Abstract
Flavonoids are low-molecular-weight phenolic compounds that are widely distributed in the plant kingdom. They have different roles in plant resistance to biotic and abiotic stresses.
The transcription factor gene MdMyb10 (Gene Bank: DQ267896) was introduced into two apple (Malus domestica Borkh.) cultivars i.e. ‘Holsteiner Cox (HC)’ and ‘Gala’ via Agrobacterium-mediated transformation. The regenerated shoots were selected on kanamycin containing media. The presence of additional MdMyb10 gene in putative shoots was confirmed by PCR, RT-PCR and Southern blotting. Expression level of introduced MdMyb10 gene was analyzed by quantitative real time PCR. The results confirmed a dramatic increase in overexpression of MdMyb10 in the transgenic plants, up to 1261 and 847-folds for cultivars Holsteiner Cox and Gala, respectively compared to non-transformed negative control plants.
HPLC-MS was used to determine the levels of different flavonoid compounds in both non-transgenic and transgenic plants. In MdMyb10 ‘HC’ transgenic plants, some of the polyphenols analyzed were enhanced while others were reduced in comparison to their levels in the non-transgenic plants. On the other hand, all of the analyzed polyphenol classes were induced in MdMyb10 ‘Gala’ transgenic plants in comparison to their levels in the non-transgenic plants.
In the present study, the flavonoid pathway was successfully modified in apple by overexpressing the MdMyb10 transcription factor to validate the hypothesis of increased effect on plant disease resistance.
Keywords: Agrobacterium, Apple, Flavonoids, MdMyb10, Metabolic engineering, Over-expression, Transcription factors
1. Introduction
Apple (Malus domestica Borkh.) originates from the area around the Himalaya mountains on the border of Western China, the former USSR and Central Asia, and has spread to most temperate regions of the world. Over the centuries, many hybrids and cultivars have been developed, yielding in around 7500 varieties in the market today. The major productions come from the temperate zone of the Northern and Southern hemispheres [1]. Nearly 84 million tons of apples were produced on almost 7.5 million hectares in 2014 (http://faostat.fao.org).
As most of the extensive crops, apple is one of the most important sources of flavonoids (approx: 1–5 mg/100 g weight), next to onion and tea [2]. The major flavonoid classes occurring in apple fruit are flavonols such as quercetin 3-glycosides, monomeric and oligomeric flavan-3-ols such as catechin, epicatechin and procyanidins, dihydrochalcones such as phloridzin. In red-colored cultivars, anthocyanins such as cyanidin 3-glycosides can be found. Treutter and Feucht [3] reported the role of flavonoids in apple trees with respect to resistance against pests and pathogens such as apple scab caused by the fungus Venturia inaequalis by accumulation at infected sites.
The flavonoid biosynthetic pathway was the first target for metabolic engineering since the early 1990s, as the pathway was well known and the results could easily be observed by changes in flower color [4], [5]. Chalcone synthase (CHS) gene, the key enzyme in the flavonoid biosynthetic pathway was first isolated from cell suspension culture of parsley (Petroselinum hortense) after UV-irradiation [6].
Flavonoids play a role in protecting plants from pests and pathogens. It is often described that pathogens induce the biosynthesis of resistance-related metabolites, but also non-pathogenic strains are capable to elicit secondary metabolism [7]. Furthermore, the flavonoid synthesis varies and is induced by other factors such as UV-light [8], [9], water stress [10], high temperature and ABA (reviewed by Chaves and Escudero [11], [12]) and ozone [13]. Thus, flavonoids are suggested as a mimic of biotic stressors [14].
The MdMyb10 gene has sequence similarity to the Myb-class of transcription factors from other species. It is known to regulate the biosynthetic steps in anthocyanin metabolism, which has been described and functionally characterized [15], [16].
Most of the previous published work focused on the role of MdMyb10 in anthocyanin biosynthesis and the association with red skin color in apple [16], [17], [18], [19]. In addition, the role of the MYB family as adaptive responses to drought stress was reviewed by Baldoni et al. [20].
The MdMyb10 gene is highly expressed in red-fleshed fruit and it induces red color when transiently expressed in tobacco leaves [16], [21].
Chagné et al. [22] have performed mapping of candidate gene as genomic approach in a fruit tree crop, providing the genetic evidence for the red color in the fruit and foliage. The red color is controlled by a single locus (Rni). They also showed that the transcription factor MdMyb10 may be the key gene underlying Rni, furthermore, they co-segregate together.
