Abstract
The cell adhesion glycoprotein E-cadherin (CDH1) is commonly inactivated in breast tumours. Precision medicine approaches that exploit this characteristic are not available. Using perturbation screens in breast tumour cells with CRISPR-Cas9 engineered CDH1 mutations, we identified synthetic lethality between E-cadherin deficiency and inhibition of the tyrosine kinase ROS1. Data from large-scale genetic screens in molecularly diverse breast tumour cell lines established that the E-cadherin/ROS1 synthetic lethality was not only robust in the face of considerable molecular heterogeneity but was also elicited with clinical ROS1 inhibitors including foretinib and crizotinib. ROS1 inhibitors induced mitotic abnormalities and multinucleation in E-cadherin defective cells, phenotypes associated with a defect in cytokinesis and aberrant p120-catenin phosphorylation and localisation. In vivo, ROS1 inhibitors produced profound anti-tumour effects in multiple models of E-cadherin defective breast cancer. This data therefore provides the pre-clinical rationale for assessing ROS1 inhibitors such as the licensed drug crizotinib in appropriately stratified patients.
Keywords: E-cadherin, ROS1, crizotinib, breast cancer, synthetic lethality
Introduction
E-cadherin defects are frequently found in breast cancer (>13 %) and gastric cancer (>14%) and are particularly prevalent in lobular breast cancers, which account for 15% of all mammary carcinomas (1). CDH1 encodes a calcium-dependent plasma membrane-bound cell-cell adhesion glycoprotein (2). In epithelial cells, E-cadherin forms homotypic adhesive complexes, known as adherens junctions that control cell-cell contact, the contractility of cells and ultimately the integrity of epithelial cell layers (3). Whilst the extracellular domain of E-cadherin interacts with E-cadherin molecules on adjacent cells, the intracellular domain interacts with, and controls, a number of proteins including p120-catenin (p120), α-catenin, γ-catenin, β-catenin, receptor tyrosine kinases, and a series of plasma membrane-associated receptors and cytoskeletal proteins (2). Loss of E-cadherin function causes a wide variety of phenotypes ranging from defects in cell migration and the orientation of the mitotic spindle, as well as dysregulation of cell-cell adhesion and anoikis resistance (reviewed in (3)).
In lobular breast cancer, loss of E-cadherin expression occurs early on in the tumourigenic process and is seen in up to 90% of cases often co-occurring with mutations in the PI3-kinase coding gene, PIK3CA (4). Lobular breast cancers tend to be estrogen receptor (ER) and progesterone receptor (PR) positive, ERBB2 amplification-negative, have a low Ki67 index and a luminal-A intrinsic subtype (1,5–7). Whilst these biomarkers might predict a favorable response to adjuvant endocrine therapy, retrospective analyses of two recent clinical trials (BIG 1-98 and ABCSG-8) suggests that a subset of invasive lobular breast cancer (ILC) patients have poorer responses to endocrine therapy when compared to those with invasive ductal carcinomas (IDCs) that display similar biomarkers (8,9). Furthermore, pathological complete response rates after neoadjuvant chemotherapy are low in ILC (10,11), suggesting that additional approaches are required to target this disease. In other breast cancer subtypes, E-cadherin expression might also influence patient outcome. For example in triple negative breast cancer, the prognosis of patients with E-cadherin-negative tumours is significantly worse than those with E-cadherin-positive disease (12,13).
At present, it is not clear whether actionable or pharmacologically tractable E-cadherin synthetic lethal effects can be identified that are likely to work clinically. Such clinically actionable synthetic lethal effects might be expected to be relatively resilient to additional molecular changes and operate in the face of a high-degree of molecular diversity that exits in cancer (i.e. hard synthetic lethal effects (14)). In the data presented below, we illustrate that the combined use of multiple, distinct, in vitro, ex vivo and in vivo model systems and the exploitation of different functional profiling modalities (genetic and chemical screens) can be used to identify robust and actionable E-cadherin synthetic lethal interactions. The most notable synthetic lethality we identified in this way was between E-cadherin and the ROS1 receptor tyrosine kinase, an effect that is clinically actionable using ROS1 inhibitors such as crizotinib or foretinib.
Results
Integrated genetic and small molecule screens identify a ROS1/E-cadherin synthetic lethal effect
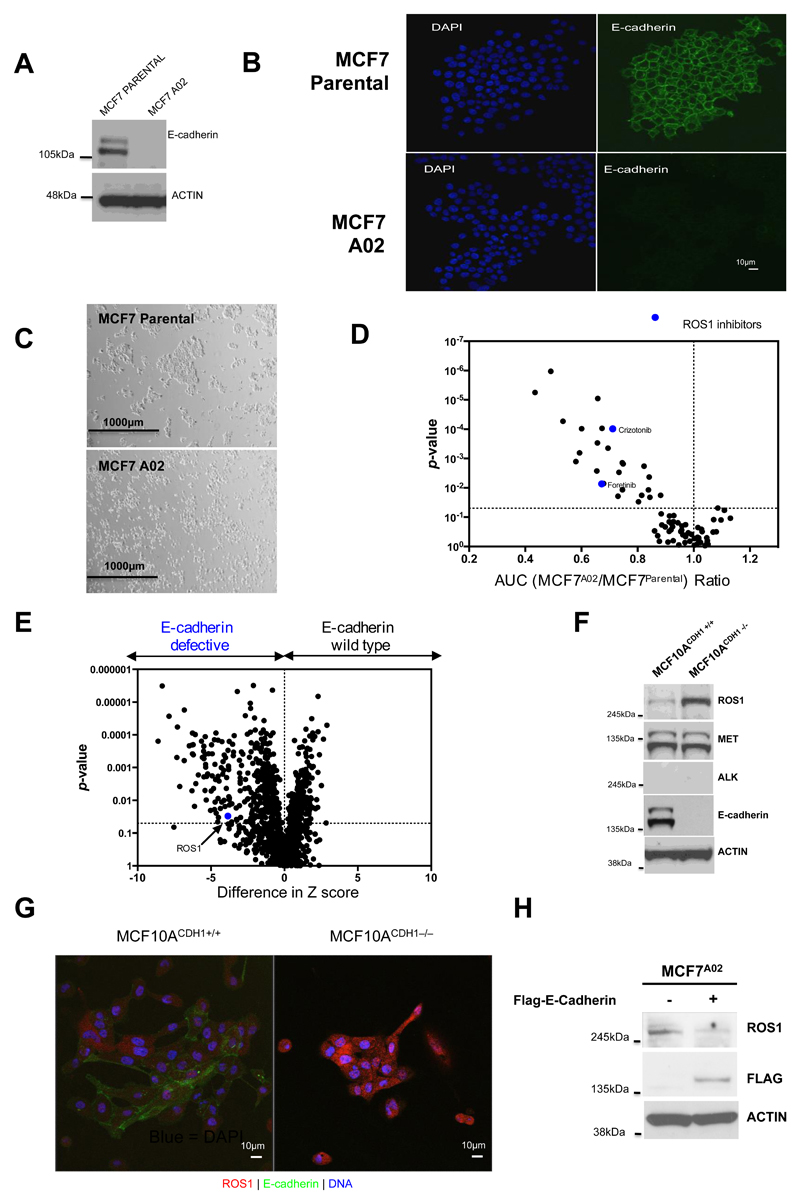
To identify candidate therapeutic targets for breast cancers with loss of E-cadherin, we used CRISPR-Cas9 mutagenesis in MCF7 breast tumour cells (ERα-positive, luminal A, PIK3CA mutant; described hereafter as MCF7Parental cells) to generate daughter clones, MCF7A02, MCF7B04 and MCF7B05, with frameshift mutations in CDH1 and loss of E-cadherin expression (Fig. 1A and B; Supplementary Fig. S1). Compared to MCF7Parental cells, E-cadherin defective cells displayed a rounded morphology also seen in breast tumour cells harbouring naturally occurring E-cadherin mutations (Fig. 1C). We used MCF7A02 and MCF7Parental cells in two parallel functional screens to identify E-cadherin synthetic lethal effects: (i) a drug sensitivity screen where we assessed the relative sensitivity of cells to an in–house curated library of 80 small-molecule inhibitors that are either in clinical use for the treatment of cancer or in late-stage clinical development (Fig. 1D; Supplementary Table S1 and S2); and (ii) a parallel siRNA sensitivity screen, using siRNA SMARTpools (four different siRNAs targeting a single gene in each well) targeting >1000 cell cycle control genes, kinase-coding genes or DNA repair related genes (see methods, Fig. 1E; Supplementary Table S3). The drug sensitivity screens identified a series of candidate E-cadherin synthetic lethal drugs, including PF-03758309 (a PAK inhibitor), PF-03814735 (an Aurora kinase inhibitor), PI3K/mTOR inhibitors (BEZ-235, PF-04691502 and Everolimus), the ROS1/MET/ALK inhibitors (15) crizotinib (PF02341066, Pfizer) and foretinib (GSK1363089, GSK) (Fig. 1D; Supplementary Table S1 and S2). In order to identify E-cadherin synthetic lethal effects from our MCF7 isogenic cell line siRNA screen, we calculated the difference in siRNA Z scores between E-cadherin defective and E-cadherin proficient cells and identified 104 E-cadherin synthetic lethal effects (p < 0.05, Fig. 1E; Supplementary Table S3). Gene ontology analysis of this 104 gene list using EnrichR (16) highlighted gene sets associated with myosin light chain kinase activity (Supplementary Table S4, adjusted p-value = 8.52 x 10-9, PLK3, AAK1, HUNK, CSNK1A1, NEK4, PAK4, CPNE3, PIM1, CLK3, RPS6KA2, SBK1, STK38L, TGFBR1 and MYLK2), gene sets related to GTP-dependent protein kinase activity (Supplementary Table S4, adjusted p-value = 1.35 x 10-8, PLK3, AAK1, HUNK, CSNK1A1, NEK4, PAK4, CPNE3 PIM1, CLK3, RPS6KA2, SBK1, STK38L, TGFBR1), and a set of candidate synthetic lethal genes associated with Rho-dependent protein serine/threonine kinase activity (Supplementary Table S4, adjusted p-value = 1.35 x 10-8, PLK3, AAK1, HUNK, CSNK1A1, NEK4, PAK4, CPNE3, PIM1, CLK3, RPS6KA2, SBK1, STK38L, TGFBR1). Other gene sets identified in this analysis included genes associated with RNA polymerase II carboxy-terminal domain kinase activity (Supplementary Table S4, adjusted p-value = 8.52E-09), protein serine/threonine kinase activity (Supplementary Table S4, adjusted p-value = 8.52 x 10-9) and calmodulin-dependent protein kinase activity (Supplementary Table S4, adjusted p-value = 8.85 x 10-9).
