Abstract
The significance and frequency of marine microorganisms as producers of bioactive metabolites-a natural source of drug discovery had varied significantly during the last decades, making marine ecosystem a huge treasure trove of novel isolates and novel compounds. Among the twelve actinomycetes isolated from marine sediment sample (Lat. 17°41′962″N, Long. 83°19′633″E), amylase, protease, lipase and cellulase activities were exhibited by 8,7,4,3 isolates respectively. Five isolates exhibited l-asparaginase activity, while 5, 6, 2 isolates exhibited antibacterial, antifungal and antimicrobial activities respectively. One isolate VMS-A10 efficiently producing alpha-amylase (25.53 ± 0.50 U/mL), protease (19.26 ± 0.25 U/mL), lipase (36.25 ± 0.10 U/mL), cellulase (14.43 ± 0.513 U/mL), l-asparaginase (0.125 ± 0.004 U/mL), antimicrobial metabolites against B. subtilis (503.33 ± 5.77 U/mL), S. aureus (536.66 ± 5.77 U/mL), E. coli (533.33 ± 5.77 U/mL), P. aeruginosa (500.00 ± 10.0 U/mL), MRSA (538.33 ± 5.77 U/mL), C. albicans (353.33 ± 11.54 U/mL) and A. niger (443.33 ± 15.27 U/mL) was selected, identified on the basis of morphological, cultural, physiological, and biochemical properties together with 16S rDNA sequence, designated as Streptomyces parvulus strain sankarensis-A10 and sequencing product (1490 bp) was deposited in the GenBank database under accession number KT906299, Culture Deposit No: NCIM-5601. Isolation and characterization of each potential actinobacteria having immense industrial and therapeutic value on an unprecedented scale from marine sediments of Visakhapatnam coast will have a burgeoning effect.
Keywords: Actinomycetes, Streptomyces, Bioactive metabolites, Phenotypic and genotypic characterization, Enzyme assays
1. Introduction
Natural products have played a pivotal role in drug discovery. The marine biosphere is a substantial sampling point, especially a huge treasure trove of actinomycetes resource for drug discovery. Within marine actinomycetes, Streptomycetes play a pivotal role in the soil environment because of their broad range of metabolic processes which include biotransformations, degradation of the insoluble remains of other organisms, such as lignocellulose and chitin one of the world’s most abundant biopolymers [1]; relevantly, there is a growing interest in the bioconversion of cellulose into fermentable sugars, which would allow the production of biofuels and chemicals through industrial fermentation processes [2], degrading complex organic materials into compost, soil or sediments by producing several biocatalysts (enzymes) such as proteases, cellulases, amylase, gelatinase, pectinases, ureases, amidases, esterases and lipases making Streptomycetes central organisms in carbon recycling thus offering green and clean solutions to chemical processes that are emerging as a challenging and revered alternative to chemical technology. Streptomycetes produce over two-thirds of the clinically useful antibiotics of natural origin such as daptomycin and Lincomycin [3] by complex secondary metabolism. Nevertheless, it is well established that each actinobacteria strain apparently has the genetic potential ability to produce 10–20 secondary metabolites [4]. Consequently, specifically targeted isolation and screening of actinomycetes producing potential antibiotics or therapeutic enzymes or industrial enzymes have been a major part of the research i.e., isolation of new streptophenazines from marine Streptomyces sp. 182SMLY active against methicillin-resistant S. aureus [5], l-asparaginase from Streptomycetes parvulus KUAP106 [6], α-glucosidase inhibitors from Streptomyces sp. OUCMDZ-3434 [7], and Cellulase and xylanase production by Streptomyces albidoflavus strain SAMRC-UFH5 [8]. The large genetic potential for primary and secondary metabolism present in the genome of most actinomycetes appears to be in contrast with the limited number of metabolites that are actually detected in a single isolate [9]. Exploitation of either unexplored or less explored ecosystems for such actinomycetes is highly necessary. Despite Indian marine ecosystem is considered as an important source for the microbial expedition, the studies on the diversity of actinomycetes and bioactive metabolite’s production from Indian peninsula are scanty and need to be explored [10].
Moreover, reports on the potential of each/every single actinomycetes strain for primary as well as secondary metabolites production in a single study are scanty. Present study was intended to isolate, screen the marine actinobacteria from the sediment sample for their potential to produce hydrolytic enzymes (amylase, protease, Lipase, cellulase), l-asparaginase, antimicrobial metabolites and characterize them up to the genetic level.
