Abstract
Klebsiella pneumoniae is the second leading causative agent of UTI. In this study, a rapid combined polymerase chain reaction and restriction fragment length polymorphism analysis was developed to identify K. pneumoniae in women, infected with urinary tract infection in the Sylhet city of Bangladesh. Analysis of 11 isolates from women at the age range of 20–55 from three different hospitals were done firstly by amplification with K. pneumoniae specific ITS primers. All of the 11 collected isolates were amplified in PCR and showed the expected 136 bp products. Then, restriction fragment length polymorphism analysis of 11 isolates were conducted after PCR amplification by 16s rRNA universal primers, followed by subsequent digestion and incubation with two restriction enzymes, Pst1 and Alu1. Seven out of 11 isolates were digested by Pst1 restriction enzymes, six isolates digested by Alu1, and while others were negative for both enzymes. Data results reveal that, women at age between 25 and 50 were digested by both enzymes. A woman aged over than 50 was negative while bellow 20 was digested by only Pst1. The results could pave the tactic for further research in the detection of K. pneumoniae from UTI infected women.
Keywords: Klebsiella pneumoniae, ITS-primer, MDR isolates, PCR-RFLP analysis
1. Introduction
Klebsiella pneumoniae is the second most potential agent of urinary tract infection after Escherichia coli, however, the pathogenicity is higher than its counterpart [1]. Approximately 12% of UTI infection caused by K. pneumoniae and the number is increasing at an alarming rate all over the world, particularly in Asia, due to spread of antibiotic resistant and extended beta-lactamase strains [2]. Women are eight times more vulnerable to UTI infection due to their position of reproductive organs and many of the infections remain asymptomatic for prolonged period [3]. The incidence rate increases with age, recurrent infections (very common for women), and during pregnancy period [4], [5]. In Bangladesh, due to geographical position, weather, food habit, early age pregnancy, and lack of awareness about UTI: the numbers of patients infected by K. pneumoniae have been proliferated in the last couple of years [6], [7]. Several researches have been conducted on Escherichia coli associated UTI, but molecular based approach for the detection and analysis of K. pneumoniae causing UTI in women has yet to be developed.
PCR alone or sometimes in combination with RFLP has been extensively used for precise detection and analysis of pathogens for many years [8]. Traditional culture based technologies are time consuming, labor intensive, and sometimes frequent use of antibiotics may affect culture positive isolates thus difficult to interpret data correctly [9]. However, PCR based molecular approaches are independent of antibiotics, more rapid, reliable, and sensitive, thus routinely used as molecular tools for pathogen identification [10]. 16s-23s internal transcribed spacer (ITS) unit of K. pneumoniae facilitating precise identification of this organism by polymerase chain reaction (PCR) [11]. Restriction endonuclease digestion of PCR products enables species determination and analysis of genome variability [12]. The sequence specific RFLP pattern of bacteria amplified from 16s rDNA primers varied widely from species to species, and the conserved sequence likely to be differentiated by PCR-RFLP method [13]. Restriction endonuclease digestion of bacterial DNA by Pst1, Alu1, and Mob1 have been used to confirm etiological agents in some earlier studies [14], [15], [16].
Multidrug resistant K. pneumoniae cause an emerging health threat worldwide, especially in least developed, and densely populated countries [17]. Current treatment practice commonly prescribe powerful antibiotics resulting spread of multidrug resistant bacteria and thereby reducing therapeutic efficacy [18]. In order to implement a successful treatment strategy for UTI, it is of great importance to know the current antibiotic resistant profile of the causative agents [17], [18]. Early detection of K. pneumoniae from UTI could minimize the widespread use of antibiotics in prevention and control programs as well as reduce the medical cost. The objective of this study was to evaluate 16s-23s ITS primer and PCR-RFLP method as a tools for the identification of multi-drug resistant (MDR) K. pneumoniae causing UTI in women.
2. Materials and methods
2.1. Collection and culture of bacterial isolates
A total of 11 bacterial isolates were collected from three different hospitals of Sylhet city of Bangladesh: Sylhet MAG Osmani Medical College and Hospitals, Popular Hospitals and Diagnostic Centre, and Jalalabad Ragib Rabeya Medical College and Hospitals. Immediately after collection, isolates were transported to USDA project laboratory of the Department of Genetic Engineering and Biotechnology of Shahjalal University of Science and Technology by maintaining cool chain. Isolates were cultured in ESBL medium and incubated overnight at 37 °C. Isolates were then numbered numerically from K1 to K11 for further studies. UTI patient’s data (Table 1) were collected from doctor’s consent form and recorded for future analysis.