Figure 1. ROS1 inhibition is synthetic lethal with E-cadherin defects in isogenic models.
A, Wild-type E-cadherin protein is lost in the CDH1 CRISPR-Cas9 mutagenised MCF7A02 clone. Western blot illustrating E-cadherin expression in parental MCF7 cells (MCF7parental) and the MCF7A02 clone is shown. Upper band is a glycosylated isoform of E-cadherin, the lower band representing the non-glycosylated form. B, Confocal microscopy images of MCF7parental and MCF7A02 cells, illustrating loss of E-cadherin expression (green) in MCF7A02 cells. Nuclei are imaged with DAPI (blue). C, Light microscopy images of MCF7parental and MCF7A02 cells, illustrating reduction in cell-cell contact in MCF7A02 cells. D, Volcano plot illustrating AUC ratios (MCF7A02 E-cadherin defective/MCF7 E-cadherin wild type parental cells) of 80 small molecule inhibitors assessed in the high-throughput screen. AUC ratio <1 indicates candidate E-cadherin synthetic lethal effects. Blue dots represent ROS1 inhibitors. E, Volcano plot of data from the siRNA SMARTpool sensitivity screens in cell lines described in (B). Blue dot highlights ROS1 siRNA identified in the screen as selectively targeting E-cadherin defective cells. F, ROS1 expression is upregulated in E-cadherin defective cells. Western blot showing expression of ROS1, MET and ALK in MCF10A CDH1+/+ and MCF10A CDH1−/− cells. ACTIN expression is used as loading control. G, Illustrative confocal microscopy images indicating an increase in ROS1 expression in E-cadherin defective MCF10A CDH1−/− cells compared to MCF10A CDH1+/+ cells. ROS1 expression is shown in red, E-cadherin in green and DNA in blue. H, Ectopic expression of E-cadherin in MCF7A02 E-cadherin defective cells reduces ROS1 expression. Western blot illustrating ROS1 expression in E-cadherin defective MCF7A02 cells transfected with FLAG epitope-CDH1 cDNA. ACTIN is used as loading control.
Amongst the most significant hits in the siRNA screen was ROS1 (ROS Proto-Oncogene 1, Receptor Tyrosine Kinase, Fig. 1E; Supplementary Table S3). We selected ROS1 for further study as: (i) the synthetic lethality observed with ROS1 siRNA was statistically significant; and (ii) the parallel small molecule inhibitor screen identified ROS1 inhibitors (foretinib, crizotinib) as candidate synthetic lethal drugs, suggesting that the ROS1 effect might be clinically tractable. The siRNA screen did not identify other targets of crizotinib or foretinib (e.g. KDR, MET, ALK and AXL) as being candidate E-cadherin synthetic lethal effects (Supplementary Fig. S2). We also noted that in both MCF7 and MCF10A isogenic systems, loss of E-cadherin expression caused upregulation of ROS1 protein but did not elicit changes in either MET or ALK expression (Fig. 1F,G; Supplementary Fig. S3), suggesting that enhanced ROS1 expression could represent a homeostatic response to E-cadherin loss; re-expression of E-cadherin in E-cadherin defective MCF7A02 cells reversed this ROS1 induction (Fig. 1H), suggesting causality.
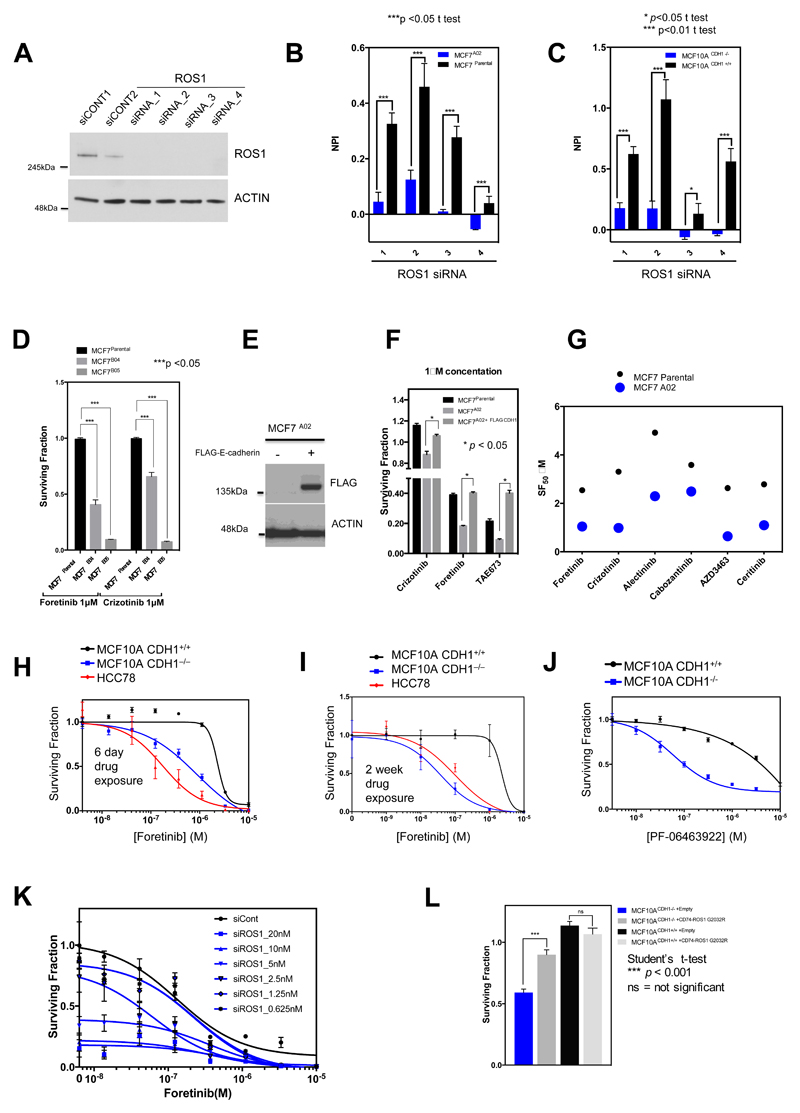
In validation experiments, we found that each of the four individual ROS1 siRNAs from the ROS1 SMARTpool caused silencing of ROS1 and preferentially inhibited the E-cadherin deficient MCF7A02 clone and also an E-cadherin defective MCF10A CDH1−/− non-tumour cell line (Fig. 2A-C). Two further E-cadherin defective clones derived by MCF7 CRISPR-Cas9 targeting, MCF7B04 and MCF7B05, were also significantly more sensitive to either foretinib or crizotinib than the parental cells (Fig. 2D; Supplementary Fig. S1), suggesting these effects were not private to MCF7A02. Restoring E-cadherin expression in MCF7A02 using a Flag-epitope tagged E-cadherin cDNA expression construct reduced foretinib sensitivity as well as sensitivity to an additional ROS1/MET/ALK inhibitor, TAE673 (Fig. 2E and F), confirming that E-cadherin influences the response to these agents. By assessing the effects of additional ROS1/MET/ALK inhibitors on E-cadherin isogenic cells, we found that foretinib and crizotinib gave the greatest difference in drug sensitivity between E-cadherin wild type and defective cells (Fig. 2G), warranting their further investigation. The sensitivity of E-cadherin defective cells to foretinib was similar to that observed in HCC78 cells (Fig. 2H and I), which harbour a SLC34A2-ROS1 gene fusion rearrangement rendering the cell line highly addicted to ROS1 kinase activity (15). We also found E-cadherin defective cells to be sensitive to a recently described ROS1 kinase inhibitor, PF-06463922 (17) (Fig. 2J). Finally, transfection of E-cadherin defective MCF7A02 cells with increasing amounts of ROS1 siRNA enhanced the cell inhibitory effects of foretinib (Fig. 2K). Conversely, expression of a crizotinib-refractory p.G2032R mutant ROS1 fusion cDNA (18) caused crizotinib resistance in E-cadherin defective cells (Fig. 2L), suggesting that ROS1 could be a critical foretinib/crizotinib target in these cells.
Figure 2. Validation of ROS1 synthetic lethality in E-cadherin defective isogenic models.
A, Western blot illustrating ROS1 silencing caused by four different ROS1 siRNAs (1, 2, 3 and 4) compared to two different non-targeting siRNAs (siCONT1, siCONT2). Uncropped western blot images are shown in Supplementary Fig. S20. B, and C, Bar charts illustrating cell inhibition caused by ROS1 siRNAs in MCF7parental and MCF7A02 cells (B) and MCF10A CDH1+/+ and MCF10A CDH1−/− cells (C). NPI = normalized percentage inhibition (compared to siCONT (NPI=1) and siPLK1 (NPI=0)). Error bars represent standard error of the mean (SEM) from three independent experiments. D, Bar chart illustrating the effect of 1 μM foretinib or crizotinib in MCF7parental compared to two additional E-cadherin defective clones, MCF7B04 and MCF7B05. Error bars represent SEM from three independent experiments. E, Western blot illustrating E-cadherin expression in MCF7A02 cell line model transfected with a FLAG epitope-CDH1 cDNA expression construct. F, Surviving fraction data from MCF7A02, MCF7parental and MCF7A02 +FLAG-CDH1 cell lines exposed to 1μM foretinib, crizotinib and TAE684. Cells were transfected with FLAG epitope-CDH1 cDNA and clones selected in G418. Clones expressing FLAG epitope-E-cadherin were exposed to 1 μM foretinib, crizotinib or TAE684 for six continuous days, at which point cell viability was assessed. *p<0.05 Student’s t-test as shown. G, Dot chart illustrating cell survival effects of ROS1/MET/ALK inhibitors in MCF7parental and MCF7A02 cells. SF50 = surviving fraction 50 (concentration required to cause 50 % reduction in survival). H, and I, Dose-response survival curves in MCF10A CDH1+/+ and MCF10A CDH1−/− cells exposed to foretinib for six days (H) or two weeks (I). Error bars represent SEM from three independent experiments. In each case, ANOVA p value MCF10A CDH1+/+ vs. MCF10A CDH1−/− cells < 0.0001. Dose-response in SLC34A2-ROS1 translocation-positive HCC78 cells is shown as a positive control. J, Dose-response survival curves in MCF10A CDH1+/+ and MCF10A CDH1−/− cells exposed to ROS1 kinase inhibitor PF-06463922 for six days. Error bars represent SEM from three independent experiments. K, Dose-response survival curves in MCF7A02 cells transfected with different concentrations of ROS1 siRNA SMARTpool and subsequently exposed to foretinib for six days. Increasing concentration of ROS1 siRNA caused a dose-dependent reduction in foretinib SF50. Error bars represent SEM from three independent experiments. L, Expression of a crizotinib-refractory p.G2032R mutant ROS1 fusion cDNA (18) causes crizotinib resistance in E-cadherin defective MCF10A CDH1-/- cells. Cells were transfected with either a p.G2032R CD74-ROS1 cDNA expression vector or a cDNA expression vector without a CD74-ROS1 insert (“empty”). Twenty-four hours after transfection, cells were exposed to crizotinib for a subsequent six days, at which point cell viability was assessed. Data shows the median effects of three independent experiments. Error bars illustrate the SEM. ***p < 0.001 Student’s t test as shown.
