) were retrieved from Genotype-Tissue Expression (GTEx) database26. To construct the tissue-specific co-expression networks, we used Multiscale Embedded Gene Co-expression Network Analysis (MEGENA, v1.3.6)27 with default parameters. Benjamini-Hochberg False Discovery Rate (FDR)28 threshold <0.05 was used to define the significant coexpression modules. For STARNET, normalized gene expression data were analyzed using block-wise weighted gene correlation network analysis (WGCNA) with two distinct beta values for within- and between-tissue correlations: 5.2 and 2.7. To infer Bayesian networks within each co-expression module, we used a Fast Greedy Equivalence Search algorithm29 implemented in the rcausal R package developed by the Center for Causal Discovery. Key driver analysis on the inferred Bayesian networks was performed using the mergeomics R package.
Statistical Analysis
Statistical analyses were carried out using R software version 3.430 and GraphPad Prism (GraphPad software).Flow cytometry data was analysed using a custom script utilizing the flowCore and flowStats Bioconductor packages for R31, 32. The number of biological replicates is shown in all figures and each experiment was carried out in technical triplicate or quadruplicate. Student’s t test were used to test for differences of the mean between specific siRNAs and the control. Groups were tested for normality using the Shapiro-Wilk test and for equal variance using the F-test to ensure validity of the t-test. . Proliferation data was analysed using a linear mixed model in R, incorporating sample group and time as fixed effects and sample replicates as random effects. Specific group mean differences were analysed at 24, 48 and 72 hours and adjusted by false discovery rate to account for multiple comparisons. All barcharts represent mean +/- standard deviation. Significance levels are as follows: n.s. p > 0.05, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
Results
JCAD knockdown alters EC function
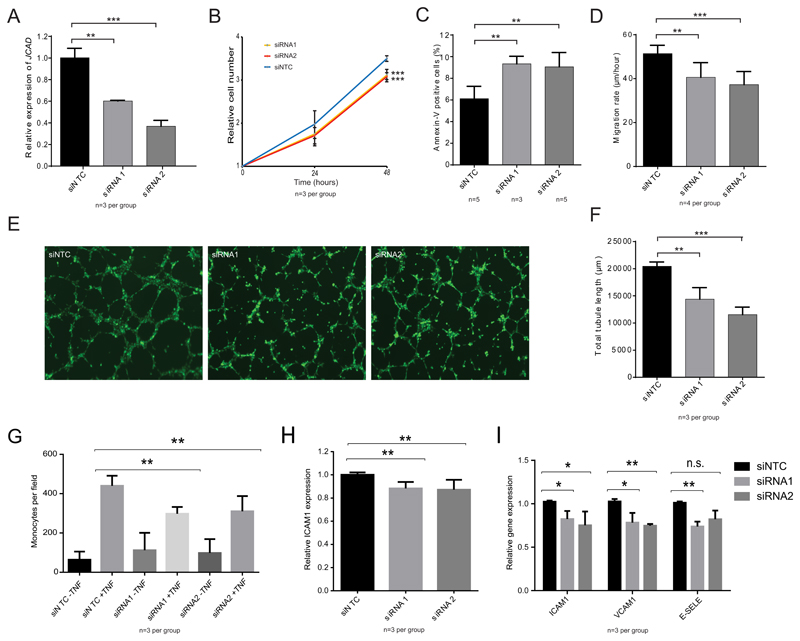
To explore the functional role of JCAD in ECs we first investigated the cellular consequences of siRNA knockdown of JCAD in human umbilical vein endothelial cells (HUVECs) (Fig. 1). We achieved knockdown of 40 to 60% in JCAD mRNA levels using 2 independent siRNAs in comparison to non-targeting control (NTC) siRNA (Fig 1A). We measured proliferation of JCAD knockdown cells compared to control and found a reduction in cell number in JCAD knockdown cells of approximately 15% after 48 hours (siRNA1 p<0.001, siRNA2 p<0.001)(Fig 1B). We found a similar decrease in proliferation in human aortic endothelial cells (HAECs) following JCAD knockdown (p=0.048) (Supplemental Figure II) suggesting a common role for JCAD throughout the vasculature. Next, we measured the proportion of cells which were apoptotic using flow cytometry with an anti-Annexin-V antibody. There was an increase in the proportion of apoptotic cells of ~50% with both siRNA1 (p=0.005) and siRNA2 (p=0.006) (Fig 1C). To test the impact of JCAD knockdown on cell migration, we performed an in vitro wound-healing assay and measured the rate at which the cells migrated to close the wound. The rate of migration was reduced in JCAD knock-down cells, with siRNA1 treated cells migrating ~20% slower in both siRNA1 (p=0.0001) and siRNA2 treated cells (p=0.0001) compared to NTC siRNA treated cells (Fig 1D). We tested the angiogenic capability of HUVECs following JCAD knockdown using a tube formation assay, which relies on the ability of endothelial cells to self-organise into a network of simple tubes when plated onto Matrigel basement membrane matrix. We measured the total length of tubes formed and found a reduction in the tubule length with JCAD knock-down of more than 25% compared to NTC siRNA (p=0.001 for siRNA and p=0.0007 for siRNA2) (Fig 1E and 1F). These phenotypes demonstrate a role for JCAD in regulating EC function – with reduced JCAD expression causing decreased proliferation, migration and angiogenic capability but increased apoptosis.
Figure 1.
Phenotypic assessment of JCAD knockdown in endothelial cells. (A) qPCR of JCAD expression in siRNA treated cells. (B) Cellular proliferation of HUVECs treated with the 3 different siRNAs. N=3 independent knock-downs across 2 independent cell lines. (C) Flow cytometry with an anti-Annexin V antibody was used to determine the proportion of siRNA-treated cells which were apoptotic. N represent independent knock-downs across 2 cell lines. (D) A scratch assay was used to measure the migration rate of siRNA-treated cells. (E and F) For a tube-formation assay, siRNA-treated cells were plated onto Matrigel and incubated for 6 hours. Calcein-AM staining was used for visualisation (D) and total tubule length determined (E). (F) Thp-1 binding to siRNA treated endothelial cells. (G) ICAM1 expression in response to TNFα was measure using flow cytometry. (H) qPCR analysis of expression of adhesion molecule expression. Significance levels: NS p > 0.05, * p < 0.05, ** p < 0.01, *** P < 0.001.
