Abstract
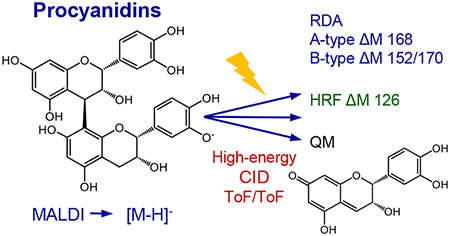
Although procyanidins constitute a unique class of polymeric plant secondary metabolites with a variety of biological properties including potent antioxidant activity, structure determination has been challenging and structures of many complex procyanidins remain uncertain. To expedite the characterization of procyanidins, negative ion matrix-assisted laser desorption ionization high-energy collision-induced dissociation tandem time-of-flight (MALDI-ToF/ToF) mass spectra of 20 isolated procyanidins containing catechin and epicatechin subunits with degrees of polymerization up to five were obtained and evaluated. Structurally significant fragmentation pathways of singly charged, deprotonated molecules were identified representing quinone methide, heterocyclic ring fission and retro-Diels-Alder fragmentation. The interpretation of the tandem mass spectra for sequencing A-type, B-type, mixed-type, linear, and branched procyanidins is explained using specific examples of each.
Keywords: procyanidin, MALDI, mass spectrometry, fragmentation
Graphical Absract
Introduction
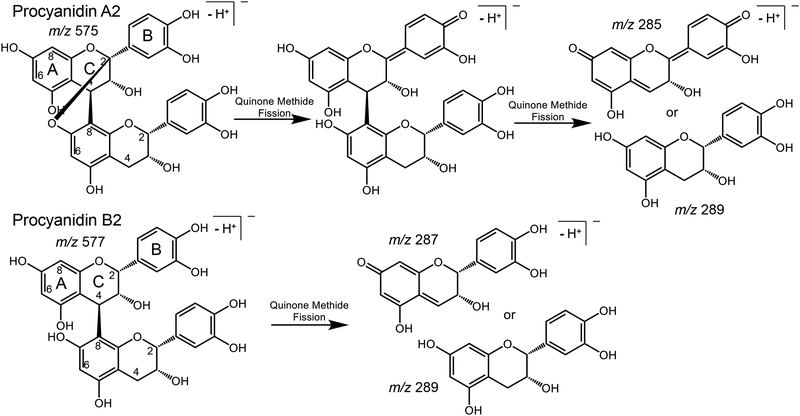
Also known as condensed tannins, proanthocyanidins occur in a wide variety of botanicals and contribute astringent flavor to foods.1 Different plant species produce distinct mixtures of gallotannins, ellagitannins and condensed tannins.2,3 Based on the hydroxylation patterns of their monomeric flavanol units and the linkages between these units in oligomers/polymers, proanthocyanidins can be divided into three types, one of the most common being procyanidins.4 The most widely studied procyanidins are dimeric procyanidin B and trimeric procyanidin C variants, both of which contain exclusively (+)-catechin and/or (−)-epicatechin monomeric units. Categorized into A-type and B-type, depending on linkages between the monomeric subunits (Figure 1), B-type procyanidins are characterized by a single carbon-carbon interflavan bond between the C-ring of one subunit and the A-ring of the next, whereas A-type procyanidins have not only a carbon-carbon interflavan bond but also a carbon-oxygen linkage (Figure 1).
Figure 1.
Quinone methide fragmentation of procyanidin dimers with A-type (procyanidin A2) and B-type (procyanidin B2) linkages.