Table 1.
Isolates with their isolation history.
| Isolates | Age | Physical status of the patient | Infection type | Hospital |
|---|---|---|---|---|
| K1 | 55 | Healthy | First time | Sylhet MAG Osmani Medical College |
| K2 | 35 | Fever, stomach pain | First time | Sylhet MAG Osmani Medical College |
| K3 | 18 | Malnutrition | First time | Jalalabad Ragib Rabeya Medical College and Hospitals |
| K4 | 22 | Secondary bacterial infection by mycoplasma | Re-current | Popular Hospital and Diagnostic Centre, Sylhet |
| K5 | 32 | Stomach pain, flatulence | Re-current | Popular Hospital and Diagnostic Centre, Sylhet |
| K6 | 28 | Healthy | First time | Sylhet MAG Osmani Medical College |
| K7 | 25 | Stomach pain ketosis (Pregnant) | First time | Popular Hospital and Diagnostic Centre, Sylhet |
| K8 | 38 | Secondary bacterial infection by chlamydia | Re-current | Popular Hospital and Diagnostic Centre, Sylhet |
| K9 | 19 | Stomach pain, flatulence | First time | Popular Hospital and Diagnostic Centre, Sylhet |
| K10 | 52 | Healthy | First time | Jalalabad Ragib Rabeya Medical College and Hospitals |
| K11 | 36 | Secondary bacterial infection | First time | Jalalabad Ragib Rabeya Medical College and Hospitals |
2.2. Genomic DNA extraction
All of the bacterial isolates were streaked in trypticase soy (TCS) agar medium for colony formation and incubated at 37 °C for overnight. A single colony was picked and grown over night at 37 °C on TCS broth in a shaker incubator for genomic DNA extraction. DNA of 11 bacteria were extracted by following the instructions of commercial genomic DNA extraction kit (Bio Basic Inc., 160 Torbay Road, Markham Ontario, Canada). Additionally, proteinase K and RNase A added after incubation step for purified DNA according to the guidelines of extraction kit. Extracted DNA were quantified by gel electrophoresis with lambda (λ-DNA marker as well as in a spectrophotometer as a ratio of DNA-protein absorbance. DNA was then stored at −20 °C for further use.
2.3. Identification of Klebsiella pneumoniae by PCR
For identification of K. pneumoniae, 16s-23s ITS primer was used to amplify DNA sequence in this study [11]. PCR master mixture was prepared in 50 µl volume containing 25 µl of 2X master mixtures (Fermentus, Gene Ruller™, USA), 2.5 µl of each forward and reverse primer (Table 2), 5 µl of template DNA (100 ng) and 15 µl of nuclease free water. PCR conditions consisted of an initial denaturation temperature of 94 °C for 4 min; denaturation step of 94 °C for 1 min, annealing for 1 min at 55 °C, and an extension at 72 °C for 1.5 min, a final extension step of 72 °C for 10 min and 4 °C for final storage. A total of 35 serial cycles of amplification reaction was performed in a MultiGene Gradient Thermal Cycler (Labnet International Inc., USA). PCR products were separated on 1.5% agarose gel followed by subsequent staining in ethidium bromide solution and visualized in a gel documentation system.
Table 2.
Primers and restriction enzymes used for the present study.