Increased YAP/TAZ activity i.e. less active Hippo signaling has recently been demonstrated to promote monocyte binding to the endothelium via regulation of adhesion molecule expression21, 22. To determine whether JCAD is involved in the regulation of monocyte adhesion, we used an in vitro assay to examine adhesion of monocytes to siRNA-treated ECs. The number of monocytes bound to the confluent layer of HUVECs was significantly reduced by approximately 30% with JCAD knock-down (p=0.0001 for siRNA1 and p=0.0007 for siRNA2) (Fig 1G). To investigate the mechanism of altered monocyte adhesion, we measured the level of Intercellular Adhesion Molecule 1 (ICAM1) protein in cells induced with TNFα using flow cytometry (Fig 1H). In cells treated with siRNA1, ICAM1 protein level was reduced by ~12% compared to control siRNA treated cells (p=0.001), in siRNA2 treated cells, ICAM1 expression was reduced by ~13% (p=0.0075). We also used qPCR to examine the expression of ICAM1 as well as VCAM1 (Vascular Cell Adhesion Molecule 1) and SELE (Selectin E), two other key adhesion genes expressed in HUVECs. ICAM1 mRNA level was reduced by ~18% in siRNA1 treated cells (p=0.022) and ~25% in siRNA2 treated cells (p=0.042). VCAM1 mRNA level was reduced by 25% in siRNA1 treated cells (p=0.022) and 24% in siRNA2 treated cells (p=0.0002). SELE mRNA level was reduced by 22% in siRNA1 treated cells (p=0.008), in siRNA2 treated cells, there was also a trend towards a reduction in expression, although this wasn’t statistically significant (p=0.092) (Fig 1I). These data show that knock-down of JCAD expression in ECs results in decreased adhesion molecule expression and monocyte adhesion.
JCAD promotes YAP activity in ECs
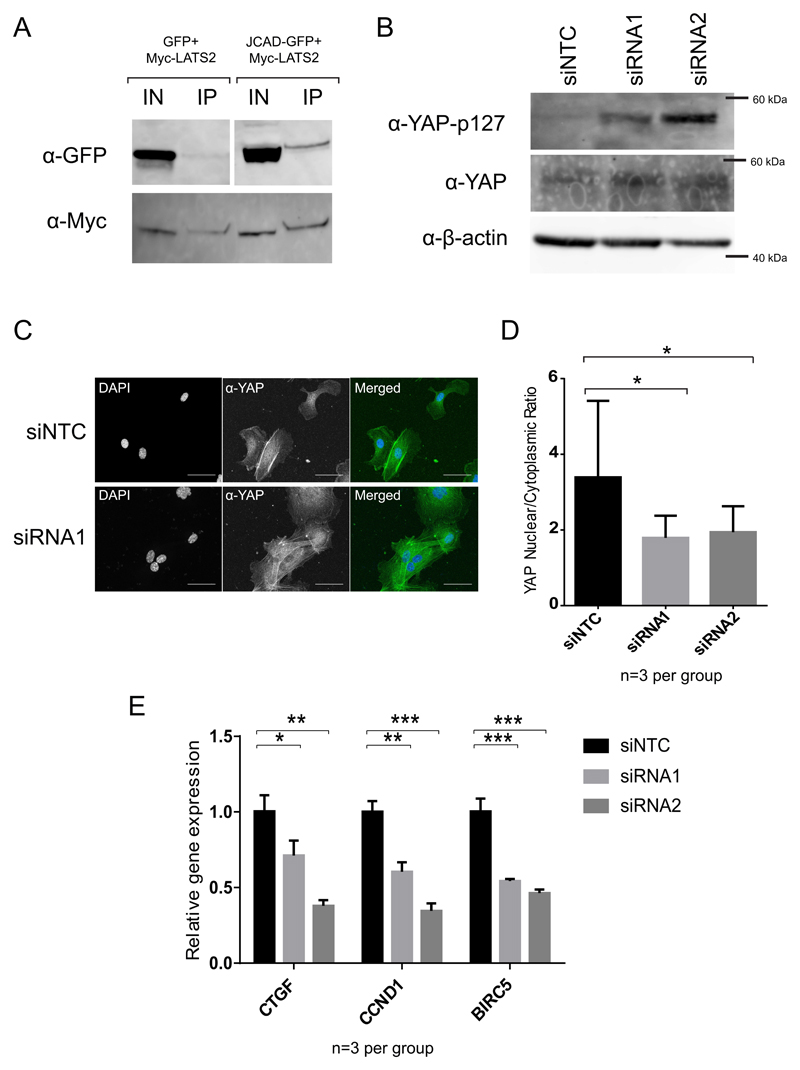
JCAD was recently found to interact with LATS2, a core kinase of the Hippo signaling pathway, in a high-throughput immunoprecipitation-mass spectrometry (IP-MS) screen13.The Hippo pathway has a key role in regulating various cell phenotypes, including proliferation, migration and apoptosis. Therefore, we sought to determine whether the regulation of EC functions by JCAD occurs via regulation of the Hippo signaling pathway. Initially, we sought to confirm the physical interaction of JCAD and LATS2 by immunoprecipitation. We overexpressed both JCAD-GFP and Myc-LATS2 in HEK293A cells, immunoprecipitated Myc-LATS2 and probed for GFP by western blot. GFP alone did not bind to the Myc-LATS2, but JCAD-GFP did (Fig 2A). To determine whether JCAD perturbs Hippo signaling, we used western blotting with a phospho-specific anti-YAP (p127) antibody, which detects inhibitory phosphorylation on a serine residue at amino acid position 127 of the Hippo pathway effector YAP. We found an increase in the level of YAP (p127) phosphorylation in JCAD knock-down cells compared to NTC control, but no change in total YAP levels (Fig 2B). When the Hippo pathway is active, phosphorylated YAP is excluded from the nucleus. We therefore examined the proportion of nuclear YAP in siRNA treated cells by immunofluorescence with an anti-YAP antibody, and found a reduction in the YAP nuclear/cytoplasmic ratio in JCAD siRNA treated cells (p=0.0001 for siRNA1 and p=0.0008 for siRNA2) (Fig 2C and 2D). The increase in phosphorylated YAP and reduction in nuclear YAP in JCAD knockdown cells demonstrate increased activity of the core Hippo pathway and would be expected to result in altered expression of Hippo regulated genes. We therefore examined the expression of 3 genes known to be activated by YAP/TAZ – CTGF (Connective Tissue Growth Factor)33, CCND1 (Cyclin D1)34 and BIRC5 (Baculoviral Inhibitor of apoptosis Repeat-Containing 5)35 by qPCR following siRNA knockdown of JCAD. All 3 genes examined showed reduced expression in JCAD knock-down cells compared to control cells (Fig 2E).