Procyanidins have mixed stereochemistry and varying degrees of polymerization which can produce different biological activities.5 For example, procyanidins are potent antioxidants and have been reported to demonstrate antibacterial, antiviral, anti-carcinogenic, anti-inflammatory, as well as vasodilatory activities.6 It has been hypothesized that the free radical scavenging properties of procyanidins provide therapeutic benefits in preventing or treating cardiovascular disease, cancer, and urinary tract infections.7
Due to their complexity, the analysis of procyanidin mixtures is by no means straightforward. Various analytical techniques have been used for the analysis of procyanidins including nuclear magnetic resonance (NMR),6 circular dichroism,6 fast atom bombardment tandem mass spectrometry spectrometry,8 thermospray liquid chromatography mass spectrometry,9 electrospray liquid chromatography mass spectrometry,10–14 and matrix-assisted laser desorption ionization (MALDI) mass spectrometry.15−22 The structural elucidation of procyanidins using NMR is limited by the large amount of material that must be isolated prior to analysis compared with mass spectrometry, which has been used to identify procyanidins in mixtures at levels that are orders of magnitude lower than those required for NMR.13
During positive ion electrospray MS/MS of procyanidins, structurally significant fragment ions have been reported that form through retro-Diels-Alder (RDA) reaction, heterocyclic ring fission (HRF), benzofuran forming (BFF) reaction, and quinone methide (QM) fission.13,14 Despite the potential for electrospray to ionize high molecular mass procyanidins, the electrospray mass spectra of tannin mixtures are dominated by the lower molecular mass components, signal intensities diminish as polymer chain length increases, and the formation of multiply-charged oligomers complicates precursor ion selection for tandem mass spectrometric analysis.13
The ease of sample preparation, tolerance toward contaminants, simultaneous determination of mixtures, and rapid analysis of both low and high mass compounds help make MALDI-TOF mass spectrometry ideal for the analysis of procyanidins.15–22 MALDI also forms predominantly singly-charged ions thereby simplifying data interpretation and facilitating the selection of precursor ions for MS/MS analysis.23,24 Although MALDI-ToF MS/MS should be ideal for procyanidin characterization and identification, few studies of procyanidins using MALDI have used tandem mass spectrometry and, among these, even fewer have used negative ion mode. Instead, most MALDI-ToF mass spectrometric studies of procyanidins have used positive ion analysis to measure adducts with sodium, potassium, silver, or cesium.13–21 In this study, we established that negative ion MALDI forms abundant deprotonated molecules of procyanidins, eliminates the need for alkali metal cationization, and in combination with tandem mass spectrometry, forms a variety of structurally significant fragment ions. Overall, negative ion MALDI-ToF/ToF mass spectrometry provides simple determination of the degree of polymerization as well as the type of linkage between subunits for the rapid characterization of procyanidins from natural sources.
Materials and Methods
Materials. Procyanidins, which had been purified from cocoa, peanut skins, cinnamon, and crab apples, were obtained from Planta Analytica (New Milford, CT). The structures of all 20 of these procyanidins had been determined using mass spectrometry and NMR.25 The MALDI matrix, 2,5-dihydroxybenzoic acid (DHB), was purchased from Sigma-Aldrich (St. Louis, MO). All solvents were HPLC-grade and were purchased from Thermo Fisher (Pittsburgh, PA).
MALDI-ToF/ToF mass spectrometry. The DHB matrix was prepared at 15 mg/mL in 50% methanol (aqueous) and 0.1% trifluoroacetic acid. Each procyanidin solution (in 50% aqueous methanol) was mixed with the matrix solution (1:9; v:v), and 1 μL was deposited onto a ground stainless steel 384-density MALDI plate and allowed to air dry. Negative ion MALDI mass spectra and tandem mass spectra were acquired using an ultrafleXtremeToF/ToF mass spectrometer (Bruker Daltonics, Billerica, MA) in reflector mode. For single-stage mass spectrometry, the ion source 1 voltage was −20 kV, the ion source 2 voltage was −17.85 kV, the lens voltage was −7.5 kV, the pulsed ion extraction delay was 150 ns, the reflector voltage 1 was −20.8 kV, and reflector voltage 2 was −10.8 kV. The UV laser power was set to 10% above the ionization threshold, and the number of accumulated laser shots ranged from 1000 to 7000 depending on signal strength at a frequency of 2000 Hz.
During MS/MS, collision-induced dissociation was carried out using argon at 9.0 × 10−6 Torr, the ion source 1 voltage was −7.5 kV, ion source 2 voltage was −6.75 kV, the lens voltage was −3.5 kV, the pulsed ion extraction delay was 200 ns, the reflector 1 voltage was −29.5 kV, and the reflector 2 voltage was −14 kV. The laser power for MS/MS was set 25% above the ionization threshold, the number of accumulated laser shots ranged from 1500 to 4000 depending on signal strength of fragment ions, and the UV laser was operated at a frequency of 1000 Hz.
Results and Discussion
Negative ion fragmentation pathways of procyanidins
Negative ion MALDI ToF mass spectrometric analysis produced abundant deprotonated molecules of all 20 procyanidins (Table 1; Figure 2). The number of catechin and epicatechin subunits (degree of polymerization) in each molecule ranged from two to five based on the measured masses of the deprotonated molecules. Negative ion MALDI was selected over positive ion mode because the signal was concentrated into one abundant deprotonated molecule instead of being distributed among mixtures of [M+H]+, [M+Na]+, and [M+K]+ adducts, which have been reported for not only positive ion MALDI21 but also for positive ion fast atom bombardment8 and electrospray.12 In addition, no doping with cations of sodium, potassium, or cesium was necessary, as had been reported for positive MALDI mass spectrometry of procyandins.23,26
Table 1.
