Abstract
Purpose
The most significant prognostic factor in early breast cancer is lymph node involvement. This stage between localised and systemic disease is key to understanding breast cancer progression, however our knowledge of the evolution of lymph node malignant invasion remains limited, as most currently available data derive from primary tumours.
Experimental design
In 11 treatment-naïve node positive early breast cancer patients without clinical evidence of distant metastasis, we investigated lymph node evolution using spatial multi-region sequencing (n=78 samples) of primary and lymph node deposits and genomic profiling of matched longitudinal circulating tumour DNA (ctDNA).
Results
Linear evolution from primary to lymph node was rare (1/11) whereas the majority of cases displayed either early divergence between primary and nodes (4/11), or no detectable divergence (6/11) where both primary and nodal cells belonged to a single recent expansion of a metastatic clone. Divergence of metastatic subclones was driven in part by APOBEC. Longitudinal ctDNA samples from 2 of 7 subjects with evaluable plasma taken peri-operatively reflected the two major evolutionary patterns and demonstrate that private mutations can be detected even from early metastatic nodal deposits. Moreover, node removal resulted in disappearance of private lymph node mutations in ctDNA.
Conclusions
This study sheds new light on a crucial evolutionary step in the natural history of breast cancer, demonstrating early establishment of axillary lymph node metastasis in a substantial proportion of patients.
Introduction
Breast cancer is characterised by high genomic and transcriptomic diversity, both between (1–3) and within patients (4–8). This inherent complexity is fully consistent with a clonal evolution model of cancer (9,10). The cancer evolution paradigm provides a biologically plausible explanation of experimental observations and may also lead to more accurate predictions of the future course of the disease, in particular prognostication and the emergence of treatment resistance (9).
Currently, clinico-pathological parameters such as age, tumour grade and stage, ER and HER2 expression have been integrated into scoring systems to estimate the probability of recurrence and death from breast cancer (11,12). Moreover, gene expression profiles provide additional prognostic and/or predictive information regarding adjuvant chemotherapy in ER-positive early breast cancer and their clinical utility is being prospectively evaluated in large randomised clinical trials (13). Large meta-analyses have indicated that in early breast cancer, the most important prognostic factor is lymph node involvement (14–16). This clinical stage represents a potentially intermediate evolutionary step between localised disease and metastatic dissemination, and it is therefore of crucial importance to understanding progression. Micro-metastases can also be present at diagnosis of some early breast cancers, and ultra-sensitive methods to analyse circulating tumour-derived DNA (ctDNA) have recently helped interrogate such deposits that can subsequently result in overt metastatic recurrence (17). Hence, combined genomic analyses of primary, lymph nodes and ctDNA are necessary to understand metastatic progression in cancer, also in light of findings in other cancer types where metastatic dissemination was found to be decoupled from lymphatic spread in a subset of cases (18,19).
Here we sought to study lymph node spread from an evolutionary perspective by analysing 78 multi-region samples taken from untreated primary tumours and lymph nodes, as well as 7 longitudinal ctDNA samples of a selected cohort of 11 primary breast cancers that had biopsy-proven ipsilateral axillary lymph node spread without clinical evidence of distant metastatic disease (see Fig. 1 and Table S1 for clinical details). We used whole-exome, whole-genome, and targeted deep sequencing data, combined with phylogenomics analysis, to understand the dynamics of lymph node spread.
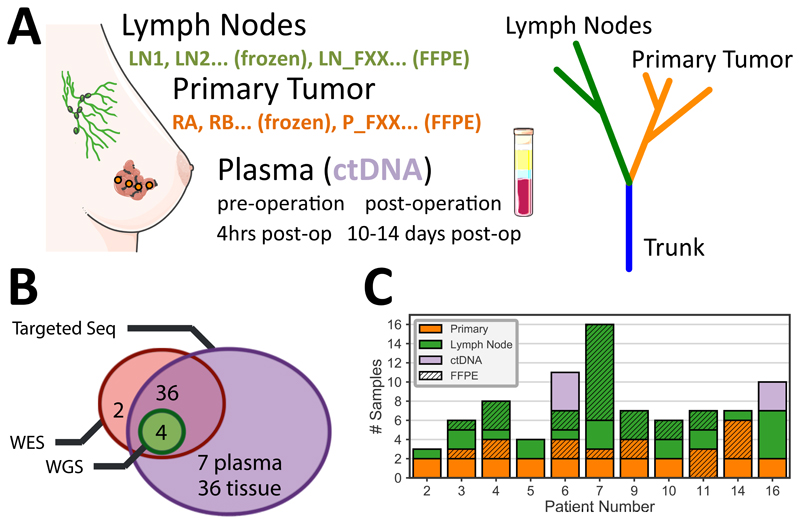
Figure 1. Spatio-temporal genomic profiling of lymph node evolution in breast cancer.