Figure 2.
JCAD knockdownreduces YAP activity. (A) Anti-GFP and Anti-Myc blots on cell lysate and immunoprecipitated Myc-LATS2. (B) Anti-phospho-YAP (p127) western blot on cell lysates from siRNA treated cells. (D) siRNA-treated cells were fixed and stained with Anti-YAP antibody and co-stained with DAPI. (E) Quantification of these images allowed determination of the Nuclear/Cytoplasmic ratio of YAP. (F) qPCR measurement of RNA expression levels of Hippo-pathway regulated genes. Significance levels: NS p > 0.05, * p < 0.05, ** p < 0.01, *** P < 0.001.
JCAD mediates Hippo pathway regulation via RhoA
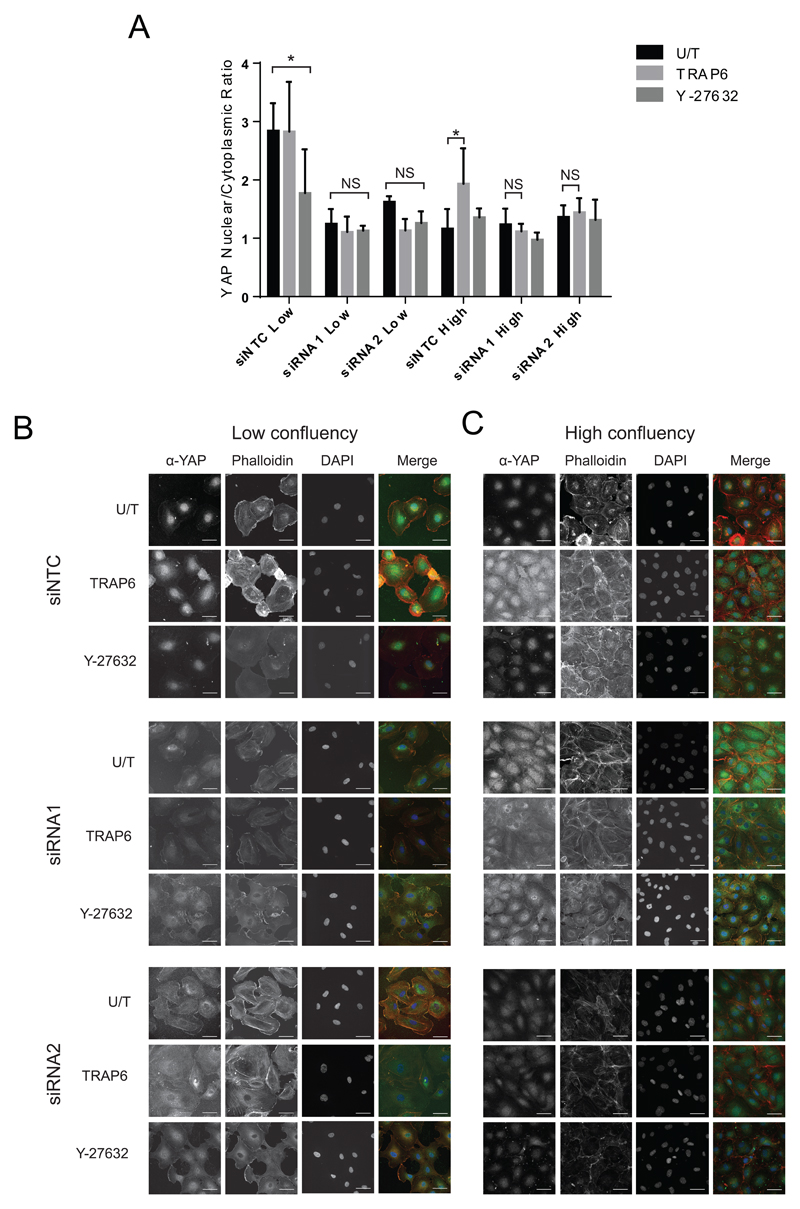
To further understand the role of JCAD in Hippo signaling, we studied the impact of JCAD knockdown on the influence of other regulators of the pathway. In cells that are at low confluency and are therefore actively growing, the Hippo pathway is inactive, and YAP is nuclear and active16. RhoA is an important regulator of Hippo signaling and responds to stimuli including cell confluency, mechanotransduction and signaling molecules such as Thrombin16, 36, 37. In low confluency cells, treatment with the ROCK (Rho-associated coiled-coil forming protein serine/threonine kinase) inhibitor Y-27632, which suppresses the effects of RhoA and promotes Hippo signaling37. In highly confluent cells, the Hippo pathway is active, YAP is excluded from the nucleus and unable to promote expression of regulated genes16. In these cells, TRAP6, a synthetic PAR1 (Protease-activated receptor1) agonist, which stimulates thrombin signaling via RhoA, down-regulates Hippo signaling and results in nuclear YAP and increased expression of regulated genes36. To explore the role of JCAD in regulating Hippo signaling we grew HUVECs to be at low confluency (40-50%) and high confluency (95-100%) 48 hours after siRNA knockdown and treated the cells with Y-27632, TRAP6, or mock treatment. We then measured the nuclear/cytoplasmic ratio of YAP by staining with α-YAP. At low confluency YAP localized to the nucleus and TRAP6 had no effect on YAP localization on NTC transfected cells (Fig 3 A and B). At high confluency, TRAP6 caused the nuclear/cytoplasmic ratio of YAP to increase by more than 60% (p=0.039) in NTC transfected cells (Fig 3 A and C). Y-27632 reduced the YAP nuclear/cytoplasmic ratio by approximately 40% (p=0.027) (Fig 3 A and B) in NTC cells at low confluency, but not in high confluency cells (Fig 3 A and C). Knockdown of JCAD resulted in exclusion of YAP from the nucleus and a loss of response to both drugs. This places JCAD in the same part of the pathway as RhoA in the Hippo signaling pathway.