Negative Ion MALDI High-Energy Collision-Induced Dissociation ToF/ToF Diagnostic Fragment Ions of Procyanidins
| Procyanidin | [M-H]− | Quinone Methide (QM) | Retro Diels-Alder (RDA) (RDA–H2O) | Heterocyclic Ring Fragmentation (HRF) | QM +RDA +HRF | Type |
|---|---|---|---|---|---|---|
| Procyanidin B1 | 577.160 | 287, 289 | 425 (407) | 451 | 1B | |
| Procyanidin B2 | 577.152 | 287, 289 | 425 (407) | 451 | 1B | |
| Procyanidin B5 | 577.153 | 287, 289 | 425 (407) | 451 | 1B | |
| Procyanidin C1 | 865.191 | 287, 289, 575, 577 | 713 (695), 425 (407) | 739, 451, 413 | 2B | |
| Cocoa tetramer D | 1153.245 | 287, 289, 575, 577, 863, 865 | 1001 (983), 713 (695), 425 (407) | 1027, 739, 451 | 3B | |
| Cocoa pentamer E | 1441.305 | 287, 289, 575, 577, 863, 865, 1151, 1153 | 1289 (1271), 1001 (983), 713 (695), 425 (407) | 1315, 1027, 739 | 4B | |
| Procyanidin A1 | 575.127 | 285, 289 | 407 | 449 | 1A | |
| Procyanidin A2 | 575.130 | 285, 289 | 407 | 449 | 1A | |
| Cinnamtannin D1 | 863.170 | 285, 289, 573, 577 | 711 (693) | 737, 451, 411 | 299 | 1A1B |
| Cinnamtannin B1 | 863.151 | 285, 289, 573, 577 | 711 (693) | 737, 451, 411 | 299 | 1A1B |
| Lindetannin | 863.145 | 285, 289, 573, 577 | 711 (693) | 737, 451, 411 | 299 | 1A1B |
| Aesculitannin B | 863.140 | 285, 289, 573, 577 | 711 (693) | 737, 451, 411 | 299 | 1A1B |
| Parameritannin A1 | 1151.235 | 287, 289, 573, 861, 863 | 999 (981), 711 (693) | 1025, 735, 451, 411 | 299 | 1A2B |
| Cassiatannin A | 1151.235 | 287, 289, 573, 861, 863 | 999 (981), 711 (693) | 1025, 735, 451, 411 | 299 | 1A2B |
| Peanut trimer B25 | 861.163 | 285, 289, 571, 575 | 693 | 735, 449, 411 | 2A | |
| Peanut trimer D25 | 863.184 | 287, 289, 573, 575 | 711 (693) | 737, 449 | 1A1B | |
| Peanut trimer A25 | 863.189 | 287, 289, 573, 577 | 711 (693) | 737, 449, 411 | 299 | 1A1B |
| Peanut trimer C25 | 863.188 | 285, 289, 573, 577 | 711 (693) | 737, 451, 411 | 299 | 1A1B |
| Peanut tetramer E25 | 1149.217 | 285, 289, 573, 575, 859, 863 | 997 (979), 711 (693) | 1023, 411 | 2A1B | |
| Peanut tetramer F25 | 1149.218 | 285, 289, 573, 575, 859, 863 | 997 (979), 711 (693) | 1023, 411 | 2A1B |
Figure 2.