(A) Multi-region sampling and genomic profiling of primary (regions RA, RB, …, 1-6cm apart) and lymph node (LN1, LN2, …) samples (total samples n=78) from a selected cohort of 11 early breast cancer patients with lymph node involvement without distant metastases. Longitudinal ctDNA samples were taken before and after surgery from 2 patients (n=7 samples). (B) Sequencing analysis was performed with whole-exome sequencing (n=42), whole-genome sequencing (n=4) and targeted deep sequencing of a cohort-specific panel (n=76) to identify mutational and copy number profiles. (C) The number of samples per location for each patient in the cohort (solid = fresh frozen, stripes = FFPE).
We found evident patterns of early divergence between primary and lymph node deposits in a subset of patients in our cohort and showed that these patterns were reflected in circulating tumour DNA. We also found that APOBEC activity contributed to such early divergence. Finally, we show proof of principle loss of ctDNA mutations private to nodes following surgical resection.
Material and Methods
Patient cohort and samples
Samples were collected from 11 breast cancer patients with positive axillary nodes. Patients had not received any treatment prior to surgery. The median age of patients in this cohort was 56 years (range 38-81). Several lymph nodes and primary tumour specimens were collected from each patient. Samples were either paraffin embedded after formalin fixation or snap frozen immediately after resection.
Whole peripheral blood was collected from each patient for germline DNA and plasma ctDNA taken at four time points: (A) intra-operatively before tumour resection, (B) intra-operatively immediately post tumour resection, (C) 4 hours postoperatively, (D) 10-14 days post-operatively, at the follow up visit. Blood at each time point (A-D) was collected into 3x10ml EDTA or STRECK tubes and was centrifuged at 1600xg for 20 minutes for a single spin. Plasma and buffy coat were collected and stored at -80C. Circulating DNA was extracted from 5ml plasma using the QIAamp circulating nucleic acid kit (Cat#55114) from Qiagen® according to manufacturer’s instructions. Briefly, plasma was lysed with proteinase K and ACL for 30 minutes at 60C with carrier RNA in AVE added. Buffer ACB was added and the sample was passed through a QIAamp Mini Spin column to bind the DNA. DNA was washed with ACW1, ACW2 and 100% Ethanol before centrifugation at 14000RPM for 3 minutes, drying for 10 minutes and elution into 50ul AVE buffer and stored at -20C. Extracted DNA was quantified by digital droplet PCR using a Taqman copy number reference assay for RNAse P (Life Technologies). ddPCR reactions were assembled using 1ul of eluate and 10ul of ddPCR SuperMix for Probes (BioRad) for a total reaction volume of 20ul. The reaction was partitioned into approximately 20,000 droplets on a BioRad QX200 droplet generator. PCR of the emulsified reaction was performed in 96 well plates on a G-Storm GS4 thermocycler for 40 PCR cycles with 60C annealing temperature. The plates were read on a BioRad QX200 droplet reader and the DNA concentration calculated using Quantasoft software (Version 1.4.0.99). At least 2 NTC wells were included in each quantification run.
Clinical and histopathological data from the patient cohort can be found in Table S1. The study protocol was approved by an Institutional Research Ethics Committee (reference number 13/LO/1015). All patients gave their written informed consent to participate before enrolling in the study. The study was carried out in accordance with the principles of the International Conference on Harmonization guidelines for Good Clinical Practice and the Declaration of Helsinki. See Supplementary Material and Methods for details on sample preparation.
Whole-exome, whole-genome and targeted sequencing
For each of the 11 patients in our cohort, 500ng of DNA from two primary breast tumour specimens and 1-5 involved lymph nodes was sent to The Broad Institute for whole exome sequencing (SureSelect Human All exon v2.0). The exome sequencing panel comprised 40 fresh frozen tissue samples (20 primary tumour and 20 lymph node), 2 FFPE specimens (both primary tumour tissues) and 11 germline samples (buffy coats). Exome sequencing data had a mean coverage of 154X. Whole-genome libraries were prepared from 30-100ng of genomic DNA with the NebNext Ultra II kit following the manufacturer’s instructions. Genomic DNAs were sheared in a Diagenode sonicator prior to library preparation. Whole-genome median coverage was 38. Further, a total of 807 exonic SNVs were selected for targeted validation. All but two samples used for whole exome sequencing were included in the targeted validation panel. In addition, we included DNA from 36 FFPE manually microdissected specimens and 7 ctDNA samples. A custom SureSelect XT2 panel (Agilent) was used to generate targeted capture libraries from these 83 samples (for 2 samples, there wasn’t enough DNA for targeted sequencing therefore information only from the exome sequencing was used) following manufacturer’s recommendations. Mean coverage for targeted sequencing was 1,813X with 98% validation. All libraries were sequenced on an Illumina HiSeq2500. See Supplementary Material and Methods for details regarding bioinformatics analysis.