Figure 3.
JCAD is required for the effect of Rho and Thrombin signaling on the Hippo pathway. (A) Quantification of YAP nuclear cytoplasmic ratio in siNTC, siRNA1 and siRNA2 treated HUVECs treated with TRAP6, Y-27632 or untreated. Representative images shown for low confluency (B) and high confluency (C) cells. Significance levels: NS p > 0.05, * p < 0.05, ** p < 0.01, *** P < 0.001.
The effects of JCAD knockdown on EC function require LATS2
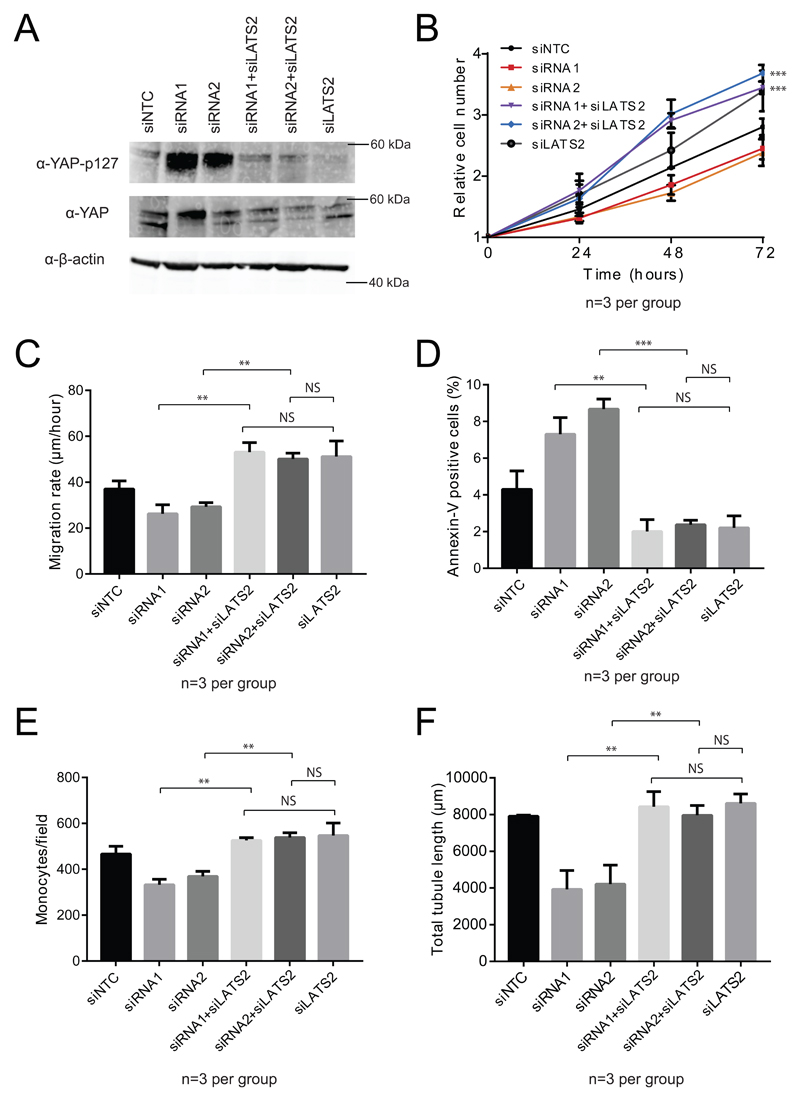
Having demonstrated that JCAD regulates Hippo signaling and that it interacts with LATS2 kinase, we sought to demonstrate that the effects of JCAD knockdown require functional LATS2 using double siRNA knockdown. Initially, we examined phospho-YAP and YAP levels in siRNA treated endothelial cells and found that the increase in YAP phosphorylation caused by JCAD siRNA was abolished by knockdown of LATS2 expression (Fig. 4A). We then used double siRNA treatments and investigated the same EC phenotypes tested previously. We examined proliferation of endothelial cells and found that where proliferation was reduced with either JCAD siRNA alone, it increased with both LATS2 knockdown and double JCAD LATS2 knockdown (for siRNA1 vs siRNA1 and siLATS2 and for siRNA2 vs siRNA2 and siLATS2, p<0.001 at 48 and 72 hour timepoints). There was no significant difference between the double siRNAs and LATS2 alone (p=0.903 for siRNA1; p=0.241 for siRNA2 at 72 hrs) (Fig. 4B). Migration rate was determined with an in vitro wound healing assay, migration rate was increased in both LATS2 and double JCAD LATS2 siRNA samples (p=0.001 for siRNA1; p=0.001 for siRNA2). There was no significant difference between the double siRNAs and LATS2 alone (p=0.700 for siRNA1; p=0.804 for siRNA2) (Fig. 4C). Apoptosis was measured by Annexin-V flow cytometry, and double siRNA treated samples had reduced Apoptosis compared to single JCAD siRNA alone (p=0.001 for siRNA1; p=0.001 for siRNA2). There was no significant difference between double siRNA and LATS2 alone (p=0.728 for siRNA1; p=0.702 for siRNA2) (Fig. 4D). Monocyte adhesion was measured and was found to be increased in LATS2 and JCAD LATS2 double siRNA treated samples compared to JCAD siRNA alone (p=0.002 for siRNA1; p=0.003 for siRNA2). There was no significant difference between double siRNA and LATS2 alone (p=0.299 for siRNA1; p=0.414 for siRNA2) (Fig 4E). Endothelial tube formation on matrigel was determined and there was increased tube formation in double siRNA treated samples compared to JCAD siRNA alone (p=0.004 for siRNA1; p=0.019 for siRNA2). Again, there was no significant difference between double siRNA and LATS2 siRNA alone (p=0.797 for siRNA1; p=0.337 for siRNA2) (Fig 4F). These data show that the effects of JCAD on EC phenotype require the action of LATS2.