2. Materials and methods
2.1. Sampling site
Sediment sample from Visakhapatnam coast at deeper region (10 m depth) of Bay of Bengal (Lat. 17°41′962″N, Long. 83°19′633″E) was collected in the month of January 2014 at the time of low tide in sterile polythene bags containing filtered and sterilized seawater (50% v/v (seawater:distilled water: 50:50)) in order to maintain moisture condition, refrigerated at 4 °C until further use for isolation of marine actinobacteria.
2.2. Measurement of physicochemical parameters of the sediment sample
The pH of the sediment sample, temperature and salinity were documented with PCSTestr 35 (Eutech PCSTEST35-01X441506/Oakton 35425-10) [11]. Dissolved Oxygen (DO) and Biological oxygen demand (BOD) of the sampling site were documented as described by Wangersky [12].
2.3. Isolation and maintenance of actinobacteria
One gram of sediment sample was aseptically transferred to 50 mL of sterile seawater (50% v/v) contained in 250 mL flask and kept on the orbital shaker (150 rpm) for 30 min at 28 ± 2 °C, keeping aside for 15–30 min to settle down the particulate matter. The suspension was serially diluted with sterilized seawater (50% v/v) up to 10−7 level. 0.1 mL of each of these dilutions was pour plated [13] in triplicates on starch casein agar medium (SCA) [composition (g/L): soluble starch 10, potassium nitrate 2, vitamin free casein 0.3, sodium chloride 2, di-potassium hydrogen phosphate 2, magnesium sulfate 0.05, calcium carbonate 0.02, ferrous sulfate 0.01, seawater (50% v/v) 1,000 mL, pH 7.0 ± 0.2] and Zobell marine agar medium (ZMA, Himedia) with the addition of 50 µg/mL of cycloheximide, 5 µg/mL of rifampicin to prevent the fungal and fastidious bacteria growth respectively, incubated for 10–21 days at 28 ± 2 °C. Individual actinobacterial colonies with chalky to leathery appearance were subcultured on SCA, ZMA, (ISP-2) slants (50% v/v seawater), incubated for 5–7 days at 28 ± 2°C for good sporulation to check the purity and then preserved at 4 °C.
2.4. Preliminary screening for bioactive metabolites
2.4.1. Amylase activity
Isolated actinomycetes were spot inoculated on starch agar media (SAM) [composition, g/L: soluble starch 10.0, Meat extract 3.0, seawater (50% v/v) 1000 mL, pH 7.0 ± 0.2, agar 15.0] and incubated for 7 days at 28 ± 2 °C, flooded with povidone–iodine solution and left for 5 min. The organisms secreting amylase produce the zone of clearance or decolorization against the blue color background [14].
2.4.2. Protease activity
The proteolytic activity was studied using milk casein agar (g/L: Peptone 1.0, sterile skimmed milk (10%), Agar 15.0, pH 7.0 ± 0.2, seawater (50% v/v) 1000 mL). Test actinobacteria were streaked and incubated for 7 days at 28 ± 2 °C. Following incubation, organisms secreting protease enzyme will exhibit a zone of proteolysis, which is demonstrated by clear zone surrounding the actinomycetes growth. The width of the hydrolyzed zone around the growth versus the width of growth was measured and recorded [15].
2.4.3. Lipase activity
Qualitative lipolytic activity of all the isolated actinobacteria was determined by streaking them on Tributyrin Agar (TA) plates [composition, g/L:Tributyrin 15 mL (v/v), peptone 5.0, agar 15.0, beef extract 3.0, seawater (50% v/v) 1000 mL, pH 7.0 ± 0.2], incubated for 7 days at 28 ± 2 °C. Lipolytic activity can be visualized by clear hydrolysis zone around the actinomycetes [16].
2.4.4. Cellulolytic activity
The isolated actinomycetes were center streaked and grown at 28 ± 2°C for 7 days on CMC agar medium [composition, g/L: carboxymethyl cellulose 10.0, potassium nitrate 2.0, potassium dihydrogen phosphate 4.0, disodium phosphate 4.0, calcium chloride 0.001, magnesium sulfate 0.2, ferrous sulfate 0.004, agar 15.0, seawater (50% v/v) 1000 mL, pH 7.0 ± 0.2]. After incubation, the plates were analyzed for cellulolytic ability by flooding with 1 mg/mL of congo red solution for 15 min. Decant the dye, and re-flood the plates with NaCl (1 M) for 15 min. Positive colonies are detected to be surrounded by a pale orange to clear zone against the red background [17].