Results
Intra-tumour heterogeneity in lymph node positive breast cancers
Using whole-exome sequencing (WES), we profiled 40 fresh-frozen (FF) and 2 formalin fixed paraffin embedded (FFPE) samples from the 11 patients, as well as matched normal (FF buffy coat), obtaining a mean depth of 154X. For each patient, we had at least two regions from the primary tumour, taken 1-6cm apart, and one lymph node (Fig. 1). Extensive intra-tumour heterogeneity (ITH) was evident our cohort, with an average of 73.5% of variants considered to be subclonal (Fig. S1 and S2). A total of 807 mutations were selected for custom targeted deep sequencing validation (mean depth of 1,813X, 98% validation rate). We also applied the same panel to 36 additional FFPE samples from the same patients (all those available with >50% tumour content – see Table S2), confirming that the original fresh frozen samples were representative of ITH both in the primary and the lymph node deposits and the observed patterns were not due to sampling bias (Fig. 2A). Estimated purity, ploidy and copy number profiles (Fig. 2B) were used to calculate cancer cell fractions for both whole-exome and targeted sequencing profiles, as presented in Figs. 2A and S2 (see Tables S3-S6 for values). For those samples where only targeted sequencing was available (i.e. additional FFPE samples), purity and ploidy estimates could not be calculated and we therefore reported presence/absence of the mutations (e.g. Fig. 2A – FFPE samples are set to cancer cell fraction =1/0). As macrodissected samples represented a small localised region of the tumour, we did not find any evident subclonal structure within each sample, although the limited mutational burden of breast cancer, combined with exome sequencing, precluded reliable subclonal analysis within such samples.
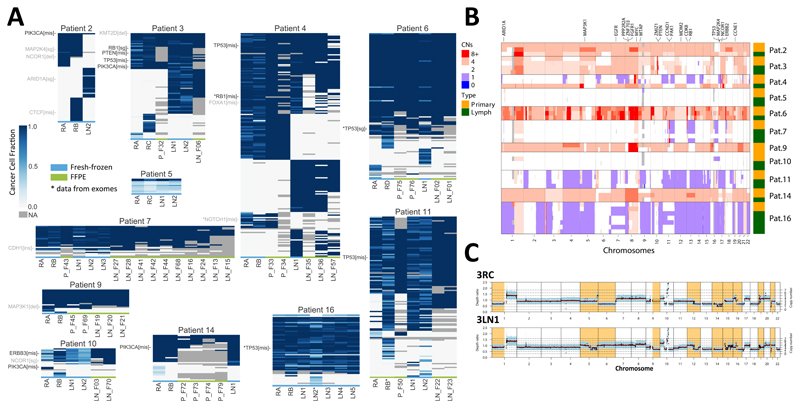
Figure 2. Distinct modes of lymph node evolution.
(A) Deep targeted sequencing of a cohort-specific panel derived from whole-exome sequencing. Heatmaps indicate cancer cell fraction (CCF) of a mutation or indel in different samples from the same patient (grey=NA, not enough coverage or variant does not overlap with a copy number segment). For FFPE samples (marked in green at the bottom) CCFs were not available and presence/absence is reported (CCF=1 or CCF=0). Tier 1 cancer genes, most likely drivers are annotated in black, Tier 2 possible drivers with uncertain pathogenicity are annotated in grey. For samples/variants marked with (*) we report exome data as targeted was not available. (B) Copy number aberrations in all samples showing differences in copy number status between primary and lymph nodes in the divergent subgroup. (C) Whole-genome sequencing for a tumour and lymph node sample of Pat. 3; divergent copy number regions shown in orange.
The mutational landscape of our cohort was consistent with previous studies (2,3,6), with TP53 and PIK3CA being the most commonly mutated drivers (Figure 2A: Tier 1 cancer genes, most likely drivers in black; Tier 2 cancer genes, possibly drivers but uncertain pathogenicity in grey). Copy number alterations (CNAs) were widespread, with patterns consistent with the profile of primary tumours(1,3), such as 1q and 8q gains and 8p loss (Fig. 2B and S3). Copy number profiles were confirmed by whole-genome sequencing (WGS performed only for Pat. 3 and 4, e.g. Fig. 2C). Mutations in tumour suppressor genes frequently co-occurred with loss of heterozygosity, consistent with the inactivation of the gene. Thus at the genomic level, our cohort was consistent with other cohorts of early breast cancers (1–3).
Distinct modes of lymph node evolution
The combination of point mutations, indels and copy number alterations clearly identified two major evolutionary patterns.