Figure 4.
Phenotype assessment of JCAD LATS2 double siRNA knockdown in endothelial cells. (A) Western blots using of anti-phospho-YAP(p127) and anti-YAP on lysates with JCAD and LATS2 siRNAs. (B) Proliferation, showing statistical comparison between siRNA1 and siRNA1+siLATS2 as well as siRNA2 and siRNA2+siLATS2, (C) Migration, (D) Apoptosis, (E) Monocyte adhesion and (F) tube formation of cells treated with JCAD and LATS2 siRNAs separately and in combination. Significance levels: NS p > 0.05, * p < 0.05, ** p < 0.01, *** P < 0.001.
CAD-associated variant in JCAD is associated with JCAD expression in vascular and atherosclerotic tissues in humans
Next, we investigated the transcriptional expression of JCAD in 672 CAD patients across 7 tissues (blood, atherosclerotic-lesion free mammary artery (MAM), atherosclerotic aortic root (AOR), subcutaneous fat (SF), visceral abdominal fat (VAF), skeletal muscle (SKLM) and liver (LIV)), measured by RNA-seq in the STARNET study (Stockholm-Tartu Atherosclerosis Reverse Networks Engineering Task)38 and all tissues in the GTEx (Genotype-tissue Expression) project dataset39. In STARNET the lead CAD-associated SNP rs2487928 is a highly significant eQTL (expression quantitative trait locus) for JCAD in the arterial wall both in atherosclerotic aortic root (AOR) (p=8.26e-18) and in atherosclerosis free internal mammary artery (MAM) (p=1.26e-34), with the protective allele associated with reduced expression of JCAD. In GTEx, rs2487928 is also a significant eQTL for JCAD in aorta (p=1.8e-7). We tested the effects of rs2487928 on endothelial tube formation in a library of HUVECs and observed a trend towards decreased tubule length in cells homozygous for the protective allele compared to risk allele homozygotes, with an intermediate effect in heterozygotes, although this was not statistically significant (Supplemental Figure IV). This is in keeping with the expected effects of reduced JCAD expression in the protective genotype. These findings suggest that the protective allele reduces CAD risk through decreased JCAD expression in the vessel wall.
Co-expression across human tissues corroborates effects of JCAD on Hippo signaling and endothelial phenotypes
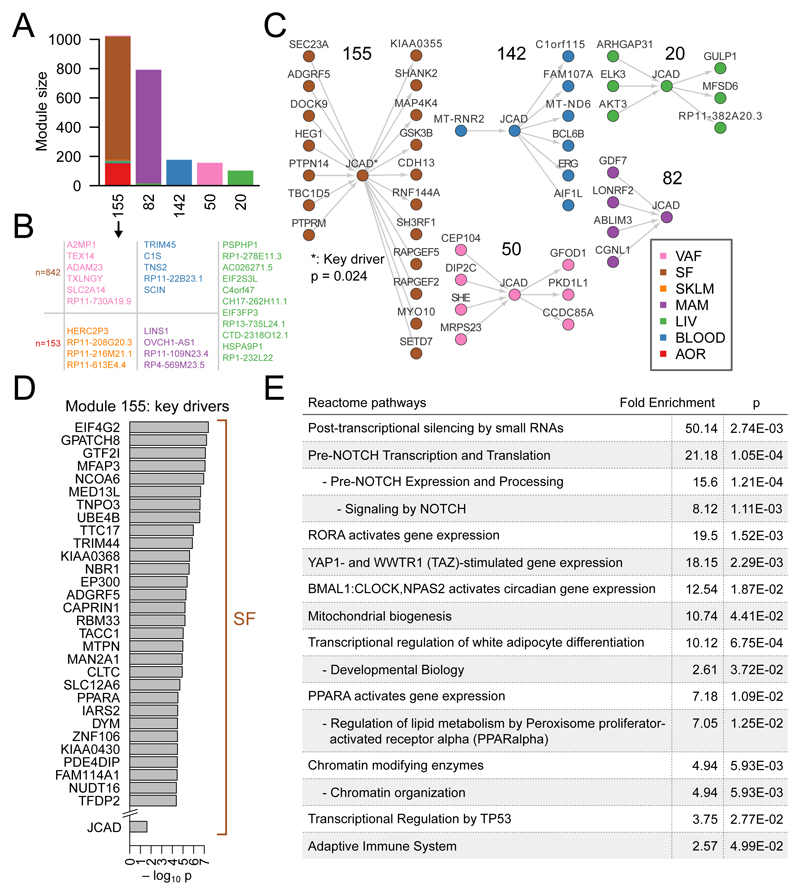
To characterize changes in the expression of other genes associated with expression of JCAD, we identified co-expression modules across all STARNET and GTEx tissues using weighted gene correlations analysis (WGCNA)38 and Multiscale Embedded Gene Co-expression Network Analysis (MEGENA)40 separately as previously described. JCAD was detected in several tissue-specific and cross-tissue modules. We inferred Bayesian networks over the transcripts within modules containing JCAD and ran weighted key-driver analysis to detect particularly influential genes. We identified JCAD as a key driver of module 155, which is a cross-tissue module composed mostly of genes from SF and AOR (Fig 5A-D). We then carried out Reactome pathway enrichment analysis on the key drivers to identify significantly over-represented pathways. Notably, “YAP1- and WWTR1 (TAZ)-stimulated gene expression”, was identified as an enriched pathway, further supporting the functional link between JCAD and Hippo signaling. Other enriched pathways included examples related to Notch signaling and transcriptional regulation by TP53, both of which are downstream of Hippo signaling (Fig 5E)41, 42. In addition, JCAD was present in coronary artery module 14 in GTEx and the predominantly MAM-specific module 82 in STARNET (Supplementary Tables I and III). Both of these modules were significantly enriched for GO categories related to the phenotypes detected in our EC experiments including angiogenesis, cell proliferation, apoptosis and adhesion (Supplementary Tables II and IV).