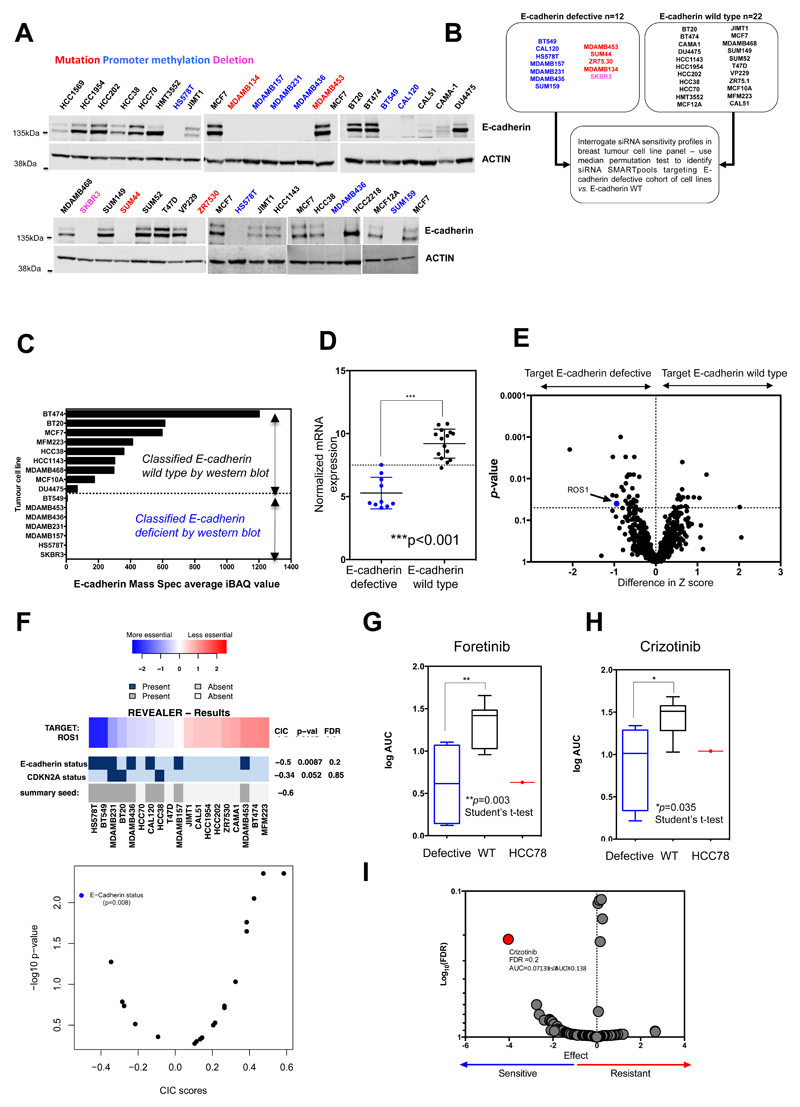
Many synthetic lethal effects are private to a small number of model systems and do not operate universally in the face of the molecular diversity found in breast and other cancers (14). To assess whether the synthetic lethal effect of ROS1 inhibition could apply more widely in breast cancer with E-cadherin loss, we interrogated recently described in-house siRNA “Achilles’ Heel” screen data describing the kinase dependencies in 34 breast tumour cell lines (19). We first determined the E-cadherin protein expression in each of these 34 models by western blotting, classifying cell lines as either “E-cadherin defective” (n=12) or “E-cadherin wild-type” (n=22), and used these classifications to identify siRNAs that selectively targeted the E-cadherin defective cohort of breast tumour cell lines (Fig. 3A and B). We found that the “E-cadherin defective” or “E-cadherin wild-type” status defined by western blotting was consistent with E-cadherin protein expression data determined by mass spectrometry (MS) (20), as well as E-cadherin mRNA expression and CDH1 mutation data (Fig. 3C and D; Supplementary Table S5). For example, each of the tumour cell line models with CDH1 truncating mutations or homozygous deletions lacked E-cadherin protein expression; similarly breast tumour cell lines with CDH1 promoter hypermethylation (1,21), also lacked full length E-cadherin protein (Fig. 3A). Using the siRNA Achilles’ Heel screen data (19) (Fig. 3B), we identified 31 siRNAs that selectively targeted the E-cadherin deficient cohort (p < 0.05, median permutation t-test), including ROS1 (Fig. 3E; Supplementary Fig. S4A and B median permutation t-test p = 0.04; Supplementary Table S6). We also assessed whether MET or ALK siRNA selectively targeted E-cadherin defective breast tumour cells, but in an analysis of 34 breast tumour cell lines, classified according to E-cadherin status (Supplementary Table S5), did not identify a significant association (Supplementary Fig. S5).
Figure 3. Synthetic lethality of ROS1 inhibition in E-cadherin deficient breast tumour cell lines.
A, Western blot illustrating E-cadherin expression in 37 breast tumour cell lines (including MCF7 as positive control). Cell line names are colour-coded according to the presence of CDH1 gene mutations, gene deletion events or CDH1 promoter hypermethylation events. Uncropped western blot images are shown in Supplementary Fig. S20. B, Schematic illustrating breast tumour cell line cohort sizes used to interrogate siRNA sensitivity screens (19). C, Bar chart illustrating E-cadherin expression of a subset of the breast tumour cell lines shown in (A), determined by mass spectrometry, described in (20). Western blot classification of E-cadherin status from (A) is shown. D, Scatter plot illustrating CDH1 mRNA expression levels in breast tumour cell lines from the CCLE dataset (51). E, Volcano plot of data from the siRNA SMARTpool sensitivity screens described in (B). Blue dot highlights ROS1 siRNA identified in the screen as selectively targeting E-cadherin defective cells. F, REVEALER analysis identifies E-cadherin status as an important determinant of ROS1 siRNA sensitivity. Heatmap illustrates the REVEALER output identifying complementary genomic alterations associated with ROS1 siRNA sensitivity in the 34 breast tumour cell line panel described in (B). IC (information correlation coefficient) scores and nominal p values with respect to the target are shown on the right side of the heatmap. E-cadherin status demonstrated the most profound IC score -0.5 from 23 molecular features examined. G, and H, Box whisker plots illustrating foretinib (G) or crizotinib (H) sensitivity in 12 breast tumour cell lines, defined by Log2 area under the curve (AUC) values. Individual AUC values are listed in Supplementary Table S8. Log2 AUC values for SLC34A2-ROS1 translocation-positive HCC78 cells are shown as a positive control. ** p value = 0.003, Student’s t-test, * p value =0.035 Student’s t-test. I, Volcano plot of drug sensitivity effects in ex vivo cultured breast cancer explants (27) annotated according to CDH1 gene copy number. Red dot highlights crizotinib as selectively targeting explants with CDH1 gene copy number loss. Median AUC in explants with CDH1 copy number loss = 0.0713, median AUC in explants without CDH1 copy number loss = 0.138, p value =0.00127. Y-axis shows adjusted p-value (-log10) from a t-test comparing AUC in ex vivo explant models with CDH1 loss vs. models with no CDH1 copy number change. X-axis shows the negative value of the t-statistic describing the difference in AUC means for ex vivo explant models with CDH1 loss vs. models with no CDH1 copy number change. Negative values suggest drug sensitivity in ex vivo explant models with CDH1 loss vs. models with no CDH1 copy number change.
We also interrogated previously published siRNA Achilles’ Heel screen data describing the kinase dependencies in an additional 69 non-breast cancer derived tumour cell lines (19). After annotating these according to E-cadherin status, we also noted a ROS1/E-cadherin synthetic lethal effect, suggesting that this effect was not necessarily specific to breast tumour models (Supplementary Fig. S6A-C; Supplementary Table S7). Validation experiments confirmed the ROS1/E-cadherin synthetic lethality in the breast tumour cell line models (Supplementary Fig. S7A-E), suggesting that this effect was not isolated to isogenic models, but also operated in molecularly diverse models of breast cancer.
We also used an orthogonal analytical approach, REVEALER (Repeated evaluation of variables conditional entropy and redundancy (22), to identify the molecular features found in breast tumour cell lines that were most associated with sensitivity to ROS1 siRNA. REVEALER uses a set of molecular features (e.g. the presence or absence of mutations or other defects in key cancer driver genes or proteins) and target inhibition data (in this case sensitivity to ROS1 siRNA) from tumour cell line profiling experiments to identify multiple, often complimentary, molecular features that correlate with target inhibition, quantifying these effects as non-linear information correlation coefficients (ICs), between 1 (perfect correlation/features associated with resistance to target inhibition) and –1 (perfect negative correlation/features associated with sensitivity to target inhibition); REVEALER ICs <–0.1 or >0.1 are regarded as profound correlations (22). We carried out a REVEALER analysis using the ROS1 siRNA Z score data in the breast tumour cell lines as the measure of target inhibition and the mutational status of the 23 recurrently mutated cancer driver genes in the breast tumour cell line panel plus the E-cadherin protein classification described above as molecular features. Using this approach, we found E-cadherin protein deficiency to have a far greater correlation with ROS1 siRNA Z score (IC -0.5) than any of the other molecular features analysed (Fig. 3F), supporting the hypothesis that E-cadherin status is an important determinant of sensitivity to ROS1 inhibition in breast cancer. We also assessed the sensitivity of the breast tumour cell line panel to foretinib or crizotinib and found that the E-cadherin defective models were more sensitive to both inhibitors (Fig. 3G and H; Supplementary Table S8). The median sensitivity to either foretinib or crizotinib in the E-cadherin defective cohort was also similar to that in the SLC34A2-ROS1 translocated HCC78 tumour cell line model (Fig. 3G and H). The responses to ROS1 inhibitors observed in the tumour cell line panel appeared to be independent of epithelial or mesenchymal status. For example, MCF10A cells do not undergo EMT upon loosing E-cadherin expression (23) but did exhibit both foretinib and crizotinib sensitivity (Fig. 2H-J). Similarly, SUM149, a mesenchymal breast tumour cell line which has wild type E-cadherin expression (24) was not sensitive to ROS1 inhibition, whereas E-cadherin defective, epithelial cells (e.g. SUM44, SKBR3) were (Supplementary Fig. S8). Furthermore, we did not find that expression of MET, ALK, pAKT, AKT, pERK or ERK correlated with E-cadherin status, nor drug sensitivity (Supplementary Fig. S9), suggesting that these were unlikely to be determinants of the synthetic lethal effects we identified. As ROS1 fusion genes are well established biomarkers of ROS1 inhibitor sensitivity we also analysed paired-end RNA-sequencing data using Chimerascan in order to identify ROS1 fusion genes in our tumour cell line panel (25,26). Whilst Chimerascan identified the SLC34A2-ROS1 fusion event in HCC78 lung tumour cells (the positive control), ROS1 fusions were not identified in any of the breast tumour cell lines (Supplementary Table S9). In addition, ROS1 copy number or ROS1 variants, although rare in our tumour cell line panel, did not appear to correlate with crizotinib/foretinib sensitivity (Supplementary Table S9). We did note an increase in ROS1 expression and phospho-ROS1 levels in E-cadherin defective cells, similar to that seen in E-cadherin isogenic systems (Fig. 1F-H; Supplementary Fig. S10), suggesting that enhanced ROS1 expression could represent a homeostatic response to E-cadherin loss. Finally, we interrogated recently published data describing drug sensitivity effects in ex vivo cultured breast cancer explants (27) to identify drug sensitivity effects associated with loss of E-cadherin. When querying this data for drug sensitivity effects associated with CDH1 gene copy number loss, we found sensitivity to crizotinib (PF-02341066 (27)) to be the most significant effect (Fig. 3I, p value 0.00127). The mean area under the curve (AUC) for crizotinib was 0.0713 in explants with CDH1 copy number loss (n = 5) and 0.138 in explants without CDH1 copy number loss (n = 12; False Discovery Rate (FDR) 0.214).