Sequential evolution where lymph node metastasis originated from a localised subclone in the primary was rare (1/11 – only Pat.11, Fig. 2A). This was confirmed using phylogenetic analysis. In this case, the tumour phylogenetic tree showed expansion of a metastatic cancer lineage which originated within region RB and spread to both LN1 and LN2. This is shown in the tree as RB, LN1 and LN2 having a recent common ancestor and forming a clade distinct from RA (Fig. 3A). Additional unique mutations within the clade are most likely passengers.
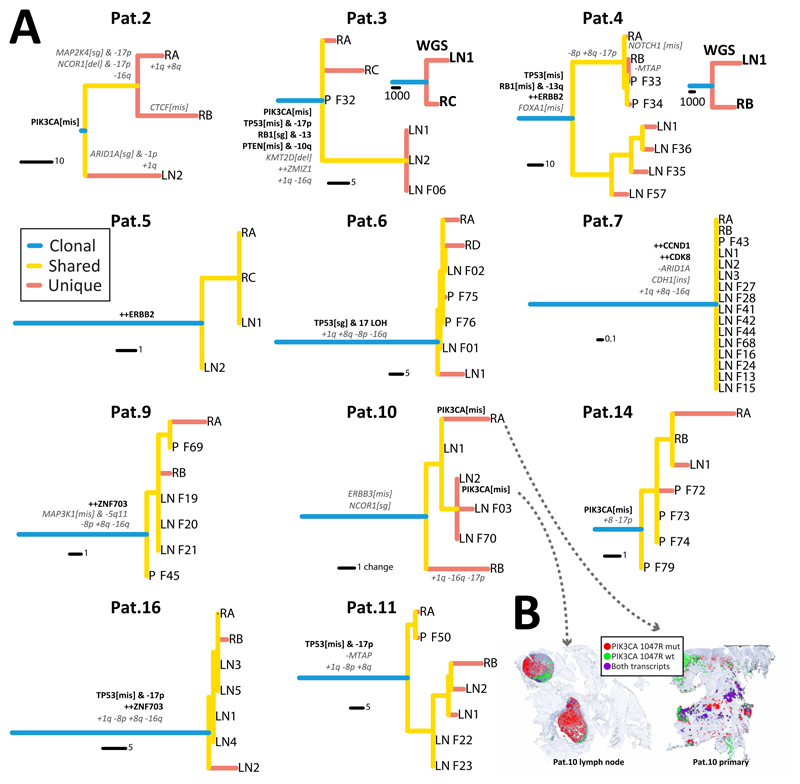
Figure 3. Evolutionary trajectories during lymph node invasion.
(A) Phylogenetic trees reconstructed with maximum parsimony for each patient illustrate the patterns of lymph node spread. For Pat. 3 and 4 results were validated with whole-genome sequencing. When multiple lymph node samples were available for a patient, those clustered together in a single clade, indicating a recent common ancestor that led to lymph node colonisation. See Figure S4 for tree bootstrap values. Putative driver genes and recurrent copy number alterations in breast cancers are annotated in the trees (Tier 1 likely driver genes in black-bold, recurrent copy number alterations and Tier 2 possible drivers of uncertain pathogenicity in grey-italic). (B) We assessed the spatial heterogeneity of subclonal mutation PIK3CA 1047R in Pat.10 at single cell resolution using in situ hybridisation of mutant vs wildtype transcripts, revealing spatial segregation of mutant and wildtype subclones (only signals from cancer cells are represented).
The two predominantly observed patterns were early divergence or in fact a complete lack of divergence. Early divergence between primary and lymph nodes was observed in Pat. 2, 3, 4 and 6. In these cases, the mutational and in part also the CNA landscapes were very different between the primary and the lymph nodes, with significant heterogeneity at the level of putative driver alterations. Phylogenetic analysis revealed that the lymph node deposits diverged very early during the evolutionary history of the tumour in these patients (Fig. 3A, see Fig. S4 for bootstrap values and Fig. S5 for WES trees). This is particularly interesting because recent breast cancer studies found similar patterns of divergence between primary and metastatic lesions (20,21). In our cohort, multiple samples from the lymph nodes were also very similar to each other, consistent with a recent common ancestor of the lymph node lesions, which indicates a clonal bottleneck. The fact that additional samples profiled with targeted sequencing corroborated the original phylogenetic topology constructed with WES (Fig. S5) confirms that divergence patterns were not due to sampling bias. This is important because phylogenetic divergence could appear simply due to undersampling of the lineages in the primary tumour (22). Furthermore, early divergence was confirmed by whole-genome sequencing in Pat. 3 and 4 (Fig. 2C and 3A). To test the impact of possible subclonal structure that may confound the phylogenies (23), we also reconstructed the phylogenetic trees with only clonal mutations in each sample with Cancer Cell Fraction (CCF)>80% (using the MRCA in each sample), and the topologies were unchanged, again highlighting early divergence in a subset of cases.