2.4.5. l-Asparaginase by rapid plate assay method (l-aspargine amidohydrolase EC 3.5.1.1)
The plate assay was performed with l-asparagine (sole nitrogen source). The isolates were center streaked on the asparagine dextrose salts agar plates (ADS agar) [composition, g/L: l-asparagine 10.0, dextrose 2.0, dipotassium phosphate 1.0, magnesium sulfate 0.5, agar 15.0, seawater (50% v/v) 1000 mL, pH 6.8 ± 0.2, phenol red 0.009%(in ethanol)]. After 7 days of incubation at 28 ± 2 °C, change in the color of the medium from yellow to pink due to the release of ammonia indicates the extracellular l-asparaginase production by the actinomycetes isolates [18].
2.4.6. Antimicrobial activity
All the actinomycete isolates were initially screened for antimicrobial activity by cross streak method [19]. Isolates that exhibited a broad spectrum of antibiotic activity were selected for agar overlay method. In agar overlay method, the purified isolates were grown by spot inoculation on the plate for 7 days at 28 ± 2 °C using nine types of media (prepared in 50% v/v seawater) namely ISP Media [20], SCA and Nutrient agar media to ascertain the medium that foster maximum antimicrobial metabolites production. After suitable growth is obtained the plates were overlayed with nutrient agar medium containing the overnight bacterial cultures and potato dextrose agar medium containing fungal cultures on the surface of isolates with less percentage of agar (0.75%), incubated again for 18–24 h at 37 °C in case of bacteria cultures and 72–120 h in case of fungi cultures.
2.5. Phenotypic and genotypic characterization of actinomycete
Physiological, Morphological, culture characteristics (such as growth, soluble pigment, color of mycelia) and biochemical parameters of isolate VMS-A10 were assessed as per ISP and compared with Bergey’s Manual® of Systematic Bacteriology [21]. The orientation of substrate and aerial mycelium, spore arrangements was observed using a trinocular microscope (LABOMED, Model-CXR3, USA) at 400× magnification. Scanning electron microscopy analysis was performed (SEM-Model-JEOL-JSM 6610 LV) for the micro-morphology and external morphology of the strain. The genotypic characterization [22] and phylogenetic tree construction were done at MTCC centre, IMTECH, Chandigarh, India.
2.6. Antibiotic sensitivity and resistance profile of the isolate VMS-A10
Antibiotic susceptibility and resistance of the isolate VMS-A10 were assayed by streak plate method toward five antibiotics, namely penicillin G, rifampicin, gentamicin, tetracycline and streptomycin incorporated in SCA medium (50% v/v seawater) and then plates were incubated for 7–10 days at 28 ± 2 °C. Based on growth, the isolate was graded as sensitive and resistant to the tested antibiotics. The MAR index was calculated as described previously [23].
2.7. Secondary screening
Out of 12 isolates, isolate VMS-A10 that exhibited broad spectrum of metabolites production ability in primary screening was tested for its extracellular bioactive metabolite’s production ability under submerged fermentation conditions (non-optimized) in 50 mL of corresponding media (as mentioned in preliminary screening excluding agar) contained in a 250 mL flask inoculated with 10% (v/v) spore suspensions of 3.0 × 107 spores/mL and incubated for 7 days at 28 ± 2 °C on an orbital shaker (150 rpm). The mycelium from the fermented broth was separated. Contents of each production flask were centrifuged (8000g) for 15 min at 4 °C to separate. All the assays were performed in triplicate using the clear supernatant.
2.7.1. Enzyme assays
DNS method was employed to analyze the amylase activity [24]. Glucose was used as a standard to estimate reducing sugars. Proteolytic activity was determined using casein as the substrate [25]. Lipase activity was measured using p-NPP as a substrate [26]. Endoglucanase activity was estimated using glucose as standard [27]. l-Asparaginase activity was analyzed by nesslerization using ammonium sulfate standard curve [28].
2.7.2. Antimicrobial assay
The isolate VMS-A10 showed the highest antimicrobial activity in starch casein agar medium (SCA) and was chosen (exclude agar) to test its extracellular antibiotic production ability under submerged, non-optimized fermentation conditions. The clear supernatant was extracted twice with ethyl acetate (1:1), concentrated by rotary evaporation and assayed for antimicrobial activity by employing potato dextrose agar medium for fungi and nutrient agar medium for bacteria cultures by agar diffusion method. The test organism suspensions as per 0.5 McFarland standards were used as inoculum. A volume of 50 μL of crude extract was dispensed to each well (6 mm in diameter) and the plates were incubated accordingly. The zone of inhibition was expressed as described earlier [29].
3. Results
3.1. Physicochemical parameters of the sediment sample
Some of the physicochemical properties of the sediment sample were determined and the data are given in Table 1. The dissolved oxygen and biological oxygen demand of the water sample collected from sediment sampling site were also recorded.
Table 1.