Introduction of the MdMyb10 transcription factor gene into Arabidopsis produced plants with enhanced tolerance to osmotic stress as well as higher levels of flavonoids, chlorophyll, malondialdehyde and proline [23].
The aim of the present study was to modify the flavonoid pathway in apple. This is done by overexpressing the MdMyb10 transcription factor involved in this pathway in order to validate the hypothesis of increased effect on plant disease resistance.
2. Materials and methods
2.1. Plant material
In vitro cultures of apple cvs. ‘Holsteiner Cox (HC)’ and ‘Gala’ were used. Shoot cultures were maintained on propagation medium consisting of Murashige and Skoog (MS) salts and vitamins [24] supplemented with 2% (w/v) sucrose, 3.1 μM 6-benzylaminopurine (BAP), 0.5 μM 1-naphthaleneacetic acid (NAA), 2.8 μM gibberellic acid (GA3) and 0.8% plant agar (Duchefa), pH was adjusted to 5.7 before autoclaving. The shoot cultures were incubated at 25 °C under a 16/8-h (day/night) photoperiod and sub-cultured every 4 weeks to fresh media.
2.2. Agrobacterium strain and binary vector
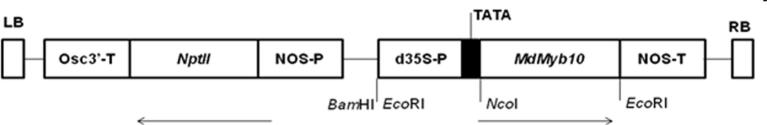
Agrobacterium tumefaciens strain GV3101 [25], harboring the binary vector pJan (gratefully provided by Dr. Julian Brüggemann, University of Bielefeld) was used for transformation. The binary vector contains the transcription factor MdMyb10 gene from apple (Gene Bank: DQ267896) under the control of cauliflower mosaic virus (CaMV 35S) promoter and Agrobacterium nopaline synthase gene (NOS) terminator. npt II gene was used as plant selectable marker gene under control of Agrobacterium nopaline synthase gene (NOS) promoter and octopine synthase 3′ region (Osc 3′) terminator [26] (Fig. 1).
Figure 1.
Functional map of the T-DNA region of transformation vector pJan-MdMyb10 (provided by Dr. Brüggemann). MdMyb10, transcription factor gene from apple (Malus domestica Borkh.); Nos-P and Nos-T, nopaline synthase gene promoter and terminator from Agrobacterium tumefaciens, respectively; Ocs 3′-T, octopine synthase 3′ region terminator from Agrobacterium tumefaciens; NptII, Neomycin phosphotransferase gene as plant selectable marker; 35 S-P, cauliflower mosaic virus (CaMV) 35 S promoter; RB and LB, the right and left borders; the arrows indicate the direction of transcription for the respective genes.
2.3. Transformation and regeneration
Four youngest unfolded leaves from 4-week-old in vitro shoots were used. Leaves were cut into strips and used for transformation.
For inoculation, 250 μl of Agrobacterium tumefaciens from a glycerol stock was grown overnight (28 °C, 240 rpm) in 25 ml yeast extract-peptone (YEP) medium consisting of 1% yeast extract, 1% bacto peptone, 0.5% NaCl and supplemented with 50 mg/l ampicillin and 50 mg/l rifampicin, after centrifugation (15 min, 4400 rpm, 4 °C) the pellet was resuspended in liquid MS medium and OD600 was adjusted to 0.8. The explants were inoculated with Agrobacterium suspension for 10 min with gentle shaking, then they were blotted dry and transferred into co-cultivation media consisting of MS macro- and microelements, MS vitamins, 3% sorbitol, 3 μM thidiazuron (TDZ), 1 μM indole-3-butyric acid (IBA) and 0.3% Gelrite for ‘Holsteiner Cox’ and MS macro- and microelements, MS vitamins, 3% sorbitol, 22 μM TDZ, 2.6 μM NAA and 0.3% Gelrite for ‘Gala’ and kept in dark in the growth room at 24 °C for 3 days, where after, the leaves were washed twice in distilled water and once in liquid MS-medium supplemented with 150 mg/l ticarcillin to eliminate the Agrobacterium from plant tissue.