The rest of the cohort (Pat. 5, 7, 9, 10, 14, 16) was characterised by a palm-tree topology, with relatively short branches and no detectable divergence between primary and lymph node lesions (Fig. 3A). Putative drivers and recurrent alterations in this subgroup were almost invariably truncal (all apart from PIK3CA in Pat.10, as also reported by others (6)). We investigated the spatial heterogeneity of PIK3CA 1047R in primary and lymph node lesions of Pat.10 further using single cell level chromogenic in situ hybridization with BaseScope™ (Fig. 3B; primary: 44.17% mutant, lymph node: 80.96% mutant; signal from cancer cells only is reported), demonstrating segregation of PIK3CA mutant and wildtype subclones. Divergent patterns were quantified using Node Cophenetic Distance(24) and confirmed significant divergence measured both on targeted exome sequencing (TES) (p=0.0043, Wilcoxon rank sum test) and WES (p=0.0079, Wilcoxon rank sum test) data (Fig. S7). Divergence was not correlated with number of samples.
Importantly, we found no evidence of genes recurrently altered in lymph nodes with respect to the primary lesions, although we cannot exclude the presence of weakly recurrent drivers that we do not have the power to detect in our cohort.
Different modes of lymph node spread are recapitulated in ctDNA
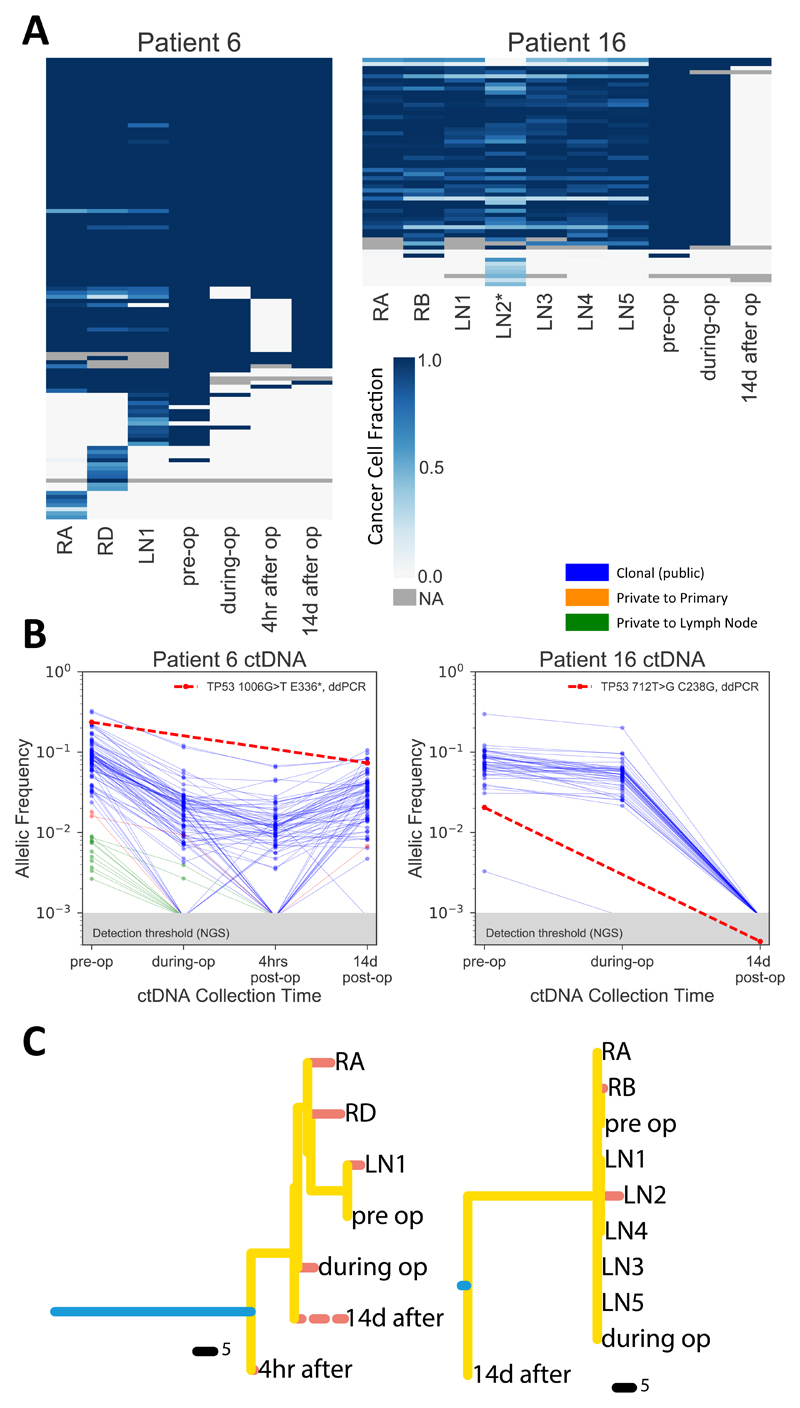
In order to follow the evolutionary dynamics of node positive early breast cancers through time, we collected cell free DNA (cfDNA) at multiple time points on 11 patients, however only 7 had enough DNA (20ng in total) to allow genomic profiling. We applied the targeted sequencing custom panel used in Figure 2A to those 7 patients, but somatic mutations were detected in only two. For these two patients, we had four time points: pre-operatively, immediately post resection, 4h post-operatively and 12 - 14 days after operation. Genomic profiling shows the dramatic impact of tumour resection on the ctDNA, as proof of principle indicating that the resected lesions were responsible for shedding detectable tumour DNA in the plasma (Fig. 4A). Indeed, the frequency of mutations dramatically drops after tumour (primary and nodes) resection (Fig. 4B). However, Pat. 6 showed mutations increasing again 14 days after the operation. Remarkably, the majority of private mutations found in the ctDNA samples before the operation were unique to the lymph nodes, corroborating the divergence patterns observed in solid samples. After lymph node resection, private mutations from the nodes disappeared from plasma, confirming the origin of the shedding. A subset of truncal mutations however persisted in the plasma 14 days after the operation. This was unlikely due to ctDNA remnants due to its short half-life and instead suggests the presence of residual micrometastatic disease shedding ctDNA in the