Physicochemical properties of the sediment sample.
| Sample | Temperature | pH | Salinity | Dissolved oxygen (D.O) | Biochemical Oxygen Demand (B.O.D) |
|---|---|---|---|---|---|
| Sediment | 26 °C | 7.8 | 32 ppta | 5.12 mg/L | 1.13 mg/L |
ppt-Parts per thousand.
3.2. Isolation and screening of marine actinomycetes for bioactive metabolites
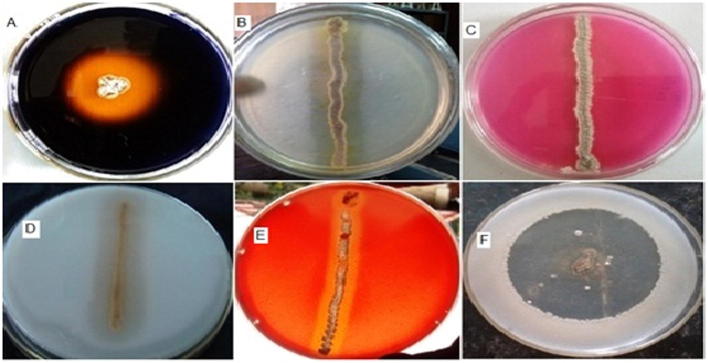
In the present study about twelve actinomycetes were isolated from Visakhapatnam coast marine sediment sample collected at a region (10 m depth) of Bay of Bengal, India, sub-cultured on SCA, ZMA agar slants (50% v/v seawater) and preserved in refrigerator for further studies. All the twelve isolated actinomycete strains were subjected to primary screening for amylase, protease, lipase, cellulose, l-asparaginase, antimicrobial metabolite’s, and the results shown in Table 2, Table 3 revealed that the isolate VMS-A10 exhibited broad spectrum of activities shown in Figs. 1A and 2. Hence subjected to secondary screening, the results are tabulated in Tables 3 and 4.
Table 2.
Primary screening of the Actinomycetes isolates for bioactive metabolites.
| S.No | Isolate | Alpha amylase | Protease | Lipase | Cellulase | l-Asparaginase | Antimicrobial activity |
|
|---|---|---|---|---|---|---|---|---|
| Anti-bacterial | Anti-fungal | |||||||
| 1 | VMS-A1 | + | + | − | − | − | + | − |
| 2 | VMS-A2 | + | − | + | − | − | − | + |
| 3 | VMS-A3 | + | − | + | − | + | − | − |
| 4 | VMS-A4 | − | − | − | + | − | + | − |
| 5 | VMS-A5 | − | + | − | − | − | − | + |
| 6 | VMS-A6 | + | + | − | + | − | + | + |
| 7 | VMS-A7 | + | − | − | − | + | + | − |
| 8 | VMS-A8 | − | + | + | − | − | − | − |
| 9 | VMS-A9 | + | + | − | − | + | − | + |
| 10 | VMS-A10 | + | + | + | + | + | + | + |
| 11 | VMS-A11 | − | + | − | − | − | − | + |
| 12 | VMS-A12 | + | − | − | − | + | − | − |
+ Showed activity, − No activity.
Table 3.
Screening of isolate VMS-A10 for bioactive metabolites.
| α-Amylase | Protease | Lipase | Cellulase | l-Asparaginase | |
|---|---|---|---|---|---|
| Zone of Clearance (mm)/Growth (mm) | 30/7 | 22/6 | 27/6 | 20/5 | 28/5 |
| Enzyme activity (U/mL)a | 25.53 ± 0.50 | 19.26 ± 0.25 | 36.25 ± 0.10 | 14.43 ± 0.513 | 0.125 ± 0.004 |
Mean ± SD, where n = 3.
Figure 1.
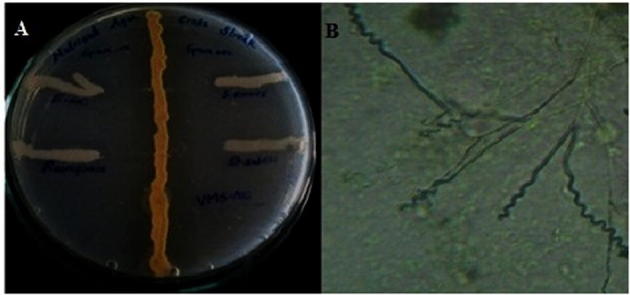
(A) Antibacterial activity (cross streak method). (B) Spore chain morphology of isolate VMS-A10 under 400× magnification (right).
Figure 2.
Bioactive metabolites produced by isolate VMS-A10 indicating (A) Alpha amylase. (B) Lipase. (C) l-Asparaginase. (D) Casein hydrolysis. (E) Cellulase. (F) Antibacterial activities.