Explants were transferred with their adaxial side in contact with the shoot induction medium supplemented with 150 mg/l ticarcillin and 50 mg/l kanamycin. The explants were kept in the dark for 2 weeks at 24 °C, then transferred to a 16/8-h (day/night) photoperiod. The explants were sub-cultured to fresh media every 2 weeks for 6 weeks on shoot-induction medium. Regenerated shoots were sub-cultured on elongation medium (MS salts and vitamins, 3% sucrose, 4.4 μM BAP, 0.28 μM GA3 and 0.8% plant agar) supplemented with 150 mg/l ticarcillin and 50 mg/l kanamycin. Elongated shoots (25–30 mm) were cultivated on root induction medium (MS salts and vitamins, 2% sucrose, 0.1% myoinositol, 7 μM IBA and 0.8% plant agar) supplemented with 150 mg/l ticarcillin and 50 mg/l kanamycin at 25 °C and 16/8-h (day/night) photoperiod. Rooted plantlets were transferred to peat moss and perlite mixture (2:1 v/v) and acclimatized in greenhouse.
2.4. Verification of the integration of the transgenes
Genomic DNA was isolated from transgenic and non-transgenic control plants using CTAB-extraction method [27]. All regenerated plants were analyzed using PCR. PCR reaction contained 50 ng DNA, 1U Taq polymerase (Promega, Germany), 1.5 mM MgCl2, 0.2 mM of each dNTPs, and 0.4 μM of each primer in a total volume of 25 μl. The PCR conditions were 94 °C for 3 min, followed by 30 cycles of 94 °C for 1 min, 62 °C (MdMyb10) or 64 °C (nptII) for 1 min, 72 °C for 1 min, and final extension at 72 °C for 10 min.
The presence of the MdMyb10 gene was confirmed using the MdMyb10 and nptII specific primers as shown in Table 1.
Table 1.
Primers used for PCR, probe preparation, RT-PCR and quantitative Real-Time PCR.
| Primer | Sequence 5′ → 3′ | Tm (°C) | Correlation coefficient | PCR efficiency % |
|---|---|---|---|---|
| 167nptII-for | CCACAGTCGATGAATCCAGA | 64 | – | – |
| 367nptII-rev | AGCACGTACTCGGATGGAAG | |||
| MdMyb10-for | ATGGAGGGATATAACGAAAAC | 62 | – | – |
| MdMyb10-rev | ATGATTCCAAAGGTCCGTGCT | |||
| Rubisco-for | GCTTGTCCAAGAGCAAGAGAAT | 62 | 0.999 | 100.3 |
| Rubisco-rev | CTCCCTCCCCTCAATTATAACC | |||
| Myb10-specific for | GCGTTGAGATTCATGGAGAGG | 62 | 0.999 | 94.7 |
| Myb10-specific rev | CTAGCAATCAATGACCACCTGTT | |||
| RNAPOL II-for | ATATGCCACCCCGTTCTCTACT | 62 | 0.999 | 90.0 |
| RNAPOLII-rev | CACGTTCCATTTGTCCAAACTT | |||
| Myb10-Probe-for | ATTAGACTTCACAGGCTTTTGGGA | 60 | – | – |
| Myb10-Probe-rev | TTCTTCCTCTAACTCAATGCTGGG | |||
| MdMyb10-for | ATGGAGGGATATAACGAAAAC | 62 | – | – |
| pJan-polyA-rev | AGCTAATTACTCATGATCAGGTAC |
2.5. Southern blot analysis
Southern blot [28] was used to study the integration pattern and copy number of introduced genes, 20 μg of genomic DNA of transgenic and non-transgenic control plants were digested with 20 U BamHI (Thermo Scientific, Germany, BamHI cuts once within T-DNA, Fig. 1) at 37 °C overnight, followed by the addition of another 10 U of the enzyme and incubated for 4–5 h to complete digestion. The digested DNA was fractionated on a 1% agarose gel and blotted on a positively charged nylon membrane (Roche, Germany) as described by Sambrook et al. [29]. The membrane was hybridized with DIG-labeled 400 bp MdMyb10 probe (Table 1) using a PCR-DIG Mix kit (Roche, Germany).
2.6. Reverse transcriptase PCR
Total RNA was isolated using PureLink™ Plant RNA Reagent (Invitrogen, Paisley, Scotland) from 100 mg of young leaves from transgenic and non-transgenic control plants. Two micrograms of RNA was treated with DNaseI (Thermo Scientific, Germany) to eliminate genomic DNA contaminations.
The treated RNA was reversed transcribed with oligo(dT)18 primer and RevertAid™ First Strand cDNA Synthesis Kit (Thermo Scientific, Germany), all steps were performed according to the manufacturer’s instructions. To check RNA contaminations with DNA, a specific primers for Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase activase mRNA, GenBank: Z21794) were used as internal control from apple.
cDNA was used as template for PCR using MdMyb10 for and pJan-polyA-rev primers (the reverse primer is located between the stop codon and the polyadenylation region) and Rubisco primers.