blood. For Pat. 16, the lack of divergence reported in the tissue was observed in plasma as well. Phylogenetic reconstruction confirmed these patterns for both patients. In particular, for Pat.6 the pre-operative ctDNA profile clustered with the lymph node sample, whereas the postoperative ctDNA sample showed an earlier divergence event of the micrometastatic disease. Since we do not know which mutations are private to the micrometastatic deposits, the post-operative ctDNA branch appears shorter in our data than it actually is. These results indicate that the patterns of lymph node spread observed in the tissue, even in this early node positive cohort, are recapitulated in the plasma.
Figure 4. Longitudinal ctDNA analysis recapitulates tissue evolution.
(A) The cohort-specific targeted panel was applied to ctDNA for two patients, heatmaps show presence (blue) and absence (yellow) of variants at four time points (pre-operation, immediately post-resection, 4 hours after operation and 14 days after operation) compared to the corresponding primary and lymph node samples per patient. (B) Variant allele frequency (VAF) changes of all mutations at different time points. (C) Phylogenetic trees reconstructed with both tissue and ctDNA data confirm these patterns. Post-operative ctDNA appears early in the tree in Pat.6, suggesting early disseminated micrometastatic disease (dashed line indicates possible additional variants not detectable with a targeted approach).
APOBEC activity is increased in lymph nodes
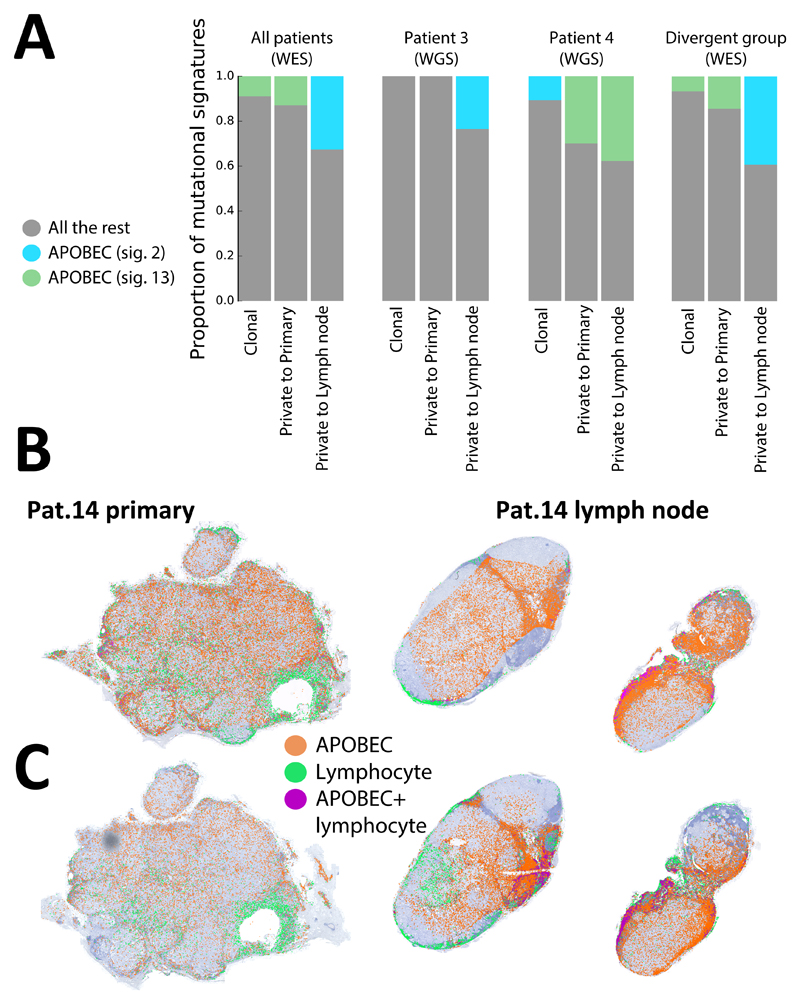
Mutational signature analysis revealed the presence of common age-related cancer signature 1, as well as signatures specific to breast cancer (25) (signatures 2, 3 and 13). Interestingly, APOBEC signatures, which were detected in 5/11 patients, were found to be increased in lymph nodes with respect to the primary tumour, especially in the divergent subgroup. These results were confirmed using WGS on Pat. 3 and Pat. 4 (Fig. 5A). RNA in situ hybridization of APOBEC3A and APOBEC3B transcripts for Pat.14 using RNAscope® (Fig. 5B,C) revealed spatial heterogeneity, with 3.17-fold higher expression of APOBEC3A and 3.72-fold higher expression of APOBEC3B in the lymph node with respect to the primary tumour (measured in dots/mm2, only signal from cancer area is reported, see Methods). Automatic identification of lymphocytes indicated that APOBEC signal came predominantly from cancer cells, although in some areas cell density was so high that the two signals overlapped (purple). This was consistent with the mutational signatures in Fig. 5A. This suggests that APOBEC may be involved in driving intra-tumour heterogeneity during metastatic spread to lymph nodes.
Figure 5. APOBEC signature is increased in lymph nodes.