Table 4.
Antimicrobial activity profile of isolate VMS-A10.
| S.No. | Microorganism | Agar overlay method zone of Inhibition (mm)a | Ethylacetate extract zone of Inhibition (Cup plate method) (U/mL)b |
|---|---|---|---|
| 1 | Bacillus subtilis (NCIM-2063) | 45.43 ± 0.40c | 503.33 ± 5.77c |
| 2 | Staphylococcus aureus (NCIM-2079) | 50.53 ± 0.30 | 536.66 ± 5.77 |
| 3 | Escherichia coli (NCIM-2065) | 48.23 ± 0.20 | 533.33 ± 5.77 |
| 4 | Pseudomonas aeruginosa (NCIM-2037) | 43.10 ± 0.10 | 500.00 ± 10.00 |
| 5 | MRSAd (Methicillin-Resistant Staphylococcus aureus) | 51.82 ± 0.71 | 538.33 ± 5.77 |
| 6 | Candida albicans (MTCC 227) | 13.30 ± 0.26 | 353.33 ± 11.54 |
| 7 | Aspergillus niger (NCIM-548) | 18.23 ± 0.25 | 443.33 ± 15.27 |
Excluding the growth diameter of VMS-A10.
Excluding the diameter of the well/cup (6 mm).
Mean ± SD where n = 3.
MRSA-clinical isolate acquired from King George Hospital, Visakhapatnam, India.
3.3. Taxonomy of isolate VMS-A10
The biochemical, physiological and morphological characteristics of the isolate VMS-A10 are shown in Table 5. Morphology of VMS-A10 isolate manifested features of Streptomyces bacteria community such as non-motile Gram-positive, cell wall chemotype-I, moderate growing, aerobic, glabrous or chalky, spore production, earthy odor, aerial and subterranean mycelium formation. Fig. 1B revealed that the aerial mycelium formed monopodial branched hyphae bearing spores with extended spiral type spore chain morphology. Different color characteristics and colony morphology displayed by the isolate on various international Streptomyces project media are shown in Table 6. The SEM analysis of the colonies in Fig. 3 showed 10–50 elliptical spores per chain with a warty surface; each elliptical spore was 0.59–1.11 µm size (width, length).
Table 5.
Phenotypic characteristics of VMS-A10.
| Properties | Strain VMS-A10 |
|---|---|
| Gram staining | Positive |
| Spore surface | Warty |
| Cell wall composition | Chemotype-I |
| Temperature (°C) | Growth |
| 12 | − |
| 25 | + |
| 37 | + |
| 42 | + |
| pH | Growth |
| 5 | + |
| 8 | + |
| 9 | + |
| 10 | + |
| % NaCl | Growth |
| 2 | + |
| 5 | + |
| 7 | + |
| 10 | − |
| Melanin production | − |
| Gelatin liquefaction | + |
| Citrate utilization | − |
| Methyl Red | − |
| Voge’s Proskauer | − |
| Nitrate reduction | + |
| Indole production | − |
| H2S production | − |
| Catalase | − |
| Oxidase | + |
| Urease | − |
| CarbonUtilization | Growth |
| Arabinose | + |
| Dextrose | + |
| Fructose | + |
| Glucose | + |
| Galactose | + |
| Mannose | + |
| Meso-inositol | − |
| Raffinose | − |
| Rhamnose | + |
| Salicin | − |
| Starch | + |
| Sucrose | − |
| Xylose | − |
| NitrogenUtilization | Growth |
| l-Valine | + |
| l-Arginine | + |
| l-Histidine | + |
| l-Threonine | + |
| l-Asparagine | + |
| KNO3 | + |
+ Positive, − Negative.
Table 6.
Growth characteristics of the isolate VMS-A10.
| S.No | Media | Growth | Aerial mycelium color | Substrate mycelium color | Pigmentation |
|---|---|---|---|---|---|
| 1. | Starch casein agar | Excellent | Light grey | Greenish yellow | Pale yellow |
| 2. | Tryptone yeast extract agar (ISP 1) | Good | Whitish grey | – | N |
| 3. | Yeast extract malt extract agar (ISP 2) | Excellent | Whitish grey | Brown | Pale yellow |
| 4. | Oat meal agar (ISP 3) | Good | Whitish grey | Yellow | |Pale yellow |
| 5. | Inorganic salt starch agar (ISP 4) | Excellent | Whitish grey | Greenish brown | N |
| 6. | Glycerol asparagine agar (ISP 5) | Excellent | Whitish grey | Greenish | N |
| 7. | Nutrient agar | Excellent | Whitish grey | Pale yellow | Pale yellow |
| 8. | Bennett agar | Excellent | Grey | Brown | Pale yellow |
N - No pigmentation.