| Target | Primer name | Sequence | Size | Reference |
|---|---|---|---|---|
| Klebsiella pneumoniae 16s-23s ITS primer | K. pneumoniae Pf | ATTTGAAGAGGTTGCAAACGAT | 130 bp | [11] |
| K. pneumoniae PrA | TTCACTCTGAAGTTTTCTTGTGTTC | |||
| Universal sequence | 27F | 5′-AGAGTTTGATCCTGGCTCAG-3′ | 1513 bp | [28] |
| 1540R | 5′-AGGAGGTGATCCAACCGCA-3′ |
| Target | Restriction enzyme | Recognition sequence | Reference |
| PCR amplified K. pneumoniae 16s rRNA | Pst1 | 5′-CTGCAG-3′ | [16] |
| Alu1 | 5′-AGCT-3′ | ||
2.4. Amplification of bacterial 16s rDNA by universal PCR
PCR master mixture was adjusted at 30 µl final volume contained 15 µl of 2X master mixtures (Fermentus, Gene Ruller™, USA), 1.5 µl of each universal 27F forward and 1540R reverse primers (Table 2), 2 µl of template DNA and 10 µl of nuclease free water. Here a total of 30 cycles of reaction was programmed in MultiGene gradient thermal cycler (Labnet International Inc. USA) with an initial denaturation temperature of 94 °C for 4 min; denaturation step of 95 °C for 1.5 min, annealing for 1.5 min at 58 °C for, an extension at 72 °C for 1.5 min, a final extension step of 72 °C for 5 min, and 4 °C for final storage.
2.5. Restriction digestion
After 16s rDNA PCR, 10 µl of PCR product was transferred to a separate eppendorf and 18 µl of nuclease free water added. Then, 2 µl of Pst1 and Alu1 restriction enzymes (Table 2) premixed with BSA were added carefully to the solution. Restriction enzyme added samples were then spin gently for few seconds and incubated at 37 °C for 2 h in a water bath [12]. Fragments then analyzed in 2% agarose on 10% TBE under UV illumination. A molecular weight marker (1kb DNA ladder, Fermentus, GeneRuller™, USA) was added for each of the gel run.
2.6. Antibiogram assay of the isolates
Antibiotic profiling of the K. pneumoniae isolates to 10 commercial antibiotic discs were performed by disc diffusion assay [17], [18]. The antibiotic discs used in this study were ampicilin (10 µg/disk), kanamycin (30 µg/disk), erythromycin (15 µg/disk), chloramphenicol (30 µg/disk), levofloxacin (5µg/disk), ciprofloxacin (30 µg/disk), cefradine (25 µg/disk), gentamicin (10 µg/disk), streptomycin (10 µg/disk), and sulphamethoxazole (25 µg/disk). Overnight bacterial culture (30 µl) was inoculated on Tryptocasein Soy Agar plates (Micromaster, India) by spreading and discs were placed aseptically onto the culture. After 24 h of incubation at 37 °C, the zone of inhibition was measured according to Sharmin et al. [31].
3. Results
3.1. Identification of K. pneumoniae isolates
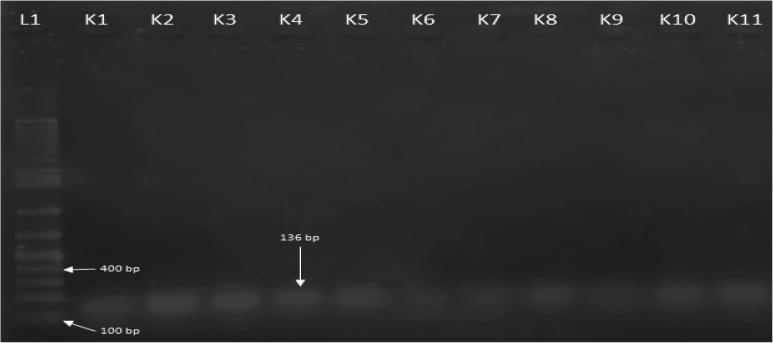
All the bacterial isolates from three different hospitals were initially supplied as K. pneumoniae grown on selective agar media supplemented with ornithine, raffinose and koser citrate [21]. However, for further confirmation, bacterial isolates were assayed for their morphological, physiological and biochemical properties according to Burgey’s manual for the identification of K. pneumoniae [22]. All of the isolates were confirmed belong to K. pneumoniae after biochemical tests. The isolates were also confirmed by amplification in PCR with the 16s-23s internal transcribed spacers (Fig. 1).
Fig. 1.
PCR amplification of K. pneumoniae from women with UTI by 16s-23s ITS primer. L1: 1 kb DNA ladder, K1-K11 amplified K. pneumoniae isolates.