ROS1 inhibitors elicit synthetic lethality in endocrine resistant, E-cadherin defective, breast tumour cells and in models of E-cadherin defective gastric cancer
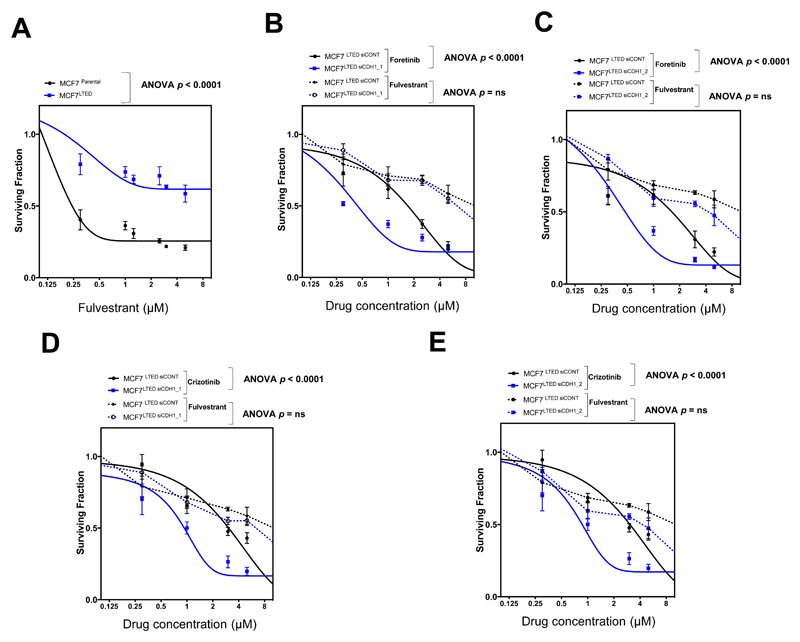
Given the central role of endocrine therapy in the treatment of ER+ve breast cancers and the frequency at which endocrine resistance develops, especially in the advanced disease setting (28), we assessed whether E-cadherin siRNA caused foretinib or crizotinib, sensitivity in previously-validated, endocrine therapy resistant, MCF7 LTED cells (29). We found that E-cadherin siRNA caused both foretinib and crizotinib sensitivity in MCF7 LTED cells (p < 0.0001, ANOVA, Fig. 4A-E) as it did in endocrine sensitive MCF7 cells (Fig. 2G), suggesting that the E-cadherin foretinib/crizotinib synthetic lethal effects were somewhat independent of endocrine therapy sensitivity. We also found that ER+ve, de novo endocrine resistant and E-cadherin defective MDAMB134VI tumour cells, which were derived from a case of invasive lobular carcinoma (30), were sensitive to ROS1 inhibitors in both in vitro (Supplementary Fig. S11A) and in vivo experiments (Supplementary Fig. S11B-D), suggesting that ROS1 inhibition could also target E-cadherin defective breast tumour cells in this particular setting.
Figure 4. E-cadherin/ROS1 synthetic lethality is independent of endocrine therapy resistance.
A, Dose-response survival curves in MCF7 and MCF7 LTED cells exposed to fulvestrant for five days. Error bars represent SEM from three independent experiments. B, and C, Dose-response survival curves in MCF7 LTED cells transfected with two independent E-cadherin (CDH1) siRNA reagents; siRNA 1 (B) and siRNA 2 (C) and a non-targeting siRNA (siCONT1) for 48 hours and subsequently exposed to foretinib or fulvestrant for five days. Error bars represent SEM from three independent experiments. ANOVA p value MCF7 LTED transfected with two independent E-cadherin siRNA reagents vs. MCF7 LTED transfected with siCONT1 cells p < 0.0001 for foretinib. No statistically significant difference was observed in the same cells exposed to fulvestrant. D, and E, Dose-response survival curves in cells as in (B) and (C) exposed to crizotinib or fulvestrant for five days. Error bars represent SEM from three independent experiments. ANOVA p value MCF7 LTED transfected with two independent E-cadherin siRNA reagents vs. MCF7 LTED transfected with siCONT1 cells p < 0.0001 for crizotinib. No statistically significant difference was observed in the same cells exposed to fulvestrant.
We next assessed whether crizotinib or foretinib sensitivity extended to models of E-cadherin defective diffuse gastric cancer, given the elevated frequency of E-cadherin defects in this cancer subtype. We found that either crizotinib or foretinib selectively targeted E-cadherin defective gastric tumour cell lines (Supplementary Fig. S12A and B). The effects of crizotinib in additional gastric tumour cell lines have also been recently assessed (27,31). We reanalyzed this data and found that those tumour cells lines with reduced E-cadherin expression (defined by CDH1 mRNA levels) were more sensitive to crizotinib than those with higher E-cadherin expression (Supplementary Fig. S12C).
ROS1 inhibition exacerbates p120 catenin, cleavage furrow and cytokinesis defects in E-cadherin defective cells
In investigating the cellular phenotypes associated with these E-cadherin selective effects, we found that exposure of E-cadherin defective cells to foretinib caused an increase in the proportion of cells with >4N DNA content (Fig. 5A and B), an increase in the frequency of cells with abnormal mitoses, particularly cells with multiple nuclei (Fig. 5C-F), an increase in expression of the G2/M DNA damage biomarker p21 (Fig. 5G) and an increase in cellular apoptosis as assessed by PARP cleavage and caspase 3/7 activity (Fig. 5H-L; Supplementary Fig. S13). These phenotypes were reminiscent of those seen in Rhabdomyosarcoma tumour cells exposed to crizotinib (32) and Chronic Myeloid Leukemia (CML) tumour cell lines, where foretinib elicits phenotypes associated with mitotic catastrophe (MC) including multinucleated giant cells, increased DNA content and apoptotic cell death (33).
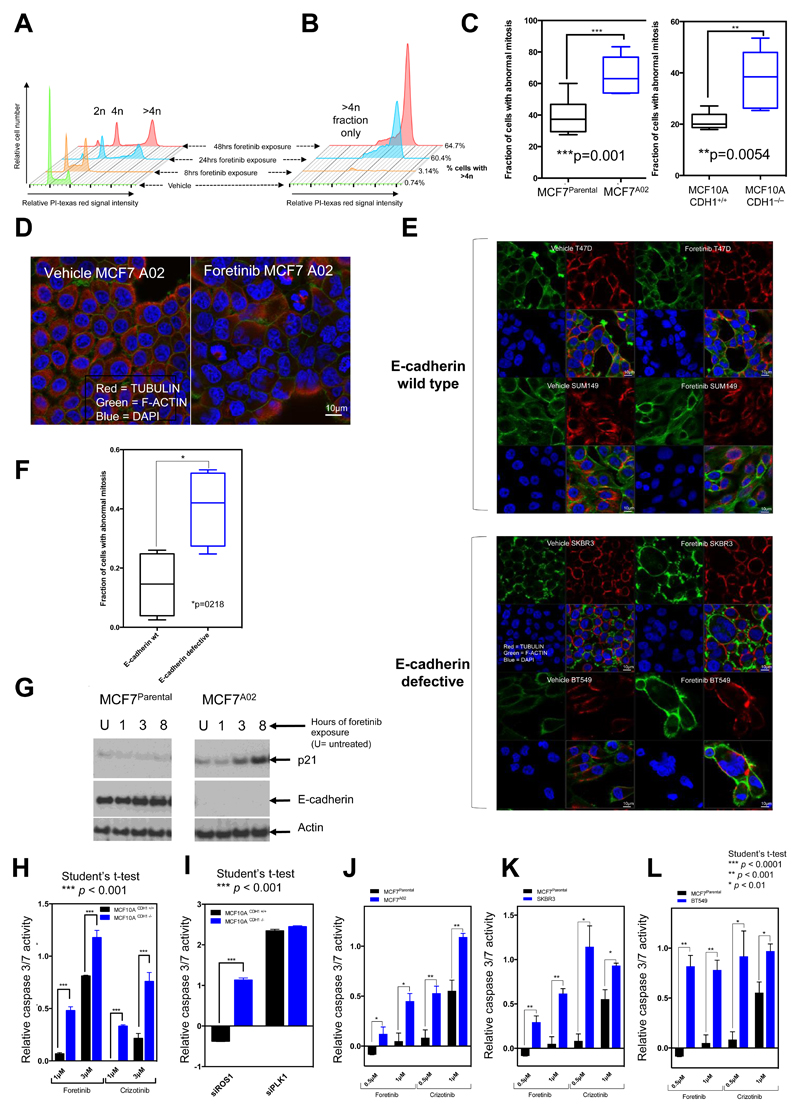
Figure 5. Cell cycle and mitotic defects in E-cadherin defective cells.