2.7. Quantitative real-time PCR (qRT-PCR)
Quantitative real-time PCR was performed using iQ SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA) and first strand cDNA on a CFX96™ Real-Time PCR Detection System (Bio-Rad). For each experiment, the cDNA sample was measured in triplicate.
The qRT-PCR reaction contains 1 μl of target cDNA, 7.5 μl iQ SYBR® Green Supermix (2X), 0.3 μl (200 mM) each of specific forward and reverse primers (Table 1) and nuclease free water to a final volume of 15 μl. The cycle conditions were set as follows: initial denaturation step at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 62 °C for 20 s, extension at 72 °C for 30 s, then one cycle at 95 °C for 10 s. This cycle was followed by a melting curve analysis, ranging from 65 °C to 95 °C, with temperature increasing by steps of 0.5 °C every 10 s. A negative control with no cDNA template (NTC) was included in each run. Fluorescence was measured at the end of each annealing step. The raw data were analyzed with the CFX software version 1.5, the expression was normalized using two reference genes, the apple Rubisco gene (Gene Bank: Z21794) and mRNA of the M. domestica cDNA clone Mdfw2033f21.y1 similar to the RNA polymerase subunit II (Gene Bank: CN579456). Amplification and correlation efficiencies of each PCR reaction were determined on cDNA serial dilutions (1:5). The PCR efficiency was used to transform the Ct-values into raw data for relative quantification.
2.8. HPLC analysis
A Bruker Daltonics esquire 3000 plus ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany) connected to an Agilent 1100 HPLC system (Agilent Technologies, Waldbronn, Germany) and equipped with a quaternary pump and a variable wavelength detector was utilized for all experiments. Components were separated with a Phenomenex (Aschaffenburg, Germany) Luna C-18 column (150 mm long × 2.0 mm inner diameter, particle size 5 μm) that was held at 25 °C. The electrospray ionization voltage of the capillary was set to −4000 V and the end plate to −500 V. Nitrogen was used as dry gas at a temperature of 300 °C and a flow rate of 10 l/min. The full scan mass spectra were measured in a scan range from 50 to 800 m/z with a scan resolution of 13000 m/z/s until the ICC target reached 20,000 or 200 ms, whichever was achieved first. Tandem mass spectrometry was carried out using helium as the collision gas (3.56 × 10–6 mbar) with the collision voltage set at 1 V. Spectra were acquired in the positive and negative ionization mode. The LC parameters went from 100% A (0.1% formic acid in water) to 50% B (0.1% formic acid in methanol) in 30 min, then in 5 min to 100% B, held for 15 min at these conditions, then returned to 100% A in 5 min at a flow rate of 0.2 ml min−1. The detection wavelength was 280 nm.
2.9. Metabolite analysis
100 mg of apple leaves were extracted with 200 μl methanol containing 0.2 mg/ml biochanin A as an internal standard. Methanol was removed in a rotary vacuum concentrator (Christ RVC 2-18, Osterode, Germany) and the extract was re-dissolved in 45 μl water for analysis by LC-ESI-MSn. Metabolites were tentatively identified by their retention times, mass spectra and product ion spectra in comparison with the data determined for authentic reference material or published data. Relative metabolite quantification was performed using the Data Analysis 4.1 and Quant Analysis 1.5 software (Bruker Daltonics, Bremen, Germany) normalizing all results to the internal standard.
Several different polyphenolic secondary metabolites were analyzed; including many of sub-groups belonging to the family of flavonoids (monomeric and polymeric flavon 3-ols, flavonols, dihydrochalcone, anthocyanidin, hydroxycinnamic acid).
3. Results
3.1. Regeneration and transformation
In the present study, we showed overexpression of MdMyb10 transcription factor gene, which is involved in the flavonoid pathway in apple cvs. ‘Holsteiner Cox’ and ‘Gala’ via Agrobacterium-mediated transformation. For further analysis, one putative ‘Holsteiner Cox’ transgenic line (HC-1) and four putative ‘Gala’ transgenic lines (Ga-1, -2, -3 and -4) were selected.
Stable transformation and T-DNA integration into genomic DNA of apple were confirmed. Figure 2, Figure 3 show PCR results using specific primers for MdMyb10 and nptII genes, respectively. These results clearly confirm the positive obtained transgenic lines.