Figure 3.
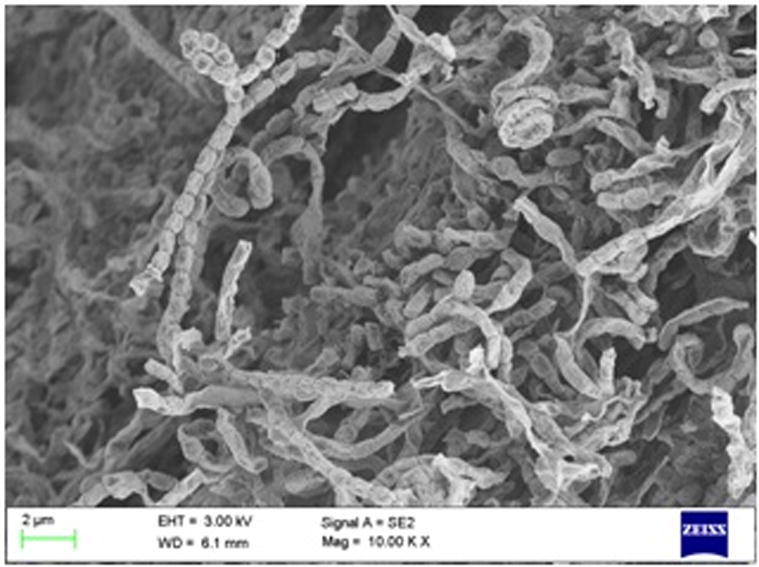
Scanning electron micrograph of VMS-A10.
3.4. Antibiotic sensitivity and resistance profile of isolates VMS-A10
Most commonly used antibiotics for bacterial infections were used to assess the antibiotic sensitivity profile (Table 7). The MAR index of the isolate VMS-A10 was found to be 0.60.
Table 7.
Antibiotic sensitivity and resistance profile of isolates VMS-A10.
| S.No. | Antibiotic (µg/mL) | Growth Response | Result | MAR Index |
|---|---|---|---|---|
| 1. | Penicillin G (10 IU) | + | Resistant | 0.60 |
| 2. | Streptomycin (100) | − | Sensitive | |
| 3. | Gentamycin (100) | − | Sensitive | |
| 4. | Rifampicin (50) | + | Resistant | |
| 5. | Tetracyclin (100) | + | Resistant |
+ Growth, − No growth, MAR - Multiple Antibiotic Resistance index.
3.5. Genotypic characterization and phylogenetic analysis of VMS-A10 strain
The 16S rDNA gene sequence of the isolate VMS-A10 was of 1490 bp. The phylogenetic tree clearly revealed its evolutionary relationship with a group of Streptomyces species (Fig. 4) generated by a neighbor-joining method in MEGA 6 [30]. Sequence analysis showed its most similarity to those of Streptomyces parvulus NBRC 13193T (AB184326), Streptomyces malachitospinus NBRC 101004T (AB249954) and Streptomyces luteogriseus NBRC 13402T (AB184379) with 99% sequence identities, exhibiting a difference in its phenotypic properties, genetic relatedness, and phylogenic analysis characteristics to the type strains that are closely related. Consequently, the isolate was designated as Streptomyces parvulus strain sankarensis-A10 (NCIM-5601), deposited under the GenBank accession number KT906299.
Figure 4.
Phylogenetic tree using neighbor-joining method for the isolate VMS-A10.
4. Discussion
Among the twelve different actinomycetes isolated from the marine sediment sample, amylase, protease, lipase, and cellulase activities were exhibited by 8, 7, 4, 3 isolates respectively. Five isolates exhibited l-asparaginase activity, while 5, 6, 2 isolates exhibited antibacterial, antifungal and antimicrobial activities respectively. One isolate VMS-A10 showed positive results for all the above activities. The results revealed that the Visakhapatnam coast marine ecosystem is a rich consortium of many potent actinobacteria.