A, FACS plots illustrating increase in DNA content in E-cadherin defective MCF7A02 cells exposed to foretinib. B, Enlarged image from (A) indicating >4n fraction. The fraction of cells with >4n DNA content is shown. C, Box whisker plots indicating fraction of cells with abnormal mitoses (e.g. multinuclear defects) in cells exposed to foretinib. *** p value = 0.0001, ** p value = 0.0054, Student’s t-test. Nuclear defects were visualized by confocal microscopy and a minimum of 200 cells were counted for each cell line. Data is representative of two independent experiments in each case. D, Illustrative confocal microscopy images indicating multinuclear phenotype in E-cadherin defective MCF7A02 cells exposed to foretinib. E, Illustrative confocal microscopy images indicating multinuclear phenotype in E-cadherin defective SKBR3 and BT549 cells and E-cadherin wild-type T47D and SUM149 cells exposed to 1μM foretinib. F, Box whisker plots indicating fraction of cells with abnormal mitoses (e.g. multinuclear defects) in cells exposed to foretinib. * p-value =0.021, Student’s t-test. Data is representative of two independent experiments in each case. G, Western blot illustrating increased p21 levels in the E-cadherin defective MCF7A02 cells exposed to 1μM foretinib vs. MCF7parental cells. H, Bar chart illustrating increased caspase 3/7 activity in E-cadherin defective cells exposed to either crizotinib or foretinib. Median effects from three independent experiments are shown, with error bars representing the SEM. I, Bar chart illustrating increased caspase 3/7 activity in E-cadherin defective cells transfected with a siRNA targeting ROS1. siPLK1 was used as a positive control. p<0.05 between E-cadherin defective vs. wild-type groups, using the Student’s t-test. Median effects from three independent experiments are shown, with error bars representing the SEM. Bar chart illustrating increased caspase 3/7 activity in E-cadherin defective MCF7A02 J BT549 K and SKBR3 L cells exposed to either crizotinib or foretinib, compared to E-cadherin wild type MCF7Parental cells. * p < 0.01, ** p < 0.001 and *** p < 0.0001, E-cadherin defective vs. MCF7Parental cells, Student’s t-test. Median effects from three independent experiments are shown, with error bars representing the SEM.
To assess what might cause such multinuclear phenotypes, we visualised cells exposed to crizotinib or foretinib using live cell microscopy. We found that in both E-cadherin wild type and defective cells, either foretinib or crizotinib extended the time cells spent in mitosis (Supplementary Fig. S14A and B). This extended mitosis had dichotomous effects, depending upon the status of E-cadherin. In E-cadherin wild type cells, exposure to a ROS1 inhibitor still resulted in the formation of two mononuclear daughter cells (Fig. 6A). In contrast, when exposed to a ROS1 inhibitor, E-cadherin defective cells initiated cytokinesis but failed to complete invagination of the cell membrane at the cleavage furrow, resulting in the formation of multinuclear cells (Fig. 6A). We noted that cytokinesis in E-cadherin defective cells exposed to foretinib or crizotinib was characterized by prolonged membrane oscillation, starting at the onset of anaphase, cleavage furrow regression resulting in multinucleated cells, and also the formation of cells with lagging chromosomes (Fig. 6A; Supplementary Fig. S15; Supplementary Video S1-S5).
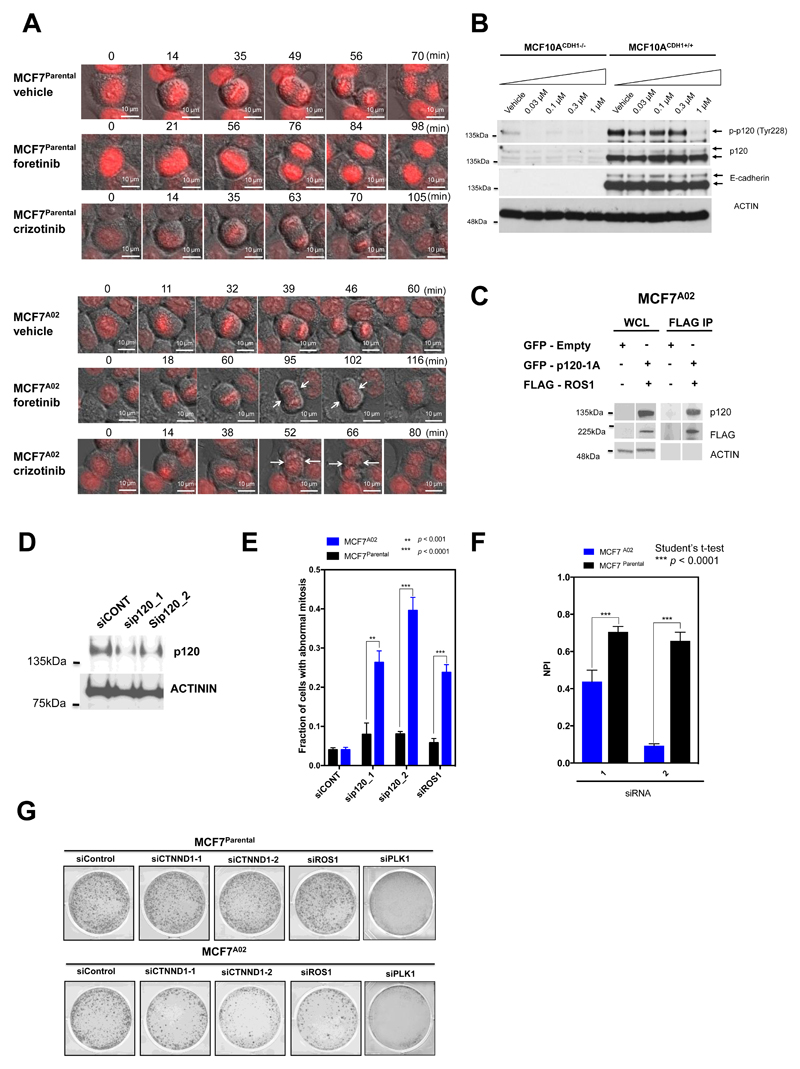
Figure 6. ROS1 inhibition exacerbates p120 catenin and cytokinesis defects in E-cadherin defective cells (previous page).
A, Time lapse microscopy images illustrating cell division in MCF7Parental (top panel) and E-cadherin defective MCF7A02 (bottom panel) cells exposed to foretinib, crizotinib or vehicle. MCF7A02 and MCF7Parental cells were first transfected with a mCherry-H2B plasmid, FACS sorted for mCherry-H2B to facilitate DNA visualization, and then exposed to foretinib, crizotinib or vehicle for a 24-hour period. Initial formation of the cleavage furrow, followed by formation of a multinuclear cell is highlighted with white arrows. Scale bar, 10 μm. B, Loss of E-cadherin is associated with a reduction in p120 levels and ROS1 inhibition causes a reduction in p120 tyrosine phosphorylation. Western blot showing phospho-p120 (Tyr228) and total p120 catenin levels in MCF10A CDH1+/+ and MCF10A CDH1−/− cells, exposed to a range of foretinib concentrations (vehicle, 0.03 μM, 0.1 μM, 0.3 μM or 1 μM) for 16 hours. C, ROS1 interacts with p120. Western blot illustrating co-immunoprecipitation of GFP-p120 and FLAG-ROS1 proteins in MCF7A02 cells. D, Western blot illustrating p120 catenin silencing caused by two different p120 siRNAs (sip120_1 and _2) compared to a non-targeting siRNA (siCONT1). E, p120 or ROS1 siRNA causes mitotic defects in E-cadherin defective cells. Bar chart indicating fraction of cells with abnormal mitoses in cells transfected with siRNAs targeting p120 or ROS1 siRNAs. *** p value < 0.0001, ** p value < 0.001, Student’s t-test. A minimum of 100 cells were analyzed for each cell line. Data is representative of three replica experiments, error bars represent SEM. F, p120 gene silencing is synthetically lethal with E-cadherin deficiency. Bar chart illustrating cell inhibition caused by two different p120 siRNAs (1 and 2) in E-cadherin wild type MCF7Parental and E-cadherin defective MCF7A02 cells. NPI = normalized percentage inhibition (compared to non-targeting siRNA, siCONT (NPI=1) and cytotoxic siRNA targeting PLK1 (NPI=0)). Error bars represent standard error of the mean (SEM) from three independent experiments. *** p value < 0.0001, Student’s t-test. G, Representative images from a colony forming assay illustrating cell inhibition caused by two different p120 siRNAs (1 and 2) or ROS1 siRNA in E-cadherin wild type MCF7Parental and E-cadherin defective MCF7A02 cells. A non-targeting siRNA, siCONT and siPLK1 are used as controls.
Cytokinesis failure and a multinuclear phenotype have previously been associated with defective p120-catenin activity (p120) (34,35). Actomyosin contractility at the ingression and cleavage furrow during anaphase and telophase is controlled by p120, which mediates these effects via binding to the centralspindlin component MKLP1 and the GTPase RhoA (35). p120 catenin also normally interacts with E-cadherin at the cell membrane where it is tyrosine phosphorylated (36–38), raising the possibility that loss of E-cadherin could impair p120 function. We found that E-cadherin defective tumour cells exhibited a reduction in p120–catenin levels (Fig. 6B), consistent with recent findings from a large-scale analysis of breast tumours with/without E-cadherin defects (39). Furthermore, as well as ROS1 immunoprecipitating with p120 (Fig. 6C), we found that ROS1 inhibition: (i) exacerbated the p120 reduction seen in E-cadherin defective cells; (ii) reduced tyrosine phosphorylation of p120; and (iii) reduced p120 levels at the cleavage furrow (Fig. 6B-C; Supplementary Fig. S16A and B). We also found that siRNA-mediated gene silencing of p120 (or ROS1) caused a multinuclear phenotype and elicited synthetic lethality in E-cadherin defective but not E-cadherin wild type cells (Fig. 6D-G). In normal epithelial cells, two opposing forces facilitate cytokinesis: (i) force provided by E-cadherin-dependent adherens junctions (AJ) at the apical membrane; and (ii) force provided by the contraction of the actomyosin ring at the cleavage furrow (40). One model to explain our observations might be that loss of one of these opposing forces in E-cadherin defective cells causes a greater reliance on processes and proteins that control cleavage furrow formation, such as p120 catenin. Our data suggest that ROS1 inhibitors exacerbate an existing p120 defect in E-cadherin defective cells and impair cytokinesis to such an extent that abnormal mitoses form; this elicits a DNA damage response (e.g. induction of p21 and γH2AX) and ultimately apoptosis. ROS1 inhibition has lesser effects when buffered by the activity of wild type E-cadherin.
Consistent with the concept that E-cadherin defects might cause a dependency upon processes controlled by actomyosin networks, such as cleavage furrow maturation, we also noted that the Rho GTPase effector kinase CDC42BPA (CDC42-binding kinase, MRCKα) which controls actomyosin function (41) was also identified as a robust synthetic lethal effect in our earlier siRNA screens (Supplementary Fig. S4A and B). We validated this synthetic lethal effect in subsequent experiments, including those using a toolbox small molecule MRCKα inhibitor (42) (Supplementary Fig. S17A-D), suggesting that additional targets associated with these processes might exist in E-cadherin defective cancers.