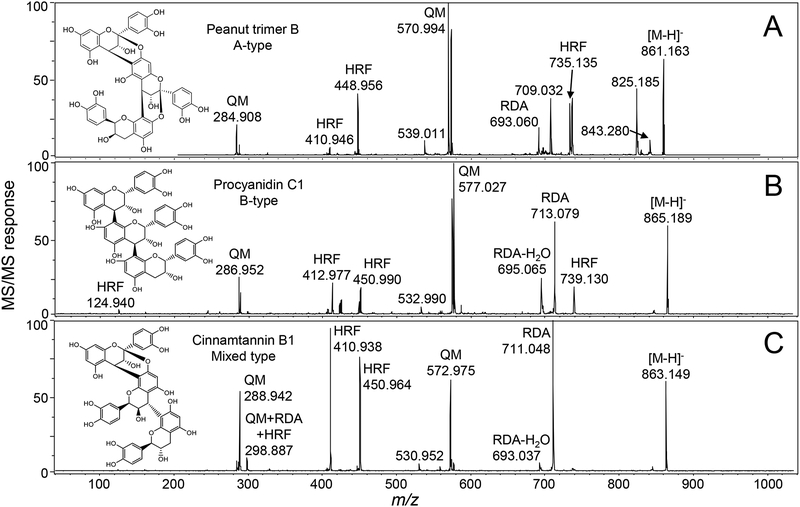
Comparison of negative ion MALDI-ToF/ToF mass spectra of procyanidin trimers with different types of bonding between the catechin/epicatechin subunits. (A) A-type procyanidin trimer with two bonds between monomeric subunits. (B) B-type procyanidin trimer with one bond between the subunits; and (C) mixed type procyanidin trimer with both A-type and B-type bonds between subunits.
Using high-energy collision-induced dissociation with negative ion tandem mass spectrometry, cleavages between monomeric subunits formed three types of class-characteristic and structurally significant product ions consisting of quinone methide (QM) (Figure 1), heterocyclic ring fission (HRF) (Figure 3), and retro-Diels-Alder (RDA) (Figure 4) fragment ions. These three procyanidin fragmentation pathways have also been reported to occur during positive ion electrospray tandem mass spectrometry of procyanidins.13,14 It should be noted that benzofuran formation, which occurs during positive ion MS/MS with collision-induced dissociation, was not observed using negative ion MALDI ToF/ToF mass spectrometry. These new data are significant because reports of MALDI ToF/ToF analysis of procyanidins are rare in the literature, and also because most previous papers utilized only positive ion mode. For example, Saldanha et al.27 used positive ion MALDI-ToF/ToF tandem mass spectrometry to characterize procyanidins up to six flavan-3-ol units in length. Sodium ions were added to enhance cationization, and although RDA fragmentation was observed, no HRF or QM fragment ions were described.
Figure 3.
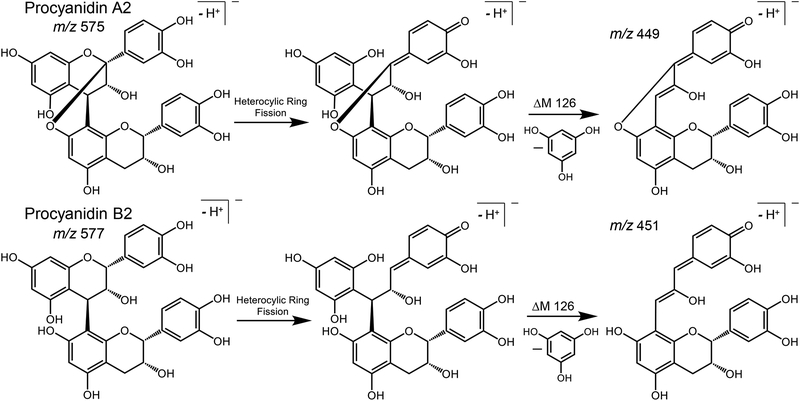
Heterocyclic ring fission fragmentation of procyanidins results in elimination of 126 Da from compounds containing A-type (procyanidin A2) and/or B-type (procyanidin B2) linkages.
Figure 4.
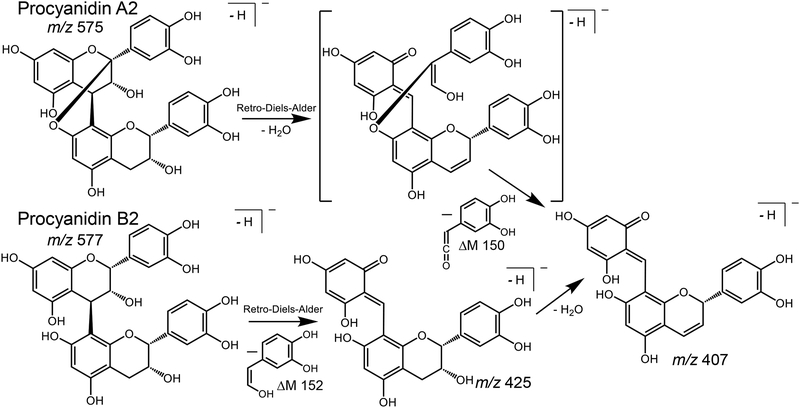
The retro-Diels-Alder fragmentation pathway for (A) A-type procyanidins; and (B) B-type procyanidins.