Figure 5.
JCAD co-expression analysis. (A) Module size and tissue composition of 5 STARNET modules with fewer than 2000 genes which contain JCAD. (B) The number of genes in the SF and AOR components and the gene names in the minority tissues in module 155. (C) Inferred bayesian networks showing directed edges involving JCAD for each module. (D) The 30 most significant key drivers of module 155 and JCAD. (E) Reactome pathway enrichment for all key drivers of module 155.
Discussion
The mechanisms by which genes at CAD associated loci identified by genome-wide association studies contribute to disease risk are largely unknown. In this study, we investigated JCAD, a largely uncharacterized CAD-associated gene and show that it encodes a LATS2 interacting protein which acts as a new negative regulator of the Hippo signaling pathway in ECs. Notably, recent studies have provided convincing evidence that the activity of YAP/TAZ, the transcriptional effectors of the Hippo pathway, is a key mediator of atherosclerosis at sites of non-linear shear stress and in response to pro-inflammatory cytokines 21, 22. Our results complement these findings and link the Hippo signaling pathway to CAD in humans via the 10p11 disease association and human gene expression analysis and JCAD function.
We have shown that knock-down of JCAD expression induces activation of the Hippo pathway, resulting in an increase in phosphorylated YAP and exclusion of YAP from the nucleus and a decrease in expression of Hippo pathway-regulated genes. This results in the perturbation of a number of important EC phenotypes consistent with known roles of Hippo signaling16, 21, 22, including increased apoptosis and decreased proliferation, migration and endothelial tube formation. This is in keeping with the known role of the Hippo pathway in these processes19, 20 and is consistent with a recent study by Hara et al., which established a role for JCAD in angiogenesis12.
The Hippo pathway responds to a number of upstream signals, including mechanical stress, confluency, and G-protein signals via RhoA16, 36, 43, 44. Notably, YAP/TAZ are required for shear stress response and promote atherogenesis in response to disturbed flow, a process which is dependent on RhoA-mediated signals21. We found that JCAD is involved in the transmission of signals to the Hippo signaling pathway via RhoA. We have also shown that JCAD knockdown reduces YAP nuclear localization in low confluency cells, but not in highly confluent cells, suggesting that JCAD may also be involved in the confluency response of YAP/TAZ, which also acts via RhoA44. In addition, we have demonstrated that the phenotypic effects of JCAD knockdown in ECs require LATS2, suggesting that these phenotypes are caused by altered regulation of Hippo signaling. Our results are supported by recent work by Ye et al who presented evidence that JCAD inhibits LATS2 in liver tumorigenesis by binding to the kinase domain of LATS215. Together, these data identify JCAD as a new regulator of Hippo signaling in both human cardiovascular disease and tumorigenesis.
JCAD is conserved across vertebrates, but lacks known functional domains and has poor homology to other proteins, although there is a short (13 amino acid) region with homology to ROCK 1 and 2 and cingulin proteins11. Further studies are needed to determine whether JCAD inhibits LATS2 in response to other signals which regulate the Hippo pathway or whether it is specific to RhoA-mediated signals. Intriguingly, in a recent GWAS10, variants in the RhoA gene have been associated with CAD risk, providing further evidence that the RhoA-Hippo pathway is involved in the development of CAD.
Co-expression analysis of RNA-seq data from both normal and atherosclerotic human blood vessels support our in vitro findings that JCAD regulates EC behavior with modules containing JCAD being enriched for GO categories related to cell proliferation, apoptosis, adhesion and angiogenesis. We also found JCAD to be a key driver of a cross-tissue module predominantly comprising genes from aorta and subcutaneous fat. Although JCAD was a weaker key driver relative to other genes in this co-expression module the identification of YAP/TAZ signaling as an over-represented pathway suggests the module captures relevant biology. It is not clear why JCAD and many of the other key drivers are from subcutaneous fat, however, this could be due to the extensive capillary network and high proportion of endothelial cells in adipose tissue45. Investigation of other key drivers of this module could potentially identify further novel regulators of Hippo signaling and YAP/TAZ activity.