ROS1/E-cadherin synthetic lethality operates in multiple in vivo models of breast cancer
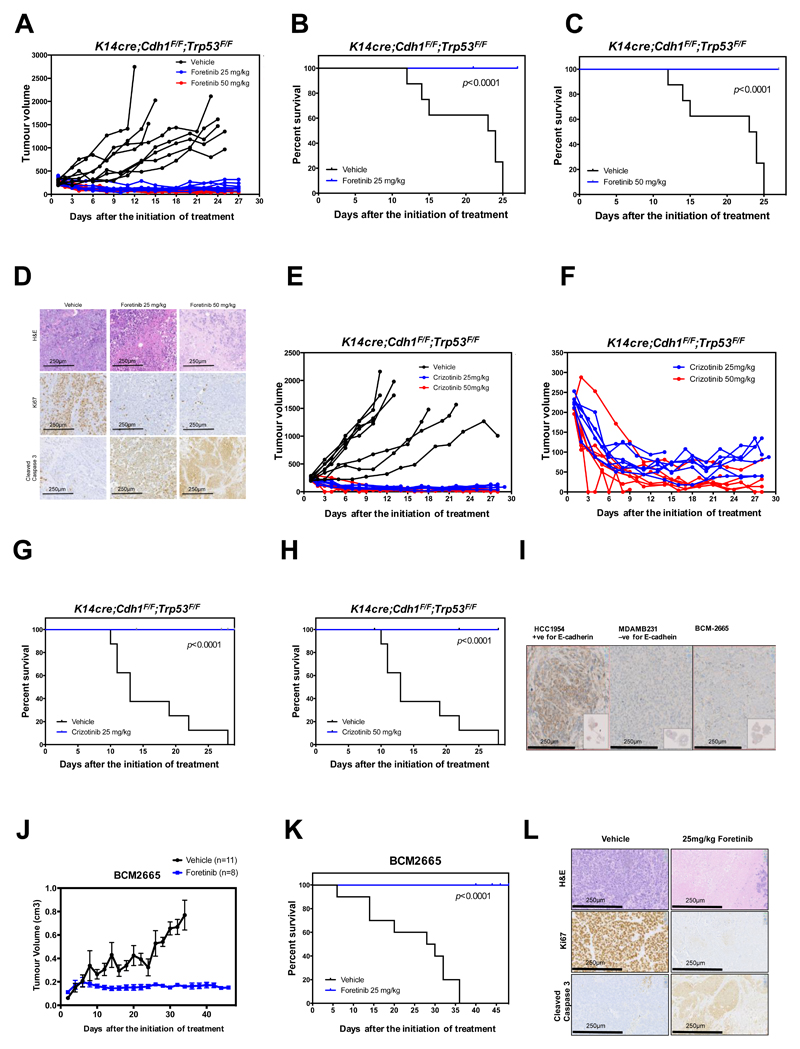
To assess the in vivo therapeutic potential of foretinib and crizotinib, we tested the ability of these drugs to affect the growth of E-cadherin defective invasive mammary carcinomas derived from the K14cre;Cdh1F/F;Trp53F/F (KEP) mouse ILC model; these mammary carcinomas show a strong resemblance to human ILC (43,44). E-cadherin defective mammary tumours from K14cre;Cdh1F/F;Trp53F/F (KEP) female mice were orthotopically transplanted into recipient mice and once established, animals were treated with foretinib, crizotinib or drug vehicle. In mice that received the drug vehicle alone, tumour growth continued unabated; in contrast, either foretinib or crizotinib treatment had a strong anti-tumour effect, reduced tumour volume and extended the survival of tumour-bearing mice (Fig. 7A-H, ANOVA p < 0.0001 in each case). Foretinib or crizotinib treatment also elicited a reduction in the proliferative index of tumours (as estimated by Ki67 immunohistochemistry) and an apoptotic response (Fig. 7D). The reduction in tumour volume in the early stages of crizotinib treatment is highlighted in Fig. 7F. Similar studies in mice bearing MCF7A02 or MCF7Parental derived xenografts also established that the anti-tumour effect of foretinib was significantly enhanced by the absence of E-cadherin (Supplementary Fig. S18A-E). We also assessed the anti-tumour effect of foretinib treatment in an E-cadherin defective patient-derived breast tumour xenograft (PDX) model, BCM2665, which was derived from a female with ER-negative, HER2-negative, basal-like breast cancer (Fig. 7I) (45). As before, foretinib significantly inhibited the growth of established tumours (p < 0.0001, ANOVA) and extended the survival of mice (Fig. 7J and K). E-cadherin defective tumours from mice treated with foretinib also consistently exhibited a profound reduction in levels of Ki67, suggesting a severe impairment in proliferative rate, showed increased levels of cleaved caspase-3, a marker of tumour cell apoptosis and had reduced hematoxylin and eosin staining, suggesting tumour necrosis (Fig. 7L). Given the availability of an immunohistochemical assay for measuring total and phosphorylated ROS1 in human tumours, we also established that foretinib treatment resulted in a decrease in pROS1 and total ROS1 (Supplementary Fig. S19).
Figure 7. E-cadherin synthetic lethal effects operate in vivo in E-cadherin defective breast tumours.
A, Therapeutic response to foretinib treatment in mice bearing E-cadherin deficient mammary tumours. Mammary tumour fragments from KEP mice were transplanted into 22 recipient mice; once tumours had established, animals were treated over a 27-day period with either drug vehicle or foretinib (25 or 50 mg/kg every other day, n=8 for vehicle-treated cohorts and n=7 for each drug treatment cohort). Tumour volumes after the initiation of treatment are shown. ANOVA p<0.0001 for both foretinib treatment regimes compared to vehicle-treated mice. B, Kaplan–Meier plot of data from (A) indicating anti-tumour efficacy of 25 mg/kg foretinib treatment. Mice were sacrificed once tumours reached a volume of 1500 mm3. C, Kaplan–Meier plot of data from (A) for vehicle vs. foretinib 50 mg/kg cohort. D, Immunohistochemistry images of tumours extracted from animals from (A) at the end of foretinib treatment are shown. Representative images of H&E, Ki67 and cleaved Caspase 3 are shown (magnification = 20x). Scale bar represents 250μm. E, Therapeutic response to crizotinib treatment in mice bearing KEP tumour allografts as in (A). ANOVA p<0.0001 for both crizotinib treatment regimes compared to vehicle-treated mice. F, Data from (E), plotted to illustrate tumour volume reduction in both crizotinib treated cohorts. G, Kaplan–Meier plot of data from (E), indicating effect of 25 mg/kg crizotinib treatment. H, Kaplan–Meier plot of data from (E) for vehicle vs. crizotinib 50 mg/kg cohort. I, Immunohistochemistry images illustrating lack of E-cadherin expression in a patient-derived xenograft (PDX) model of breast cancer (BCM2665) compared to positive (HCC1954 breast tumour cells) and negative controls (MDAMB231 breast tumour cells). J, Therapeutic response to foretinib treatment in mice bearing BCM2665 PDX. BCM2665 was transplanted into 19 recipient mice; once tumours had established, animals were treated over a 47-day period with either drug vehicle (n=11), or foretinib (25 mg/kg every other day, n= 8) as shown. ANOVA = p<0.0001. K, Kaplan–Meier plot of data from (I). L, Representative images of FFPE tumours from animals in (I) stained with H&E, Ki67 and cleaved caspase 3 are shown (magnification = 20x). Scale bar represents 250μm.
Discussion
E-cadherin defects are a common characteristic of human tumours. Here we show that ROS1 inhibition constitutes an E-cadherin synthetic lethal interaction that can be elicited with clinical inhibitors such as crizotinib or foretinib. These effects appear to be robust in the face of considerable molecular heterogeneity that exist between different isogenic models, distinct tumour cell lines, ex vivo breast tumour explants and two different mouse models of E-cadherin defective breast cancer. The profound anti-tumour effects seen in mice with E-cadherin defective tumours suggests that the observed synthetic lethal effect might also have translational utility. Several distinct approaches can be used to assess the potential of ROS1 inhibitors in E-cadherin defective cancers. For example, clinical trials in advanced forms of ILC, where E-cadherin defects are common, might seem appropriate. In such trials, absence of E-cadherin using immunohistochemical analysis could be used to pre-select appropriate patients for treatment. As crizotinib is already used in the treatment of non-small cell lung cancer, and has been the subject of prior clinical safety assessment, this drug seems a reasonable candidate for testing in such trials.
Mechanistically, our data suggests that loss of E-cadherin imparts upon cells a dependency upon ROS1 that is likely related to the ability to undergo cytokinesis. Our work suggests that p120 defects could play a role in these phenotypes, but it also seems possible that other proteins contribute to the synthetic lethal phenotype. For example, we also found that other proteins associated with actomyosin control such as MRCKα are also synthetic lethal with E-cadherin defects; these could conceivably play a part in the ROS1/E-cadherin synthetic lethal effect. Whilst drug-like inhibitors of Rho effector kinases such as MRCKα are still in development (46), licenced drugs such as crizotinib, might provide an actionable approach to targeting E-cadherin defective cancers. Similarly, although we were able to elicit E-cadherin synthetic lethal effects with multiple different ROS1 siRNA reagents and to partially reverse crizotinib sensitivity with a p.G2032R mutant ROS1 fusion cDNA (Fig. 2), the anti-tumour therapeutic effect of drugs such as crizotinib or foretinib could also be influenced by inhibition of kinases other than ROS1. Although we were unable to elicit synthetic lethality with siRNAs for MET, ALK, AXL and KDR in isogenic or non-isogenic systems (Supplementary Fig. S2 and S5), we cannot formally exclude the possibility that other kinase targets of foretinib or crizotinib also contribute to the E-cadherin synthetic lethality. It seems possible that clinical studies where mechanisms of resistance to crizotinib in E-cadherin defective tumours are assessed will inform this area.
Materials and Methods
Materials and cell lines
Small-molecule inhibitors were obtained from SelleckChem. siRNAs were obtained from GE Dharmacon. Cell lines were obtained from American Type Culture Collection (ATCC) and European Collection of Cell Cultures (ECACC) in 2010-2011 and maintained according to the supplier’s instructions as described in (19,47). The MCF10A CDH1-/- and MCF10A CDH1+/+ isogenic cell lines, were obtained in 2014 from Sigma Aldrich, and maintained according to the supplier’s instructions. At sixth-monthly intervals and prior to storage, the identity of each cell line was confirmed by short tandem repeat (STR) profiling of 10 loci using the GenePrint 10 system (Promega). At monthly intervals, mycoplasma testing of cell cultures was carried out using MycoAlert Mycoplasma Detection Kit (Lonza).