3.2. PCR-RFLP analysis
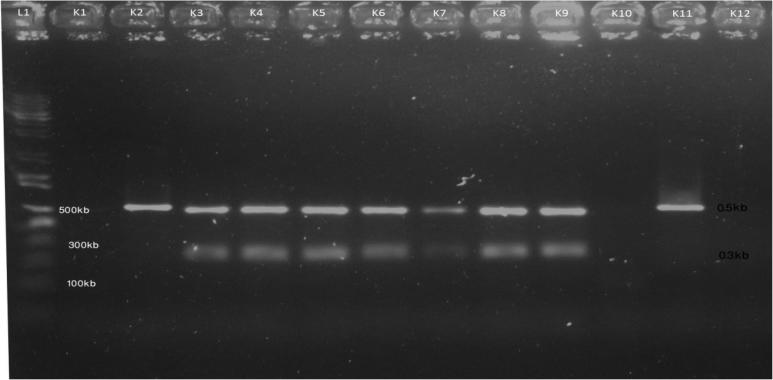
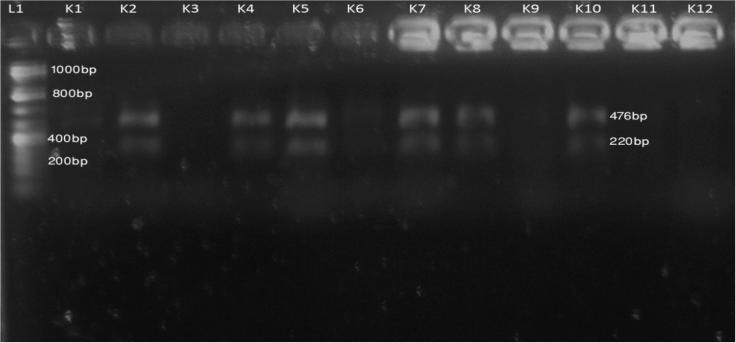
Among the 11 isolates, seven (K3, K4, K5, K6, K7, K8 and K9) were digested by Pst1 after 2 h incubation, two isolates (K2 and K11) were amplified in PCR but not digested by Pst1 and two others were negative in PCR amplification (Fig. 2). In other experiment, six isolates (K2, K4, K5, K7, K8 and K10) were fragmented by Alu1, two isolates (K1 and K6) gave negative Alu1 digestion while three others were negative in PCR amplification (Fig. 3).
Fig. 2.
PCR-RFLP digestion pattern of 16s rDNA sequence of K. pneumoniae by Pst1. L1: 1 kb DNA ladder, K1-negative control, K2-K12 digested K. pneumoniae isolates after incubation.
Fig. 3.
PCR-RFLP digestion pattern of 16s rDNA sequence of K. pneumoniae by Alu1. L1: 1 kb DNA ladder, K1-negative control, K2-K12 digested K. pneumoniae isolates after incubation.
3.3. Antibiotic sensitivity of K. pneumoniae isolates
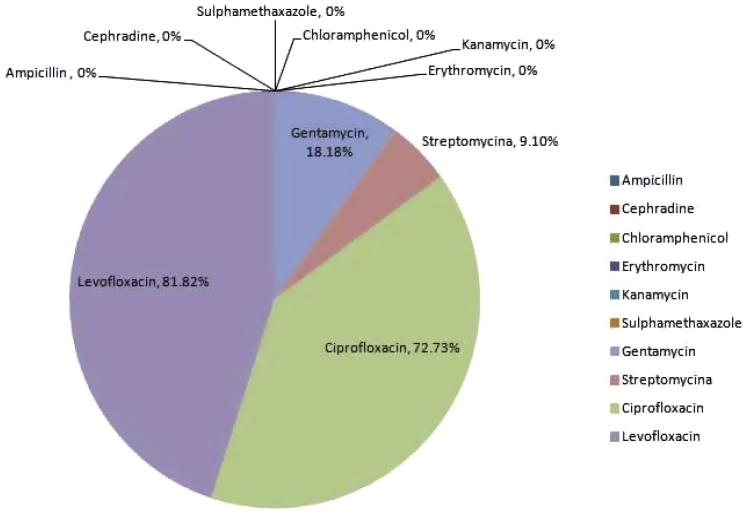
To know the antibiogram profile of the isolates, we screened ten antibiotics representing different antibiotic groups. All of the isolates showed resistant to ampicillin, erythromycin, chloramphenicol, cephradine, kanamycin and sulphamethaxazole. Sensitivity (approximately 80%) only observed for two antibiotics, ciprofloxacin and levofloxacin. Streptomycin and gentamycin showed sensitivity to only K3 isolate (younger patient), but neither showed 100% sensitivity to all of the tested isolates. Therefore, all the K. pneumoniae isolates were resistant to multiple antibiotics tested in this study (Fig. 4).