Every procyanidin tested formed fragment ions via the QM pathway that facilitated the identification of A-type and B-type linkages (Table 1). QM fragmentation cleaves both of the inter-subunit bonds of A-type procyanidins and the single bond between subunits in B-type procyanidins, so that the resulting fragment ions may be used to indicate the number of A-type and B-type linkages between monomeric subunits (Figure 1). During QM fragmentation, a diquinone will form in A-type procyanidins generating two possible product ions (Figure 1). B-type procyanidins form a single quinone resulting in two possible product ions, but the QM product ion containing the quinone will differ from the corresponding A-type diquinone ion by 2 Da (Figure 1). Examples of QM fragmentation include the pairs of ions of m/z 285/575 and m/z 571/289 for the A-type procyanidin peanut trimer B (Figure 2A) and m/z 287/577 and m/z 575/289 for the B-type procyanidin C1 (Figure 2B). Mixed-type procyanidins such as the trimer cinnamtannin B1 form combinations of QM fragment ions corresponding to cleavages of both A-type and B-type linkages (Table 1).
A second structurally significant fragmentation pathway for deprotonated procyanidins is heterocyclic ring fission (HRF), which results in the elimination of 1,3,5-trihydroxybenzene, [M-H-126]−, from both A-type and B-type compounds (Figure 3). Examples of HRF fragmentation include the ions of m/z 739, m/z 451 and m/z 413 in the product ion tandem mass spectra of procyanidin C1 (B-type), m/z 735, m/z 449 and m/z 411 for the peanut trimer B (A-type), and m/z 737, m/z 451, and m/z 411 for the mixed-type procyanidin cinnamtannin B1 (Figure 2). In some cases, deprotonated 1,3,5-trihydroxybenzene was detected at m/z 125 (Figure 2). Additional examples of HRF fragmentation are summarized in Table 1.
Retro-Diels-Alder (RDA) reactions are a third structurally significant procyanidin fragmentation pathway occurring during negative ion MALDI tandem mass spectrometry (Figure 4). For these B-type procyanidins composed of catechin and epicatechin subunits, RDA fragmentation was characterized by elimination of hydroxyvinyl benzenediol, [M-H-152]−, as well as loss of an additional molecule of water, [M-H-152–18]− (Table 1 and Figure 4). A pair of RDA fragment ions 18 u apart were observed for each B-type linkage in the procyanidin. For example, procyanidin C1, which contained two B-type linkages, formed ions of m/z 713/695 through RDA fragmentation and m/z 425/407 by RDA fragmentation of the QM product ion of m/z 577 (Figure 2B). Instead of forming pairs of RDA fragment ions 18 Da apart, A-type procyanidins formed single fragment ions, [M-H-168]− (Figure 4). For example, the A-type procyanidin dimers procyanidin A1 and procyanidin A2 formed a single RDA fragment ion of m/z 407, and the A-type peanut trimer B formed the single RDA fragment ion of m/z 693. RDA fragmentation of mixed-type procyanidins formed pairs of RDA ions for each B-type linkage but no RDA fragment ions for A-type linkages. As an example, mixed-type cinnamtannin B1 eliminated hydroxyvinyl benzenediol to form an abundant ion of m/z 711 plus a less abundant ion of m/z 693 corresponding to an additional loss of water (Figure 2C and Table 1).
Combinations of three fragmentation pathways (QM, RDA and HRF) contributed to the formation of the product ion of m/z 299. Observed in abundance for most mixed-type procyanidins (Table 1), this ion was formed by a QM A-type elimination of 286 u, RDA B-type loss of 152 u, and HRF elimination of 126 u. Examples included cinnamtannin B1 (Figure 2C), cinnamtannin D1, lindetannin, aesculitannin B, parameritannin A1, cassiatannin A, peanut trimer A, and peanut trimer C. The product ion of m/z 299 was below the limit of detection for the A-type and B-type procyanidins, and for the mixed-type procyanidins peanut trimers D, E and F.
Sequencing procyanidins using negative ion MALDI ToF/ToF
Based on the mass of the singly-charged deprotonated procyanidin formed during negative ion MALDI mass spectrometry, the total number of catechin and epicatechin subunits and the number of A-type and B-type linkages plus branching may be established. Next, determining the A-type or B-type bonding between procyanidin subunits may be accomplished by inspecting the QM fragment ions. Confirmatory data may be obtained by inspecting the HRF and RDA fragmentation of A-type and B-type procyanidins, which also show significant differences as noted above. By combining all this information, A-type, B-type, mixed-type, and branched procyanidins may be sequenced.