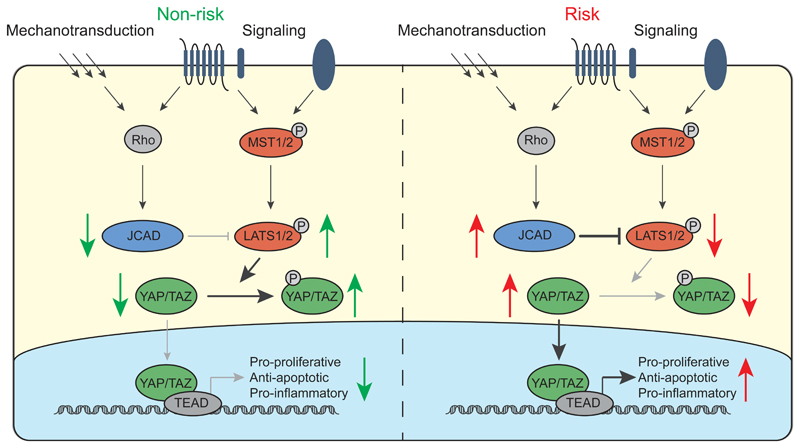
How CAD-associated variants in JCAD affect its function and contribute to disease will require further analysis. We identified an eQTL between the protective allele of the lead SNP, rs2487928, and decreased JCAD expression in both atherosclerotic and atherosclerosis-free arterial tissue, suggesting altered transcriptional regulation of JCAD as the causal mechanism. Based on our data, we propose a model of the relationship between JCAD and the core Hippo pathway and the how the risk and non-risk alleles alter Hippo signaling (Fig 6). We would expect decreased JCAD expression caused by the rs2487928 protective allele to contribute to decreased endothelial dysfunction in response to pro-atherosclerotic signals and decreased monocyte recruitment, resulting in lower macrophage content in atherosclerotic plaques, which could reduce plaque growth and instability. Additionally, promotion of angiogenesis by JCAD could result in increased neovascularization in plaques, higher neovessel density and contribute to plaque growth and instability in this way 46. In vivo models, such as a JCAD knockout or overexpressing mouse, will be required to fully determine the role of JCAD in disease pathogenesis. In addition, while we have demonstrated a role for JCAD in ECs, but we cannot rule out a role in other cell types and further work should be carried out to investigate this role. In addition endothelial specific targeting of JCAD in mice would also improve our understanding of the role of JCAD and Hippo signaling in CAD and the contribution of Hippo signaling.
Figure 6.
A model for the effect of JCAD CAD risk genotype on the Hippo signaling pathway. The Hippo signaling pathway consists of a core kinase cascade comprising MST1/2 and LATS1/2 and is regulated by multiple extracellular signals. Our data suggests that JCAD acts downstream of RhoA to inhibit LATS2. We propose that in individuals with the non-risk genotype, lower JCAD expression results in more active LATS1/LATS2, increased phosphorylation of YAP/TAZ and its exclusion from the nucleus. Conversely, in individuals with the risk genotype, higher JCAD expression results in lower LATS1/2 activity, reduced YAP/TAZ phosphorylation and more active nuclear YAP, resulting in increased expression of pro-proliferative, anti-apoptotic and pro-inflammatory genes, promoting endothelial dysfunction and atherogenesis.
In conclusion, we have demonstrated that the CAD associated gene JCAD encodes a novel regulator of Hippo signaling in ECs and suggest that variants at the 10p11.23 CAD locus act through JCAD to regulate EC function via YAP/TAZ. Future work should assess the therapeutic potential of JCAD and other Hippo pathway components in treating CAD.
Supplementary Material
Highlights.
JCAD, the candidate gene at the 10p11 locus affects endothelial cell function via the Hippo pathway.
JCAD acts via LATS2 to regulate YAP activity.
The protective allele of the lead CAD associated SNP, rs2487928, associates with decreased JCAD expression in arteries.
Acknowledgements
Sources of funding
This work was supported by a Transatlantic Networks of Excellence Award (12CVD02) from The Leducq Foundation. The research leading to these results has received funding from the European Union Seventh Framework Programme FP7/2007-2013 under grant agreement number HEALTH-F2-2013-601456. H.W., K.M.C., N.J.S. and T.R.W. are funded by the British Heart Foundation. K.M.C., H.W. and N.J.S. are UK National Institute for Health Research (NIHR) Senior Investigators. Work was supported by BHF grant PG/15/35/31403 to K.M.C.. Y.Z. and X.Y. are partially supported by the American Heart Association. The DNA genotyping and RNA sequencing in STARNET of which J.L.M.B. is P.I. were in part performed by the SNP&SEQ technology platform at Science for Life, the National Genomics Infrastructure (NGI) in Uppsala and Stockholm supported by Swedish Research Council (VR-RF1), Knut and Alice Wallenberg Foundation and UPPMAX. STARNET has also been supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Non-standard abbreviations and acronyms
- CAD
coronary artery disease
- EC
endothelial cell
- HUVEC
human umbilical vein endothelial cells
- ICAM1
intercellular adhesion molecule 1
- JCAD
junctional protein associated with coronary artery disease
- LATS2
large-antigen tumor suppressor
- NTC
non-targeting control
- TAZ
transcriptional coactivator with PDZ-binding motif
- YAP
yes-associated protein
Footnotes
Author Contributions: P.D.J., G.D., T.K., H.W., K.M.C., S.Y., N.J.S. and T.R.W conceived and designed the experiments. P.D.J., M.A.K., M.G.N., G.D., T.K., S.A. and R.R. carried out the experiments. S.K., Y.Z., X.Y. and J.L.M.B. contributed network data and carried out network and expression analyses. P.D.J., N.J.S. and T.R.W. wrote the manuscript. All authors contributed and commented on the manuscript. We also thank Dr. Mintu Nath for statistical review of the manuscript.
Disclosures
J.L.M.B. is the founder and chairman of Clinical Gene Networks.