The potential of the strain VMS-A10 was determined by its alpha-amylase (25.53 ± 0.50 U/mL), protease (19.26 ± 0.25 U/mL), lipase (36.25 ± 0.10 U/mL), cellulase (14.43 ± 0.513 U/mL), l-asparaginase (0.125 ± 0.004 U/mL) and antimicrobial activity against B. Subtilis (503.33 ± 5.77 U/mL), S. aureus (536.66 ± 5.77 U/mL), E. coli (533.33 ± 5.77 U/mL), P. aeruginosa (500.00 ± 10.0 U/mL), MRSA (538.33 ± 5.77 U/mL), C. albicans (353.33 ± 11.54 U/mL), A. niger (443.33 ± 15.27 U/mL). Based on phenotypic and genotypic characteristic variations, isolate VMS-A10 was designated as Streptomyces parvulus strain sankarensis-A10 (NCIM-5601, GenBank-KT906299). Similarly, Streptomyces fradiae strain RSU15 has been reported to produce hydrolytic enzymes namely amylase, protease, lipase, and cellulose [31]. A terrestrial actinomycete Streptomyces corchorusii strain UCR3-16 was found to be positive for the production of chitinase, β-1,3-glucanase, β-1,4-glucanase, lipase and protease [32]. Species of S. parvulus are known as producers of polypeptide antibiotic actinomycins and have been considered for industrial applications [33]. The outcomes of the present study serve as evidence in an attempt of connecting unexplored genes of omnipresent species of actinobacteria to molecules. The isolate was also tolerant to various physiological conditions, thus indicating its biodiversity, its ecological relative importance in the marine environment (disposing cellulosic wastes that are continuously added to the marine environment), its tremendous potential as a sustainable source of the robust enzymatic system and natural bioactive compound(s).
Many research publications have reported the isolation of Streptomyces parvulus species and its type strains indicating its frequency of isolation and their targeted screening for antibiotic production, such as production of actinomycin D by Streptomyces parvulus CBJ1 [34], anti Staphylococcus antibiotic by Streptomyces parvulus [35], and antimicrobial metabolites by Streptomyces parvulus DOSMB-D105 [36]. But their potential to produce other bioactive metabolites under submerged fermentation conditions has not been evaluated in a single study. This is the first time to report such an isolate (VMS-A10) which could produce multiple bioactive metabolites from Visakhapatnam coast, Bay of Bengal (India).
5. Conclusion
Despite some serious impediment to scientific progress in past few decades in search (isolation) of novel microorganisms producing novel metabolites from either unexplored or less explored marine or terrestrial ecosystems, the current awareness regarding the exploitation of potential of each/every single microbial (bacteria, fungi, actinomycetes, etc.) strain for primary as well as secondary metabolites deserves attention that might lead to the isolation of novel compounds from diversified biological source revealing their importance in biotechnology.
It is expected that the attempt for the isolation and identification of potential actinomycetes from Visakhapatnam coast marine sediments, which could synthesize broad spectrum bioactive compound’s that could be safely used for the human therapeutic purpose as well as for multiple industrial applications would be of great use. However, further experimental studies to optimize, isolate and identify individual bioactive metabolite(s) that will give us a broader range of perception about its (isolate VMS-A10) potency under various marine environmental conditions and its exact nature of interaction with marine ecosystem are under progress.
During the screening study the moderately active isolates that were neglected demand more investigational studies under optimized culture conditions.
Culture deposition
The isolate VMS-A10 was deposited in NCIM, Pune, India, under the deposit number NCIM-5601.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgment
The authors would like to acknowledge Maulana Azad National Fellowship for Minority Students, Ministry of Minority Affairs, UGC, New Delhi, Government of India, for providing the fellowship. The authors are also thankful to Scientists of CMFRI, Visakhapatnam, Mr. Loveson Edward and Mr. N. Rajendra Naik for providing the marine sediment samples.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Bentley S., Chater K. Nature. 2002;417(6885):141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 2.Saratale G.D., Saratale R.G., Oh S.E. Biomass Bioenergy. 2014;47:302–315. [Google Scholar]
- 3.Eustáquio A.S., Gust B., Galm U., Chater K.F., Heide L., Eusta A.S., Li S. Appl. Environ. Microbiol. 2005;71(5):2452–2459. doi: 10.1128/AEM.71.5.2452-2459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valan Arasu M., Duraipandiyan V., Agastian P., Ignacimuthu S. J. Mycol. Med. 2009;19(1):22–28. [Google Scholar]
- 5.Y. Liang, L. Chen, X. Ye, K. Anjum, X. Lian, 6419(April) (2016), doi:http://dx.doi.org/10.1080/14786419.2016.1169419.