Sequencing linear procyanidins with only A-type or B-type linkages is straightforward, mixed-type linear procyanidins are more complicated, branched procyanidins are even more challenging, and sequencing mixed-type branched procyanidins are the most challenging. Beginning with a mixed-type linear example, the negative ion MALDI ToF/ToF mass spectrum of peanut tetramer E formed a deprotonated molecule of m/z 1149.217, which, based on mass alone, corresponds to four catechin/epicatechin subunits connected by 2 A-type and 1 B-type linkages. The RDA fragment ion of m/z 997 and its corresponding ion formed by an additional loss of water (m/z 979) confirm that peanut tetramer E contains one B-type linkage. The HRF fragment ion of m/z 411 indicates that the first linkage (connecting the middle A-ring of the first subunit to the C-ring of the second subunit) is A-type. Based on a deprotonated molecule of m/z 1149 and the QM series of fragment ions of m/z 863, m/z 575 and m/z 289, the initial linkage is confirmed to be A-type (ΔM 286), the middle linkage is B-type (ΔM 288), and the terminal linkage is A-type (ΔM 286). The complementary QM fragment ion series of m/z 285, m/z 573 and m/z 859 confirm that the initial linkage is A-type, the middle linkage is B-type (ΔM 288), and the terminal linkage is A-type (ΔM 286). Additional product ions in the negative ion MALDI tandem mass spectrum of peanut tetramer E included m/z 449/447 and m/z 737, which were formed by a combination of HRF and QM fragmentation.
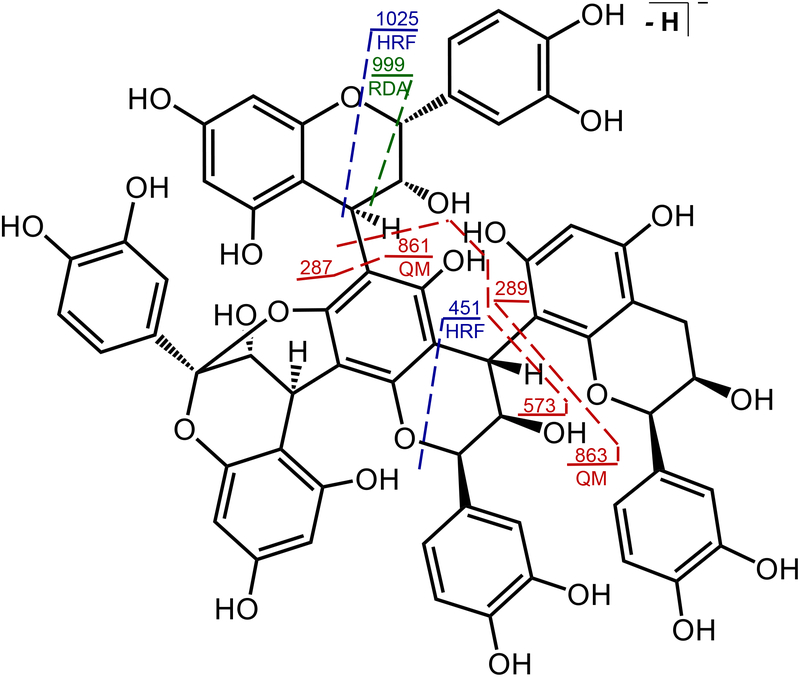
As a more complicated example, the deprotonated, mixed-type branched procyanidin parameritannin A1 (Figure 5), was measured at m/z 1151.235, which for a linear procyanidin would correspond to four catechin/epicatechin subunits and perhaps two A-type and one B-type linkage. However, the observation of two pairs of RDA fragment ions of m/z 999/981 and m/z 711/693 indicates the presence of two B-type linkages instead of just one. Therefore, this procyanidin must be branched to account for the missing two hydrogen atoms and contains two B-type linkages and one A-type linkage. The HRF ion of m/z 1025 is consistent with a mixed procyanidin containing one A-linkage and two B-linkages, and the HRF fragment ion of m/z 451 indicates that the bottom linkage is B-type. The most abundant QM ion of m/z 863 (ΔM 288) is consistent with loss of a B-type fragment. However, the next most abundant QM ion of m/z 573 is too low in mass to be a typical A-type or B-type fragment ion, and no ions of m/z 575 or m/z 577 were detected. Therefore, the second catechin/epicatechin subunit is in the middle and serves as a branch point (Figure 5).