CRISPR-Cas9 targeting
MCF7 cells were targeted using the Edit-R-CRISPR-CAS9-gene engineering kit (GE Dharmacon) according to the supplier’s instructions. The following crRNA sequence was used: 5’GCUGAGGAUGGUGUAAGCGAGUUUUAGAGCUAUGCUGUUUUG 3’. MCF7 cells were transfected in 24 well plates (100,000 cells/well) with tracerRNA, crRNA and Cas9 plasmid. 72 hours after transfection, cells were plated in 15 cm dishes and continuously cultured until colonies formed. Colonies were recovered and profiled using PCR and Sanger sequencing to determine the presence of CDH1 mutations.
RNAi and small-molecule synthetic lethal screens
siRNA screens and small-molecule screens were performed as described in (19,48). See also Supplementary Materials and Methods for details.
Cell survival and apoptosis assays
Cell survival analysis was conducted as previously describes (49). See also Supplementary Materials and Methods for details.
Protein analysis
Whole-cell protein extracts were prepared from cells lysed in NP250 buffer (20 mM Tris pH 7.6, 1 mM EDTA, 0.5 % NP40, 250 mM NaCl); supplemented with protease inhibitor cocktail tablets (Roche, Burgess Hill, UK). Protein concentrations were measured using BioRad Protein Assay Reagent (BioRad, Hemel Hempstead, UK). For Western blot analysis, 50 μg of whole cell lysates were electrophoresed on Novex 4–12% gradient bistris pre-cast gels (Invitrogen) and immunoblotted overnight at 4°C with antibodies listed in Supplementary Table S11.
Confocal microscopy
Cells were plated on coverslips and exposed to drugs the following day. After drug exposure, cells were fixed in 4% (v/v) paraformaldehyde for 10 minutes, washed, permeabilized in 0.2% (v/v) Triton X-100 in PBS for 20 minutes, washed, and blocked in IFF (1% (w/v) bovine serum albumin, 2% (v/v) FBS in PBS) then immunostained with primary antibodies targeting F-Actin conjugated to Alexa Fluor 488 and Tubulin (Santa Cruz Biotech) and detected with a Texas red conjugated secondary antibody (Supplementary Table S11). DAPI staining was used to detect nuclei. Mitotic and nuclear phenotypes of at least 200 cells per condition were scored in each replicate experiment. Confocal experiments were imaged using Zeiss CLM700.
Time lapse microscopy
Time-lapse microscopy was performed in 6 well plates using a Diaphot inverted microscope (Nikon), in a humidified CO2 chamber at 37°C, using a motorized stage (Prior Scientific), controlled by Simple PCI software (Compix). Cells were first transfected with a mCherry-H2B plasmid, FACS sorted for mCherry-H2B to facilitate DNA visualization, and then exposed to foretinib or crizotinib for a 24 hour period, over which they were microscopically imaged. Sorting of cells is described in Supplementary Materials and Methods.
Cell cycle analysis
Measurements took place on a BD LSR II SORP flow cytometer (BD Biosciences, USA) equipped with a 488 nm blue laser, a 561 nm yellow laser, a 633 nm red laser and a 404 nm violet laser. Propidium Iodide was measured with 600 LP 610/20 BP. Cell population was gated in a FSC/SSC dot plot and doublets were gated out based on a DNA dye area/width dot plot. This cell population was further analyzed regarding its cell distribution (G1, S and G2/M phase) using Flow Jo V software.
In vivo assessment of foretinib and crizotinib efficacy
In vivo efficacy studies were conducted using K14cre;Cdh1F/F;Trp53F/F (KEP) or BCM2665 PDX tumour bearing mice as previously described (45,50). Experiments on K14cre;Cdh1F/F;Trp53F/F (KEP) tumour bearing mice were carried out at the Netherlands Cancer Institute according to local and international regulations and ethical guidelines, and were approved by the local animal experimental committee at the Netherlands Cancer Institute (DEC-NKI AvD:30100 2015 407 appendix 1, WP 5791 and WP 5900). Balb/c nudes were implanted with K14cre;Cdh1F/F;Trp53F/F (KEP) tumour sections in their 4th mammary gland on the right side as described previously (50). Once tumours reached 200 mm3 in volume, mice were randomized into cohorts who received either crizotinib/foretinib 25mg/kg or 50mg/kg a day via oral gavage for 28 days or the drug vehicle. Operators were blinded to which cohort received foretinib or crizotinib and which received vehicle. A surrogate of animal survival was used and defined by the amount of time taken for tumours to reach a pre-agreed volume of 1500mm3, at which point mice were sacrificed. Experiments using the BCM 2665 PDX model were conducted at the ICR according to local (UK Home Office) and international regulations and ethical guidelines, and were approved by the local animal experimental committee at the Institute of Cancer Research. Viably frozen fragments of the BCM2665 model (45) were obtained from Dr. Michael Lewis, Baylor College of Medicine, USA, under material transfer agreement and propagated in SCID-Beige and NSG hosts. SCID-beige host mice were obtained from Charles River at 21-28 day of age and were implanted with BCM2665 tumour fragments (~2mm in length) subcutaneously into the inguinal mammary fat pad following standard procedures. Once tumours measured 2 mm in diameter (assessed by palpation and caliper measurement), mice were randomized into cohorts who received either foretinib (25mg/kg) or the drug vehicle (1% hydroxypropylmethylcellulose, 0.2% SDS in H2O) every other day (4 days per week) by oral gavage. Operators were blinded to which cohort received foretinib and which received vehicle. Tumour growth was monitored over time (assessed by palpation and caliper measurement) and a surrogate of animal survival defined by the amount of time taken for tumours to reach a pre-agreed 12mm diameter, at which point mice were sacrificed and tumours excised. Tumour volume was calculated using the formula: V= (π x length x width2/6), where the length is the largest tumour diameter and width is the perpendicular diameter. Statistical analysis was performed using Prism. Tumours were formalin-fixed paraffin-embedded and slides were stained with H&E, or immunohistochemistry with antibodies against Ki-67 and cleaved caspase-3 was undertaken using standard procedures.
Supplementary Material
Statement of significance.
E-cadherin defects are common in breast cancer but are currently not targeted with a precision medicine approach. Our pre-clinical data indicate that licensed ROS1 inhibitors, including crizotinib, should be repurposed to target E-cadherin defective breast cancers, thus providing the rationale for the assessment of these agents in molecularly–stratified phase 2 clinical trials.
Acknowledgments
We thank the late Chris Marshall FRS and members of his laboratory at The Institute of Cancer Research, London for helpful discussions. We thank members of the Breast Cancer Now Toby Robins Breast Cancer Research Centre Core Pathology Laboratory for pathology support. We also thank members of the Preclinical Intervention Unit of the Mouse Clinic for Cancer and Ageing (MCCA) at the Netherlands Cancer Institute and members of the Biological Services Unit at the ICR for in vivo study support. This work was funded by Breast Cancer Now as part of programme funding to CJL. We acknowledge NHS funding to the NIHR Royal Marsden Hospital Biomedical Research Centre.
Grant Support
This study was funded by Programme Grant funding to CJL from Breast Cancer Now (CTR-Q4-Y2) as part of funding to the Breast Cancer Now Toby Robins Research Centre and Programme Grant funding to CJL from CRUK (C30061/A24439). PWBD is supported by a grant from the Royal Dutch Cancer Society (KWF UU 2011-5203). MJLC is funded by a pre-doctoral grant from the Carlos III Health Institute (PFIS16/000246) and CIBERONC, an initiative of the Carlos III Health Institute (CB16/12-00473). CJR is a Sir Henry Wellcome Fellow jointly funded by Science Foundation Ireland, the Health Research Board, and the Wellcome Trust (103049/Z/13/Z) under the SFI-HRB-Wellcome Trust Biomedical Research Partnership.
Footnotes
Additional information. This work was funded by a Programme Grant from Breast Cancer Now to CJL as part of their funding to the Breast Cancer Now Toby Robins Breast Cancer research Centre
Disclosure of Potential Conflicts of Interest
We have no conflicts of interest to declare.