Fig. 4.
Antibiotic sensitivity pattern of K. pneumoniae isolates.
4. Discussion
K. pneumoniae cause a wide variety of diseases in both humans and animals. Among these diseases, urinary tract infection is one of the common that cause serious health threat to women, especially to the pregnant and immunocompromised person [23], [24]. In recent years, the prevalence of UTI infection caused by K. pneumoniae has been increased in Asia including Bangladesh [6]. Data results also suggest that women during pregnancy witnessed recurrent UTI infection are also suffered from other bacterial infections (Chlamydia and Mycoplasma) [5]. Although pronounce effects, no experimental data available for molecular detection of UTI causing K. pneumoniae and its 16s rRNA restriction digestion analysis in infected women in Bangladesh. Therefore, this study could be used as a platform for rapid detection, virulence properties, and drug sensitivity pattern of K. pneumoniae associated UTI in women.
Biochemical characterization and other media based identification of K. pneumoniae often give false positive results and required considerable amount of time for confirmation [25]. Therefore PCR method has been widely used for precious detection of pathogens and analysis of their genetic diversity [26], [27]. Previous PCR based researches for the detection of K. pneumoniae by 16s-23s internal transcribed spacer although successful for most of the isolates but didn’t produce perfection [2], [11], [26]. The PCR in this study was outstanding in amplification of all the 11 isolates and was so sensitive that it produced reproducible results in repetitive experiments.
Sequence specific enzymatic cleavage of amplified 16s rRNA allows precise and early diagnosis of diseases [28]. In this study, RFLP digestion of 16 rRNA produced a distinct pattern of cleavage, size ranging from 0.5 kb to 0.75 kb. Restriction enzyme Pst1 developed three types of banding pattern; seven isolates were fragmented and produced bands of the same sizes. Enzyme Alu1 produced digestion pattern had close proximity to Pst1; six isolates were cleaved consistently at the same length. Sharma et al. conducted a study on the detection of E. coli and K. pneumoniae from tertiary care hospital of India and performed restriction digestion analysis with EcoR1 and Pst1, where 60% of isolates were positive for Pst1, and gave bands at molecular weight of 150 bp to 750 bp [28]. In present study, we found 64% digestion of K. pneumoniae 16s rRNA, a slightly more sensitivity to Pst1 digestion than the Sharma’s study in 2010. This is probably due to similar circulating strains spreading over the South-East Asia [28]. For Alu1, Kalghatgi et al. performed an experiment to differentiate K. pneumoniae from other pathogenic bacteria using some restriction enzymes where 60% of K. pneumoniae isolates were digested by Alu1 and showed band at 476 bp, 220 bp and 65 bp [29]. In this study, 63.4% of isolates were sensitive to Alu1 and displayed similar banding pattern like earlier study (476 bp and 220 bp) [29].
Another significant finding of this study was the homogenous banding pattern of isolates from pregnant (K7) women and recurrent UTI (K4, K5 and K8) patients, suggesting a common evolutionary origin for all of these isolates [28]. In addition, patient’s data reveals that these samples came from the same hospital (Popular Hospitals and Diagnostic Centre) and community (slum). Moreover, the isolates were resistant to all of the commercial antibiotics tested. Therefore, environmental factors and food habit might play some role for recurrent infection and drug resistant pattern [27], [31].
The growth curve for antibiotic resistant pattern has been constantly increased at an alarming rate in Bangladesh. In 2012, 60% of UTI causing K. pneumoniae were resistant to commercial drugs [31]. However, in 2016, resistancy pattern increased by 20% to common antibiotics used to treat UTI [27]. Present study found over 90% of the isolates were resistant to multiple antibiotics tested. The availability and frequent use of antibiotics possibly responsible for this upward resistance pattern in Bangladesh [7].
Finally, rather treating K. pneumoniae associated UTI by antibiotics, we have to put more emphasis on early detection methods. Antibiotics based common treatment strategy add further complicacy to UTI patients [31]. Therefore, a rapid and precise molecular method needed to address the problem, and PCR-RFLP here could be a simple, selective, and cost effective alternative of the traditional culture based method in Bangladesh.