Figure 5.
The connectivity of catechin/epicatechin subunits within the mixed, branched procyanidin tetramer parameritannin A1.
Requiring no chromatographic separation step, MALDI ToF/ToF mass spectrometry is inherently fast and provides structural information for procyanidins that includes not only the number of A-type and B-type linkages but also their locations in the oligomer. Negative ion MALDI procyanidin ToF and ToF/ToF mass spectra are easier to interpret than positive ion mass spectra, due to the signals being concentrated in the deprotonated molecules instead of distributed among several cationized species,13,14 and because only QM, RDA and HRF pathways but no benzofuran forming product ion species are produced. Negative ion MALDI ToF/ToF mass spectrometry facilitates the rapid and nearly complete structure determination of oligomeric procyanidins and need only be complemented by specific NMR experiments to determine stereochemistry and confirm structure.
Supplementary Material
Acknowledgments
Funding Sources
Funding for this research was provided by National Institutes of Health grants R01 AT007659 and F31 AT009039 from the National Center for Complementary and Integrative Health.
Abbreviations Used
- HRF
heterocyclic ring fission
- MALDI
matrix-assisted laser desorption ionization
- QM
quinone methide
- RDA
retro-Diels-Alder
- ToF
time-of-flight
Footnotes
Conflict of Interest
The authors declare no competing financial interest.
References
- (1).USDA Database for the Proanthocyanidin Content of Selected Foods. Nutrient Data Laboratory, Beltsville Human Nutrition Research Center, ARS, USDA: Beltsville, MA: 2004, p 33. [Google Scholar]
- (2).Hernes PJ; Hedges JI Tannin signatures of barks, needles, leaves, cones, and wood at the molecular level. Geochim. Cosmochim. Acta 2004, 68, 1293–1307. [Google Scholar]
- (3).Okuda T; Yoshida T; Hatano T Correlation of oxidative transformations of hydrolyzable tannins and plant evolution. Phytochemistry 2000, 55, 513–529. [DOI] [PubMed] [Google Scholar]
- (4).Hellström JK; Törrönen AR; Mattila PH Proanthocyanidins in common food products of plant origin. J. Agric. Food Chem 2009, 57, 7899–7906. [DOI] [PubMed] [Google Scholar]
- (5).Xie DY; Dixon RA Proanthocyanidin biosynthesis--still more questions than answers? Phytochemistry. 2005, 66, 2127–2144. [DOI] [PubMed] [Google Scholar]
- (6).Esatbeyoglu T; Jaschok-Kentner B; Wray V; Winterhalter P Structure elucidation of procyanidin oligomers by low-temperature 1H NMR spectroscopy. J. Agric. Food Chem 2011, 59, 62–69. [DOI] [PubMed] [Google Scholar]
- (7).Han XZ; Shen T; Lou HX Dietary polyphenols and their biological significance. Int. J. Mol. Sci 2007, 8, 950–988. [Google Scholar]
- (8).Karchesy JJ; Hemingway RW; Foo YL; Barofsky E; Barofsky DF Sequencing procyanidin oligomers by fast atom bombardment mass spectrometry. Anal. Chem 1986, 58, 2563–2567. [Google Scholar]
- (9).Kiehne A; Lakenbrink C; Engelhardt UH Analysis of proanthocyanidins in tea samples I. LC-MS results. Z. Lebensm. -Unters. -Forsch. A 1997, 205, 153–157. [Google Scholar]
- (10).Gu L; Kelm MA; Hammerstone JF; Zhang Z; Beecher G; Holden J; Haytowitz D; Prior RL Liquid chromatographic/electrospray ionization mass spectrometric studies of proanthocyanidins in foods. J. Mass Spectrom 2003, 38, 1272–1280. [DOI] [PubMed] [Google Scholar]
- (11).Friedrich W; Eberhardt A; Galensa R Investigation of proanthocyanidins by HPLC with electrospray ionization mass spectrometry. Eur. Food Res. Technol 2000, 211, 56–64. [Google Scholar]
- (12).Hammerstone JF; Lazarus SA; Mitchell AE; Rucker R; Schmitz HH Identification of procyanidins in cocoa (Theobroma cacao) and chocolate using high-performance liquid chromatography/mass spectrometry. J. Agric. Food Chem 1999, 47, 490–496. [DOI] [PubMed] [Google Scholar]