Authors' Contributions
Conception and design: Ilirjana Bajrami, Alan Ashworth, Christopher J. Lord
Development of methodology: Ilirjana Bajrami, Rebecca Marlow, Marieke van de Ven, Rachel Brough, Helen N. Pemberton, Jessica Frankum, Fei Fei Song, Rumana Rafiq, Asha Konde, Malini Menon, James Campbell, Aditi Gulati, Rahul Kumar, Dragomir B. Krastev, Colm Ryan, Patrick W. Derksen, Jos Jonkers, Andrew N.J. Tutt, Alan Ashworth, Christopher J. Lord
Acquisition of data (provided animals, provided facilities, etc.): Ilirjana Bajrami, Rebecca Marlow, Marieke van de Ven, Rachel Brough, Helen N. Pemberton, Jessica Frankum, Fei Fei Song, Rumana Rafiq, Asha Konde, Malini Menon, James Campbell, Aditi Gulati, Rahul Kumar, Stephen J. Pettitt, Mark D. Gurden, Marta Llorca Cardenosa, Irene Chong, Lesley-Ann Martin, Mitch Dowsett, Patrycja Gazinska, Fredrik Wallberg, Spiros Linardopoulos, Colm Ryan, Patrick W. Derksen, Jos Jonkers, Andrew N.J. Tutt, Alan Ashworth, Christopher J. Lord
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Ilirjana Bajrami, Rebecca Marlow, Marieke van de Ven, James Campbell, Aditi Gulati, Rahul Kumar, Rachael Natrajan, Colm Ryan, Christopher J. Lord
Writing, review, and/or revision of the manuscript: All authors
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Ilirjana Bajrami, Rachel Brough, Helen N. Pemberton, Jessica Frankum, Fei Fei Song, Rumana Rafiq, Asha Konde, Malini Menon, James Campbell, Aditi Gulati, Rahul Kumar, Stephen J. Pettitt, Elinor J. Sawyer, Christopher J. Lord
Study supervision: Alan Ashworth, Christopher J. Lord
References
- 1.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–19. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–9. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lecuit T, Yap AS. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol. 2015;17:533–9. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- 4.Christgen M, Derksen P. Lobular breast cancer: molecular basis, mouse and cellular models. Breast Cancer Res. 2015;17:16. doi: 10.1186/s13058-015-0517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desmedt C, Zoppoli G, Gundem G, Pruneri G, Larsimont D, Fornili M, et al. Genomic Characterization of Primary Invasive Lobular Breast Cancer. J Clin Oncol. 2016;34:1872–81. doi: 10.1200/JCO.2015.64.0334. [DOI] [PubMed] [Google Scholar]
- 6.Michaut M, Chin SF, Majewski I, Severson TM, Bismeijer T, de Koning L, et al. Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci Rep. 2016;6:18517. doi: 10.1038/srep18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sikora MJ, Jankowitz RC, Dabbs DJ, Oesterreich S. Invasive lobular carcinoma of the breast: patient response to systemic endocrine therapy and hormone response in model systems. Steroids. 2013;78:568–75. doi: 10.1016/j.steroids.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Metzger Filho O, Giobbie-Hurder A, Mallon E, Gusterson B, Viale G, Winer EP, et al. Relative Effectiveness of Letrozole Compared With Tamoxifen for Patients With Lobular Carcinoma in the BIG 1-98 Trial. J Clin Oncol. 2015;33:2772–9. doi: 10.1200/JCO.2015.60.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knauer MGC, Dietze O, Greil R, Stöger H, Rudas M, Bago-Horvath Z, Mlineritsch B, Kwasny W, Singer C, Dubsky P, Jakesz R, et al. Survival advantage of anastrozol compared to tamoxifen for lobular breast cancer in the ABCSG-8 study. Cancer Res. 2015;75 nr S2–06. [Google Scholar]
- 10.Christgen M, Steinemann D, Kuhnle E, Langer F, Gluz O, Harbeck N, et al. Lobular breast cancer: Clinical, molecular and morphological characteristics. Pathol Res Pract. 2016;212:583–97. doi: 10.1016/j.prp.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Cristofanilli M, Gonzalez-Angulo A, Sneige N, Kau SW, Broglio K, Theriault RL, et al. Invasive lobular carcinoma classic type: response to primary chemotherapy and survival outcomes. J Clin Oncol. 2005;23:41–8. doi: 10.1200/JCO.2005.03.111. [DOI] [PubMed] [Google Scholar]
- 12.Kashiwagi S, Yashiro M, Takashima T, Nomura S, Noda S, Kawajiri H, et al. Significance of E-cadherin expression in triple-negative breast cancer. Br J Cancer. 2010;103:249–55. doi: 10.1038/sj.bjc.6605735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen T, Zhang K, Siegal GP, Wei S. Prognostic Value of E-Cadherin and beta-Catenin in Triple-Negative Breast Cancer. Am J Clin Pathol. 2016;146:603–10. doi: 10.1093/ajcp/aqw183. [DOI] [PubMed] [Google Scholar]
- 14.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–8. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 15.Davare MA, Saborowski A, Eide CA, Tognon C, Smith RL, Elferich J, et al. Foretinib is a potent inhibitor of oncogenic ROS1 fusion proteins. Proc Natl Acad Sci U S A. 2013;110:19519–24. doi: 10.1073/pnas.1319583110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou HY, Li Q, Engstrom LD, West M, Appleman V, Wong KA, et al. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc Natl Acad Sci U S A. 2015;112:3493–8. doi: 10.1073/pnas.1420785112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Awad MM, Katayama R, McTigue M, Liu W, Deng YL, Brooun A, et al. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med. 2013;368:2395–401. doi: 10.1056/NEJMoa1215530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell J, Ryan CJ, Brough R, Bajrami I, Pemberton HN, Chong IY, et al. Large-Scale Profiling of Kinase Dependencies in Cancer Cell Lines. Cell Rep. 2016;14:2490–501. doi: 10.1016/j.celrep.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence RT, Perez EM, Hernandez D, Miller CP, Haas KM, Irie HY, et al. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015;11:630–44. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollestelle A, Nagel JH, Smid M, Lam S, Elstrodt F, Wasielewski M, et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat. 2010;121:53–64. doi: 10.1007/s10549-009-0460-8. [DOI] [PubMed] [Google Scholar]
- 22.Kim JW, Botvinnik OB, Abudayyeh O, Birger C, Rosenbluh J, Shrestha Y, et al. Characterizing genomic alterations in cancer by complementary functional associations. Nat Biotechnol. 2016;34:539–46. doi: 10.1038/nbt.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen A, Beetham H, Black MA, Priya R, Telford BJ, Guest J, et al. E-cadherin loss alters cytoskeletal organization and adhesion in non-malignant breast cells but is insufficient to induce an epithelial-mesenchymal transition. BMC Cancer. 2014;14:552. doi: 10.1186/1471-2407-14-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollestelle A, Peeters JK, Smid M, Timmermans M, Verhoog LC, Westenend PJ, et al. Loss of E-cadherin is not a necessity for epithelial to mesenchymal transition in human breast cancer. Breast Cancer Res Treat. 2013;138:47–57. doi: 10.1007/s10549-013-2415-3. [DOI] [PubMed] [Google Scholar]
- 25.Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27:2903–4. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–12. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 27.Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016;167:260–74 e22. doi: 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston SR, Leary A, Martin LA, Smith IE, Dowsett M. Enhancing endocrine response with novel targeted therapies: why have the clinical trials to date failed to deliver on the preclinical promise? Cancer. 2008;112:710–7. doi: 10.1002/cncr.23190. [DOI] [PubMed] [Google Scholar]
- 29.Chan CM, Martin LA, Johnston SR, Ali S, Dowsett M. Molecular changes associated with the acquisition of oestrogen hypersensitivity in MCF-7 breast cancer cells on long-term oestrogen deprivation. J Steroid Biochem Mol Biol. 2002;81:333–41. doi: 10.1016/s0960-0760(02)00074-2. [DOI] [PubMed] [Google Scholar]
- 30.Sikora MJ, Cooper KL, Bahreini A, Luthra S, Wang G, Chandran UR, et al. Invasive lobular carcinoma cell lines are characterized by unique estrogen-mediated gene expression patterns and altered tamoxifen response. Cancer Res. 2014;74:1463–74. doi: 10.1158/0008-5472.CAN-13-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu YJ, Shen D, Yin X, Gavine P, Zhang T, Su X, et al. HER2, MET and FGFR2 oncogenic driver alterations define distinct molecular segments for targeted therapies in gastric carcinoma. Br J Cancer. 2014;110:1169–78. doi: 10.1038/bjc.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Megiorni F, McDowell HP, Camero S, Mannarino O, Ceccarelli S, Paiano M, et al. Crizotinib-induced antitumour activity in human alveolar rhabdomyosarcoma cells is not solely dependent on ALK and MET inhibition. J Exp Clin Cancer Res. 2015;34:112. doi: 10.1186/s13046-015-0228-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dufies M, Jacquel A, Robert G, Cluzeau T, Puissant A, Fenouille N, et al. Mechanism of action of the multikinase inhibitor Foretinib. Cell Cycle. 2011;10:4138–48. doi: 10.4161/cc.10.23.18323. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Moreno M, Song W, Pasolli HA, Williams SE, Fuchs E. Loss of p120 catenin and links to mitotic alterations, inflammation, and skin cancer. Proc Natl Acad Sci U S A. 2008;105:15399–404. doi: 10.1073/pnas.0807301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van de Ven RA, de Groot JS, Park D, van Domselaar R, de Jong D, Szuhai K, et al. p120-catenin prevents multinucleation through control of MKLP1-dependent RhoA activity during cytokinesis. Nat Commun. 2016;7:13874. doi: 10.1038/ncomms13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukumoto Y, Shintani Y, Reynolds AB, Johnson KR, Wheelock MJ. The regulatory or phosphorylation domain of p120 catenin controls E-cadherin dynamics at the plasma membrane. Exp Cell Res. 2008;314:52–67. doi: 10.1016/j.yexcr.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roura S, Miravet S, Piedra J, Garcia de Herreros A, Dunach M. Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J Biol Chem. 1999;274:36734–40. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- 38.Ozawa M, Ohkubo T. Tyrosine phosphorylation of p120(ctn) in v-Src transfected L cells depends on its association with E-cadherin and reduces adhesion activity. J Cell Sci. 2001;114:503–12. doi: 10.1242/jcs.114.3.503. [DOI] [PubMed] [Google Scholar]
- 39.Ryan CJ, Kennedy S, Bajrami I, Matallanas D, Lord CJ. A Compendium of Co-regulated Protein Complexes in Breast Cancer Reveals Collateral Loss Events. Cell Syst. 2017;5:399–409 e5. doi: 10.1016/j.cels.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guillot C, Lecuit T. Adhesion disengagement uncouples intrinsic and extrinsic forces to drive cytokinesis in epithelial tissues. Developmental cell. 2013;24:227–41. doi: 10.1016/j.devcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Z, Manser E. Myotonic dystrophy kinase-related Cdc42-binding kinases (MRCK), the ROCK-like effectors of Cdc42 and Rac1. Small GTPases. 2015;6:81–8. doi: 10.1080/21541248.2014.1000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan I, Lai J, Yong J, Li SF, Leung T. Chelerythrine perturbs lamellar actomyosin filaments by selective inhibition of myotonic dystrophy kinase-related Cdc42-binding kinase. FEBS Lett. 2011;585:1260–8. doi: 10.1016/j.febslet.2011.03.054. [DOI] [PubMed] [Google Scholar]
- 43.Derksen PW, Braumuller TM, van der Burg E, Hornsveld M, Mesman E, Wesseling J, et al. Mammary-specific inactivation of E-cadherin and p53 impairs functional gland development and leads to pleomorphic invasive lobular carcinoma in mice. Dis Model Mech. 2011;4:347–58. doi: 10.1242/dmm.006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–49. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73:4885–97. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unbekandt M, Croft DR, Crighton D, Mezna M, McArthur D, McConnell P, et al. A novel small-molecule MRCK inhibitor blocks cancer cell invasion. Cell Commun Signal. 2014;12:54. doi: 10.1186/s12964-014-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brough R, Frankum JR, Sims D, Mackay A, Mendes-Pereira AM, Bajrami I, et al. Functional viability profiles of breast cancer. Cancer Discov. 2011;1:260–73. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller RE, Brough R, Bajrami I, Williamson CT, McDade S, Campbell J, et al. Synthetic Lethal Targeting of ARID1A-Mutant Ovarian Clear Cell Tumors with Dasatinib. Mol Cancer Ther. 2016;15:1472–84. doi: 10.1158/1535-7163.MCT-15-0554. [DOI] [PubMed] [Google Scholar]
- 49.Bajrami I, Kigozi A, Van Weverwijk A, Brough R, Frankum J, Lord CJ, et al. Synthetic lethality of PARP and NAMPT inhibition in triple-negative breast cancer cells. EMBO Mol Med. 2012;4:1087–96. doi: 10.1002/emmm.201201250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rottenberg S, Nygren AO, Pajic M, van Leeuwen FW, van der Heijden I, van de Wetering K, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci U S A. 2007;104:12117–22. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.