References
- 1.GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specifi c mortality for 249 causes of death, 1980–2015: A systematic analysis for the global burden of disease study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 3.Tabas I, Garcia-Cardena G, Owens GK. Recent insights into the cellular biology of atherosclerosis. The Journal of cell biology. 2015;209:13–22. doi: 10.1083/jcb.201412052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McPherson R, Tybjaerg-Hansen A. Genetics of coronary artery disease. Circulation research. 2016;118:564–578. doi: 10.1161/CIRCRESAHA.115.306566. [DOI] [PubMed] [Google Scholar]
- 5.CARDIoGRAMplusC4D Consortium. Deloukas P, Kanoni S, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature genetics. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nature genetics. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myocardial Infarction Genetics Consortium. Kathiresan S, Voight BF, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nature genetics. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. The New England journal of medicine. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature genetics. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson CP, Goel A, Butterworth A, et al. Support for 243 loci associated with coronary artery disease with data from uk biobank. Under review. 2017 [Google Scholar]
- 11.Akashi M, Higashi T, Masuda S, Komori T, Furuse M. A coronary artery disease-associated gene product, jcad/kiaa1462, is a novel component of endothelial cell-cell junctions. Biochemical and biophysical research communications. 2011;413:224–229. doi: 10.1016/j.bbrc.2011.08.073. [DOI] [PubMed] [Google Scholar]
- 12.Hara T, Monguchi T, Iwamoto N, et al. Targeted disruption of jcad (junctional protein associated with coronary artery disease)/kiaa1462, a coronary artery disease-associated gene product, inhibits angiogenic processes in vitro and in vivo. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:1667–1673. doi: 10.1161/ATVBAHA.117.309721. [DOI] [PubMed] [Google Scholar]
- 13.Couzens AL, Knight JDR, Kean MJ, Teo G, Weiss A, Dunham WH, Lin Z-Y, Bagshaw RD, Sicheri F, Pawson T, Wrana JL, et al. Protein interaction network of the mammalian hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signaling. 2013;6:rs15. doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- 14.Wang W, Li X, Huang J, Feng L, Dolinta KG, Chen J. Defining the protein-protein interaction network of the human hippo pathway. Molecular & cellular proteomics : MCP. 2014;13:119–131. doi: 10.1074/mcp.M113.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye J, Li TS, Xu G, Zhao YM, Zhang NP, Fan J, Wu J. Jcad promotes progression of nonalcoholic steatohepatitis to liver cancer by inhibiting lats2 kinase activity. Cancer research. 2017;77:5287–5300. doi: 10.1158/0008-5472.CAN-17-0229. [DOI] [PubMed] [Google Scholar]
- 16.Yu FX, Guan KL. The hippo pathway: Regulators and regulations. Genes & development. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng Z, Moroishi T, Guan KL. Mechanisms of hippo pathway regulation. Genes & development. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen CG, Moroishi T, Guan KL. Yap and taz: A nexus for hippo signaling and beyond. Trends in cell biology. 2015;25:499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Freire Valls A, Schermann G, et al. Yap/taz orchestrate vegf signaling during developmental angiogenesis. Developmental cell. 2017;42:462–478 e467. doi: 10.1016/j.devcel.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Choi HJ, Zhang H, Park H, Choi KS, Lee HW, Agrawal V, Kim YM, Kwon YG. Yes-associated protein regulates endothelial cell contact-mediated expression of angiopoietin-2. Nature communications. 2015;6:6943. doi: 10.1038/ncomms7943. [DOI] [PubMed] [Google Scholar]
- 21.Wang KC, Yeh YT, Nguyen P, Limqueco E, Lopez J, Thorossian S, Guan KL, Li YJ, Chien S. Flow-dependent yap/taz activities regulate endothelial phenotypes and atherosclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:11525–11530. doi: 10.1073/pnas.1613121113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Luo JY, Li B, et al. Integrin-yap/taz-jnk cascade mediates atheroprotective effect of unidirectional shear flow. Nature. 2016 doi: 10.1038/nature20602. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Wu Y, Wang H, et al. Interplay of mevalonate and hippo pathways regulates rhamm transcription via yap to modulate breast cancer cell motility. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E89–E98. doi: 10.1073/pnas.1319190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, Manfrin A, Ingallina E, Sommaggio R, Piazza S, Rosato A, et al. Metabolic control of yap and taz by the mevalonate pathway. Nature cell biology. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 25.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: An open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonsdale J, Thomas J, Salvatore M, et al. The genotype-tissue expression (gtex) project. Nature genetics. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song WM, Zhang B. Multiscale embedded gene co-expression network analysis. Plos Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 29.Chickering DM. Optimal structure identification with greedy search. Journal of Machine Learning Research. 2002;3:507–554. [Google Scholar]
- 30.R Core Team. R: A language and environment for statistical computing. R foundation for statistical computing; vienna, austria: 2014. [Google Scholar]
- 31.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahne F, LeMeur N, Brinkman RR, Ellis B, Haaland P, Sarkar D, Spidlen J, Strain E, Gentleman R. Flowcore: A bioconductor package for high throughput flow cytometry. BMC bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. Tead mediates yap-dependent gene induction and growth control. Genes & development. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao X, Pfaff SL, Gage FH. Yap regulates neural progenitor cell number via the tea domain transcription factor. Genes & development. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the hippo-yap pathway by protease-activated receptors (pars) Genes & development. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 38.Franzen O, Ermel R, Cohain A, et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353:827–830. doi: 10.1126/science.aad6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GTEx Consortium. The genotype-tissue expression (gtex) project. Nature genetics. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song WM, Zhang B. Multiscale embedded gene co-expression network analysis. PLoS Comput Biol. 2015;11:e1004574. doi: 10.1371/journal.pcbi.1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tschaharganeh DF, Chen X, Latzko P, et al. Yes-associated protein up-regulates jagged-1 and activates the notch pathway in human hepatocellular carcinoma. Gastroenterology. 2013;144:1530–1542 e1512. doi: 10.1053/j.gastro.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furth N, Ben-Moshe NB, Pozniak Y, Porat Z, Geiger T, Domany E, Aylon Y, Oren M. Down-regulation of lats kinases alters p53 to promote cell migration. Genes & development. 2015;29:2325–2330. doi: 10.1101/gad.268185.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M. Yap/taz as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS letters. 2014;588:2663–2670. doi: 10.1016/j.febslet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, et al. Role of yap/taz in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 45.Eto H, Suga H, Matsumoto D, Inoue K, Aoi N, Kato H, Araki J, Yoshimura K. Characterization of structure and cellular components of aspirated and excised adipose tissue. Plastic and reconstructive surgery. 2009;124:1087–1097. doi: 10.1097/PRS.0b013e3181b5a3f1. [DOI] [PubMed] [Google Scholar]
- 46.Moulton KS. Plaque angiogenesis: Its functions and regulation. Cold Spring Harb Symp Quant Biol. 2002;67:471–482. doi: 10.1101/sqb.2002.67.471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.