Acknowledgments
Acknowledgement
The present study was conducted under the research project titled “Virulent gene targeting and analysis of virulence factors of K. pneumoniae causing pneumonia and urinary tract infection (UTI) in Bangladesh” funded by Ministry of Science and Technology, Bangladesh (MOST).
Conflict of interest
The author declares no conflict of interest.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
Contributor Information
Golam Mahmudunnabi, Email: mnrabbee@gmail.com.
Al Nahian Khan Majlish, Email: nahiankm@gmail.com.
Farhana Momtaz, Email: sharnafarhana@gmail.com.
Md Javed Foysal, Email: mjfoysal-geb@sust.edu.
Md Mahbubur Rahman, Email: mahbub-biotech@bsmrau.edu.bd.
Kamrul Islam, Email: kamrul-gen@sust.edu.
References
- 1.Farajnia S., Alikhani M.Y., Ghotaslou R., Naghili B., Nakhlband A. Causative agents and antimicrobial susceptibilities of urinary tract infections in the northwest of Iran. Int J Infect Dis. 2009;13(2):140–144. doi: 10.1016/j.ijid.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Lopes A.C.S., Rodrigues J.F., Clementino M.B.M., Miranda C.A.C., Nascimento A.P.A., De Morais M.A. Application of PCR ribotyping and tDNA-PCR for Klebsiella pneumoniae identification. Mem Inst Oswaldo Cruz. 2007;102(7):827–832. doi: 10.1590/s0074-02762007005000113. [DOI] [PubMed] [Google Scholar]
- 3.Al-Badr A., Al-Shaikh G. Recurrent urinary tract infections management in women: a review. Sultan Qaboos Univ Med J. 2013;13(3):359–367. doi: 10.12816/0003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Josip C., Sheikh A. Recurrent urinary tract infection in women: can cranberry prevent it? BMJ. 2003;327(7425) doi: 10.1136/bmj.327.7425.1204. 1204–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matuszkiewicz-Rowińska J., Małyszko J., Wieliczko M. Urinary tract infections in pregnancy: old and new unresolved diagnostic and therapeutic problems. Arch Med Sci. 2015;11(1):67–77. doi: 10.5114/aoms.2013.39202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begum N., Aba M., Hossain M., Begum N., Sa C., Mf R. UTI among female workers in a selected garment industry of Dhaka city: a cross sectional study. 2006;23(January):325–327. [Google Scholar]
- 7.Rahman S.R., Ahmed M.F., Begum A. Occurrence of urinary tract infection in adolescent and adult women of shanty town in Dhaka City, Bangladesh. Ethiop J Health Sci. 2014;24(2):145–152. doi: 10.4314/ejhs.v24i2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalghatgi A.T., Praharaj A.K., Sahni A.K., Pradhan D., Kumaravelu S., Prasad P.L. Detection of bacterial pathogens in cerebrospinal fluid using restriction fragment length polymorphism. Med J Armed Forces India. 2008;64(1):29–32. doi: 10.1016/S0377-1237(08)80141-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gholoobi Aida, Masoudi-Kazemabad Ali, Meshkat Mojtaba, Meshkat Zahra. Comparison of culture and PCR methods for diagnosis of Mycobacterium tuberculosis in different clinical specimens. Jundishapur J Microbiol. 2014;7(2):e8939. doi: 10.5812/jjm.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarridge, Jill E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17(4):840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y., Liu C., Zheng W. PCR detection of Klebsiella pneumoniae in infant formula based on 16S–23S internal transcribed spacer. Int J Food Microbiol. 2008;125(3):230–235. doi: 10.1016/j.ijfoodmicro.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Okhravi N., Adamson P., Matheson M.M., Towler H.M.A., Lightman S. PCR-RFLP-mediated detection and speciation of bacterial species causing endophthalmitis. Investig Ophthalmol Vis Sci. 2000;41(6):1438–1447. [PubMed] [Google Scholar]
- 13.Rahmani S., Mosavp M., Rezaeej A. Detection of bacteria by amplifying the 16S rRNA gene with universal primers and RFLP. Med J Islam Repub Iran. 2006;19(4):333–338. [Google Scholar]