- (13).Yang Y; Chien M Characterization of grape procyanidins using high-performance liquid chromatography/mass spectrometry and matrix-assisted laser Desorption/Ionization time-of-flight mass spectrometry. J. Agric. Food Chem 2000, 48, 3990–3996. [DOI] [PubMed] [Google Scholar]
- (14).Oliveira J; Alhinho Da Silva M; Teixeira N; De Freitas V; Salas E Screening of anthocyanins and anthocyanin-derived pigments in red wine grape pomace using LC-DAD/MS and MALDI-TOF techniques. J. Agric. Food Chem 2015, 63, 7636–7644. [DOI] [PubMed] [Google Scholar]
- (15).Li HJ; Deinzer ML Tandem mass spectrometry for sequencing proanthocyanidins. Anal. Chem 2007, 79, 1739–1748. [DOI] [PubMed] [Google Scholar]
- (16).Pérez-Jiménez J; Torres JL Analysis of proanthocyanidins in almond blanch water by HPLC–ESI–QqQ–MS/MS and MALDI–TOF/TOF MS. Food Res. Int 2012, 49, 798–806. [Google Scholar]
- (17).Perez-Gregorio MR; Mateus N; de Freitas V Rapid screening and identification of new soluble tannin-salivary protein aggregates in saliva by mass spectrometry (MALDI-TOF-TOF and FIA-ESI-MS). Langmuir 2014, 30, 8528–8537. [DOI] [PubMed] [Google Scholar]
- (18).Hurst WJ; Stanley B; Glinski JA; Davey M; Payne MJ; Stuart DA Characterization of primary standards for use in the HPLC analysis of the procyanidin content of cocoa and chocolate containing products. Molecules 2009, 14, 4136–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).De Marchi F; Seraglia R; Molin L; Traldi P; Dalla Vedova A; Gardiman M; De Rosso M; Flamini R Study of isobaric grape seed proanthocyanidins by MALDI-TOF MS. J. Mass Spectrom 2014, 49, 826–830. [DOI] [PubMed] [Google Scholar]
- (20).Van Huynh A; Bevington JM MALDI-TOF MS analysis of proanthocyanidins in two lowland tropical forest species of Cecropia: a first look at their chemical structures. Molecules 2014, 19, 14484–14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Li H-J; Deinzer ML The mass spectral analysis of isolated hops A-type proanthocyanidins by electrospray ionization tandem mass spectrometry. J. Mass Spectrom 2008, 43, 1353–1363. [DOI] [PubMed] [Google Scholar]
- (22).Ohnishi-Kameyama M; Yanagida A; Kanda T; Nagata T Identification of catechin oligomers from apple (Malus pumila cv. Fuji) in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and fast-atom bombardment mass spectrometry. Rapid Commun. Mass.Spectrom 1997, 11, 31–36. [DOI] [PubMed] [Google Scholar]
- (23).Monagas M; Quintanilla-López J; Gómez-Cordovés C; Bartolomé B; Lebrón-Aguilar R MALDI-TOF MS analysis of plant proanthocyanidins. J. Pharm. Biomed. Anal 2010, 51, 358–372. [DOI] [PubMed] [Google Scholar]
- (24).Rue EA; Rush MD; van Breemen RB Procyanidins: a comprehensive review encompassing structure elucidation via mass spectrometry. Phytochem. Rev 2018, 17, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dudek MK; Gliński VB; Davey MH; Sliva D; Kaźmierski S; Gliński JA Trimeric and tetrameric A-type procyanidins from peanut skins. J. Nat. Prod 2017, 80, 415–426. [DOI] [PubMed] [Google Scholar]
- (26).Jerez M; Sineiro J; Guitián E; Núñez MJ Identification of polymeric procyanidins from pine bark by mass spectrometry. Rapid Commun. Mass Spectrom 2009, 23, 4013–4018. [DOI] [PubMed] [Google Scholar]
- (27).Saldanha AA; de Siqueira JM; Castro AH; de Azambuja Ribeiro RI; de Oliveira FM; de Oliveira Lopes D; Pinto FC; Silva DB; Soares AC Anti-inflammatory effects of the butanolic fraction of Byrsonima verbascifolia leaves: Mechanisms involving inhibition of tumor necrosis factor alpha, prostaglandin E(2) production and migration of polymorphonuclear leucocyte in vivo experimentation. Int. Immunopharmacol 2016, 31, 123–131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.