(A) Mutational signature analysis applied to whole-exome sequencing mutations from the whole cohort and whole-genome sequencing for Pat.3 and Pat. 4 showed an increase in APOBEC signatures in mutations private to the lymph nodes. Spatial heterogeneity in expression of APOBEC3A (B) and APOBEC3B (C) was assessed at single-cell level using in situ hybridisation for Pat.14, revealing higher expression of both APOBEC3A and APOBEC3B in the lymph node lesion (only signals from cancer cells are represented).
Discussion
Breast cancer can spread from one organ system to another via haematogenous and lymphatic routes. Understanding lymph node spread from an evolutionary perspective is crucial to improve the understanding of progression to metastatic disease. In this study, we focused on untreated lymph node positive patients without evidence of distant deposits and performed a spatio-temporal analysis of the evolution of lymph node invasion. We found striking patterns of early divergence in a significant proportion of patients. Remarkably, ctDNA analysis identified the divergent lymph nodes as the main contributor to circulating tumour DNA at resection, thus reflecting the evolutionary patterns identified in the tissue. This implies that ctDNA may partially inform on the biology of axillary lymph node spread. The divergent lesions were highly distinct in terms of mutations and partly also in terms of copy number changes, suggesting a clonal bottleneck during lymph node spread in these patients. Importantly, our data are consistent with a model of punctuated evolution in breast cancer, where tumorigenesis is driven by relatively rare but dramatic selection events{Gao:2016du}. Moreover, from a therapeutic perspective, inhibiting APOBEC may prevent or slow down metastatic evolution. The question on whether evolutionary patterns such as lymph node divergence, have prognostic and/or predictive value remains open and will require testing in larger cohorts.
This study has several limitations, for example the limited number of patients and the lack of ctDNA longitudinal tracking beyond day 14 post-surgery. Moreover, the follow-up is relatively short and, to date, we have not had the opportunity to profile distant metastatic deposits to understand the representation of the clones from primary and nodes in the three subgroups. Further efforts on larger cohorts of patients are needed to validate the three subgroups, and to determine their utility in determining patients’ prognosis in addition to already established prognostic factors, and potentially direct more effective treatment strategies.
Data and Materials Availability
Sequence data have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001002947. Further information about EGA can be found on https://ega-archive.org. High-resolution images have been deposited in BioStudies with accession number S-BSST110.
Supplementary Material
Translational Relevance.