- 6.Usha R., Mala K.K., Venil C.K., Palaniswamy M. Polish J. Microbiol. 2011;60(3):213–221. [PubMed] [Google Scholar]
- 7.Chen Z., Hao J., Wang L., Wang Y., Kong F., Zhu W. Nat. Publ. Gr. 2016:1–9. [Google Scholar]
- 8.Fatokun E., Nwodo U., Okoh A. Appl. Sci. 2016;6(10):286. [Google Scholar]
- 9.Sosio M., Bossi E., Bianchi A., Donadio S. Mol. Gen. Genet. 2000;264(3):213–221. doi: 10.1007/s004380000336. [DOI] [PubMed] [Google Scholar]
- 10.Suthindhiran K., Jayasri M.A., Dipali D., Prasar A. J. Basic Microbiol. 2014;54(10):1098–1109. doi: 10.1002/jobm.201300563. [DOI] [PubMed] [Google Scholar]
- 11.Ramesh S., Mathivanan N. World J. Microbiol. Biotechnol. 2009;25(12):2103–2111. [Google Scholar]
- 12.P.J. Wangersky, Methods of seawater analysis, Vol. 7, 1978.
- 13.Shetty P.R., Buddana S.K., Tatipamula V.B., Naga Y.V.V., Ahmad J. Brazilian J. Microbiol. 2014;45(1):303–312. doi: 10.1590/S1517-83822014005000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S., Lin C., Liu Y., Shen Z., Jeyaseelan J., Qin W. Int. J. Biochem. Mol. Biol. 2016;7(1):1–10. [PMC free article] [PubMed] [Google Scholar]
- 15.Suganthi C., Mageswari A., Karthikeyan S., Anbalagan M., Sivakumar A., Gothandam K.M. J. Genet. Eng. Biotechnol. 2013;11(1):47–52. [Google Scholar]
- 16.Ugur A., Sarac N., Boran R., Ayaz B., Ceylan O., Okmen G. ISRN Biochem. 2014;2014:1–7. doi: 10.1155/2014/289749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teather R.M., Wood P.J. Appl. Environ. Microbiol. 1982;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulati R., Saxena R.K., Gupta R. Lett. Appl. Microbiol. 1997;24(1):23–26. doi: 10.1046/j.1472-765x.1997.00331.x. [DOI] [PubMed] [Google Scholar]
- 19.Waksman S.A., Reilly H.C., Harris D.A. J. Bacteriol. 1948;56(3):259–269. doi: 10.1128/jb.56.3.259-269.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirling E.B., Gottlieb D. Int. J. System. Bacterilogy. 1966;16(3):313–340. [Google Scholar]
- 21.Goodfellow Michael, Kämpfer Peter. Springer; New York: 2012. Bergey’s Manual® of Systematic Bacteriology, Volume Five: The Actinobacteria, Part A and B. [Google Scholar]
- 22.Kumar P.S., Duraipandiyan V., Ignacimuthu S. Kaohsiung J. Med. Sci. 2014;30(9):435–446. doi: 10.1016/j.kjms.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krumperman P.H. Appl. Environ. Microbiol. 1983;46(1):165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John L.M.P., Harley P. Lab. Exerc. Microbiol. 2002:117–124. [Google Scholar]
- 25.Kathiresan S.M.k. Res. J. Environ. Sci. 2007;1(4):173–178. [Google Scholar]
- 26.Boran R., Ugur A. Prep. Biochem. Biotechnol. 2010;40(4):229–241. doi: 10.1080/10826068.2010.488929. [DOI] [PubMed] [Google Scholar]
- 27.Nitisinprasert S., Temmes A. J. Appl. Bacteriol. 1991;71(2):154–161. doi: 10.1111/j.1365-2672.1991.tb02972.x. [DOI] [PubMed] [Google Scholar]
- 28.El-Naggar N.E., Moawad H., El-Shweihy N.M., El-Ewasy S.M. Biomed. Res. Int. 2015;2015:627031. doi: 10.1155/2015/627031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxwell P.W., Chen G., Webster J.M., Dunphy G.B. Appl. Environ. Microbiol. 1994;60(2):715–721. doi: 10.1128/aem.60.2.715-721.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asnani A., Ryandini D., Suwandri IOP Conf. Ser. Mater. Sci. Eng. 2016;107:12056. [Google Scholar]
- 32.Tamreihao K., Ningthoujam D.S., Nimaichand S., Singh E.S., Reena P., Singh S.H., Nongthomba U. Microbiol. Res. 2016;192:260–270. doi: 10.1016/j.micres.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 33.T. Nishizawa, 4(May) (2011) 2014–5. doi:http://dx.doi.org/10.1128/JB.05177-11.
- 34.Cibi R., Nair A.J. Int. J. Curr. Microbiol. Appl. Sci. 2016;5(7):461–467. [Google Scholar]
- 35.S.V. Pereira, 2016, 10.1101/060392. [DOI]
- 36.Baskaran R., Mohan P.M., Sivakumar K., Kumar A. Acta Microbiol. Immunol. Hung. 2016;63(1):27–46. doi: 10.1556/030.63.2016.1.2. [DOI] [PubMed] [Google Scholar]