- 14.Pavlik I., Horvathova A., Dvorska L. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J Microbiol Methods. 1999;38(1–2):155–167. doi: 10.1016/s0167-7012(99)00091-3. [DOI] [PubMed] [Google Scholar]
- 15.Ko W.Y., David R.M., Akashi H. Molecular phylogeny of the Drosophila melanogaster species subgroup. J Mol Evol. 2003;57(5):562–573. doi: 10.1007/s00239-003-2510-x. [DOI] [PubMed] [Google Scholar]
- 16.Chen H.J., Tsai J.C., Chang T.C. PCR-RFLP assay for species and subspecies differentiation of the Streptococcus bovis group based on groESL sequences. J Med Microbiol. 2008;57(4):432–438. doi: 10.1099/jmm.0.47628-0. [DOI] [PubMed] [Google Scholar]
- 17.Kang C.I., Song J.H. Antimicrobial resistance in Asia: current epidemiology and clinical implications. Infect Chemother. 2013;45(1):22–31. doi: 10.3947/ic.2013.45.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouamri M.C., Arsalane L., Kamouni Y.E., Zouhair S. Antimicrobial susceptibility of urinary Klebsiella pneumoniae and the emergence of carbapenem-resistant strains: a retrospective study from a university hospital in Morocco, North Africa. Africal J Urol. 2014;21(1):36–40. [Google Scholar]
- 21.Bruce S.K., Schick D.G., Tanaka L., Jimenez E.M., Montgomerie J.Z. Selective medium for isolation of Klebsiella pneumoniae. J Clin Microbiol. 1981;13(6):1114–1116. doi: 10.1128/jcm.13.6.1114-1116.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breed R.S., Murray E.G.D., Smith N.R. 7th ed. Williams & Wilkins Company; New York: 1957. Bergey’s manual of determinative bacteriology. [Google Scholar]
- 23.Chacon M.R., Figueras M.J., Castro-Escarpulli G., Soler L., Guarro J. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie Van Leeuwenhoek. 2003;84(4):269–278. doi: 10.1023/a:1026042125243. [DOI] [PubMed] [Google Scholar]
- 24.Clegg S., Murphy C.N. Epidemiology and virulence of Klebsiella pneumoniae. Microbiol Spectr. 2016;4(1):1–17. doi: 10.1128/microbiolspec.UTI-0005-2012. [DOI] [PubMed] [Google Scholar]
- 25.Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turton J.F., Perry C., Elgohari S., Hampton C.V. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59(5):541–547. doi: 10.1099/jmm.0.015198-0. [DOI] [PubMed] [Google Scholar]
- 27.Al-Marzooq F., Mohd Yusof M.Y., Tay S.T. Molecular analysis of antibiotic resistance determinants and plasmids in Malaysian isolates of multidrug resistant Klebsiella pneumoniae. PLoS ONE. 2015;10(7):1–21. doi: 10.1371/journal.pone.0133654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma J., Sharma M., Ray P. Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res. 2010;132(September):332–336. [PubMed] [Google Scholar]
- 29.Kalghatgi C.A.T., Cmde S., Praharaj A.K., Sahni C.A.K., Pradhan D., Kumaravelu B.S. Detection of bacterial pathogens in cerebrospinal fluid using restriction fragment length polymorphism. Med J Armed Forces India. 2008;64(1):29–32. doi: 10.1016/S0377-1237(08)80141-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharmeen R., Hossain M.N., Rahman M.M., Foysal M.J., Miah M.F. In-vitro antibacterial activity of herbal aqueous extract against multi-drug resistant Klebsiella sp. isolated from human clinical samples. Int Curr Pharm J. 2012;1(6):133–137. [Google Scholar]
Further reading
- 19.Bauer A.W., Kirby W.M., Sherris J.C.T.M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. [PubMed] [Google Scholar]
- 20.Rahman M.M., Richardson A., Sofian-Azirun M. Antibacterial activity of propolis and honey against Staphylococcus aureus and Escherichia coli. African J Microbiol Res. 2010;4(16):1872–1878. [Google Scholar]
- 30.Brisse S., Issenhuth-Jeanjean S., Grimont P.A.D. Molecular serotyping of Klebsiella species isolates by restriction of the amplified capsular antigen gene cluster. J Clin Microbiol. 2004;42(8):3388–3398. doi: 10.1128/JCM.42.8.3388-3398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]