Lymph node involvement is an important prognostic factor in breast cancer, but the patterns of cancer evolution during lymph node spread are poorly understood. Furthermore, circulating tumour DNA as a potential biomarker to guide treatment is relatively unexplored in this context. In this prospective study, we used multi-region sequencing of multiple samples of primary tumours and multiple matched lymph nodes and showed remarkable differences in the patterns of lymph node infiltration between subsets of patients. Specifically, one subgroup of cases showed early lymph node divergence. Those patterns were reflected in the circulating tumour DNA of the same patients and demonstrated (to our knowledge for the first time) disappearance of private nodal mutations after surgical resection. Divergence was also associated to APOBEC activity. Together, these results suggest that evolutionary patterns of lymph node infiltration may be important to predict the course of the disease in individual patients and combined with circulating tumour DNA sampling, could aid patient stratification and personalised, precision medicine.
Acknowledgements
A.S. is supported by the Chris Rokos Fellowship in Evolution and Cancer, by Wellcome Trust (202778/B/16/Z) and by Cancer Research UK (A22909). Y.Y. is supported by Cancer Research UK (A21808). This work was also supported by Wellcome Trust funding to the Centre for Evolution and Cancer (105104/Z/14/Z). We also acknowledge funding from Breast Cancer Now and the Royal Marsden and ICR NIHR Biomedical Research Centre. C.C.M. was supported in part by NIH grants U54 CA217376, P01 CA91955, R01 CA170595, R01 CA185138 and R01 CA140657 as well as CDMRP Breast Cancer Research Program Award BC132057 and an Arizona Investigator Grant ADHS18-198847. We thank Advanced Cell Diagnostics for providing the BaseScope™ PIK3CA probes. Image from Fig. 1A was taken from Servier Medical Art (licensed under a Creative Commons Attribution 3.0 Unported License).
References
- 1.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–60. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21:751–9. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nature Genetics. 2016 doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–6. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGranahan N, Swanton C. Cell. Vol. 168. Elsevier; 2017. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future; pp. 613–28. [DOI] [PubMed] [Google Scholar]
- 11.Blamey RW, Pinder SE, Ball GR, Ellis IO, Elston CW, Mitchell MJ, et al. Reading the prognosis of the individual with breast cancer. Eur J Cancer. 2007;43:1545–7. doi: 10.1016/j.ejca.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Wishart GC, Bajdik CD, Azzato EM, Dicks E, Greenberg DC, Rashbass J, et al. A population-based validation of the prognostic model PREDICT for early breast cancer. Eur J Surg Oncol. 2011;37:411–7. doi: 10.1016/j.ejso.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of Oncology. 2015:v8–30. [Google Scholar]
- 14.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Davies C, Godwin J, Gray R, Clarke M, Cutter D, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–84. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Peto R, Davies C, Godwin J, Gray R, Pan HC, et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379:432–44. doi: 10.1016/S0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.EBCTCG Early Breast Cancer Trialists' Collaborative Group. Effect of radiotherapy after mastectomy and axillary surgery on 10-year recurrence and 20-year breast cancer mortality: meta-analysis of individual patient data for 8135 women in 22 randomised trials. The Lancet. 2014;383:2127–35. doi: 10.1016/S0140-6736(14)60488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Science Translational Medicine. Vol. 7. American Association for the Advancement of Science; 2015. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer; pp. 302ra133–3. [DOI] [PubMed] [Google Scholar]
- 18.Naxerova K, Reiter JG, Brachtel E, Lennerz JK, van de Wetering M, Rowan A, et al. Science. Vol. 357. American Association for the Advancement of Science; 2017. Origins of lymphatic and distant metastases in human colorectal cancer; pp. 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olmeda D, Cerezo-Wallis D, Riveiro-Falkenbach E, Pennacchi PC, Contreras-Alcalde M, Ibarz N, et al. Nature. Vol. 546. Nature Publishing Group; 2017. Whole-body imaging of lymphovascular niches identifies pre-metastatic roles of midkine; pp. 676–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F, et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. New England Journal of Medicine. 2013;368:1199–209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 21.Ng CKY, Bidard F-C, Piscuoglio S, Geyer FC, Lim RS, de Bruijn I, et al. Clin Cancer Res. Vol. 23. American Association for Cancer Research; 2017. Genetic Heterogeneity in Therapy-Naïve Synchronous Primary Breast Cancers and Their Metastases; pp. 4402–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong WS, Shpak M, Townsend JP. Inferring the Origin of Metastases from Cancer Phylogenies. Cancer Res. 2015;75:4021–5. doi: 10.1158/0008-5472.CAN-15-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alves JM, Prieto T, Posada D. Trends in Cancer. Vol. 3. Elsevier; 2017. Multiregional Tumor Trees Are Not Phylogenies; pp. 546–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paradis E, Claude J, Strimmer K. Bioinformatics. Vol. 20. Oxford University Press; 2004. APE: Analyses of Phylogenetics and Evolution in R language; pp. 289–90. [DOI] [PubMed] [Google Scholar]
- 25.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.