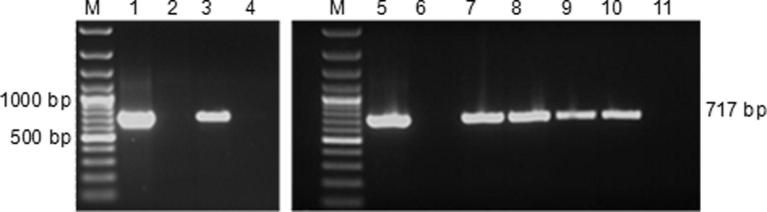
Figure 2.
Gene specific PCR analysis for the detection of MdMyb10 apple transgenic lines from genomic DNA isolated from leaves of putative apple transformants of Holsteiner Cox (HC) and Gala using MdMyb primers (PCR product of 717 bp). M, 100 bp plus DNA molecular weight marker; Lanes 1 & 5, pJan-Myb10 plasmid as positive control; lane 2, HC negative control; Lane 3, Holsteiner Cox transgenic line HC-1; Lanes 4 & 11, water controls; Lane 6, Gala negative control; Lanes 7, 8, 9 and 10, Gala transgenic lines Ga-1, Ga-2, Ga-3 and Ga-4, respectively.
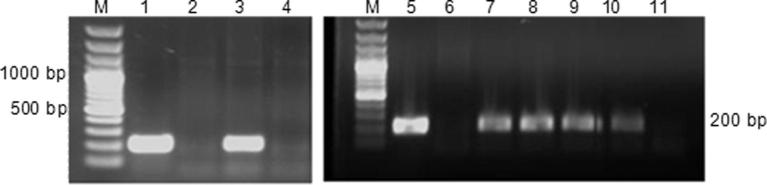
Figure 3.
PCR of different transgenic lines of Holsteiner Cox (HC) and Gala using nptII primers (spanning a 200 bp DNA fragment); M, 100 bp plus DNA molecular weight marker; Lanes 1 & 5, pJan-Myb10 plasmid as positive control; lane 2, HC negative control; Lane 3, Holsteiner Cox transgenic line HC-1; Lanes 4 & 11, water control; Lane 6, Gala negative control; Lanes 7, 8, 9 and 10, Gala transgenic lines Ga-1, Ga-2, Ga-3 and Ga-4, respectively.
Southern blot hybridization was performed using a DIG labeled PCR amplified probe (Roche, Germany). The result showed that one copy of the MdMyb10 gene was found in all analyzed plants (non-transgenic and transgenic plants), which refers to the native MdMyb10 gene. Introduced gene was presented either as one additional copy in lines Ga-1 and Ga-4 or two copies in lines Ga-2 and Ga-3 compared to non-transgenic control plant (Fig. 4).
Figure 4.
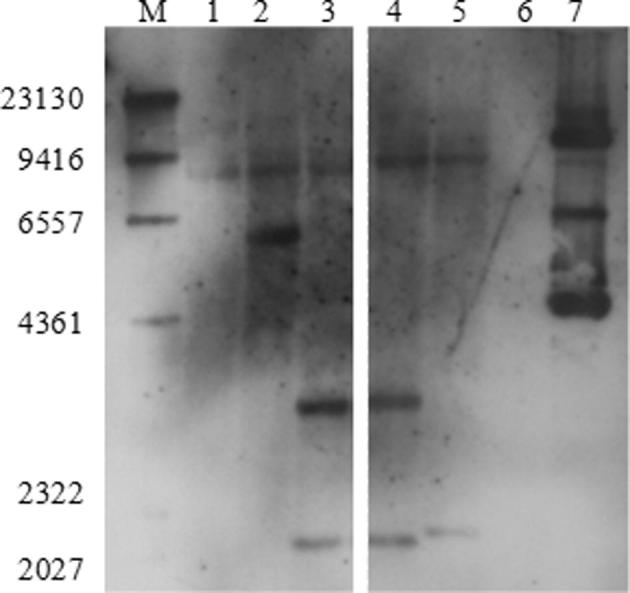
Southern blot analysis of gDNA digested with BamHI from Gala transgenic plants transformed with pJan-MdMyb10, the membrane was hybridized with MdMyb10 probe. M, Dig labeled DNA Marker; lane 1, Gala non-transgenic control plant; lanes 2, 3, 4 and 5, Gala transgenic lines Ga-1, Ga-2, Ga-3 and Ga-4, respectively; lane 6, blank; lane 7, pJan-MdMyb10 plasmid as positive control.
3.2. Reverse transcriptase PCR
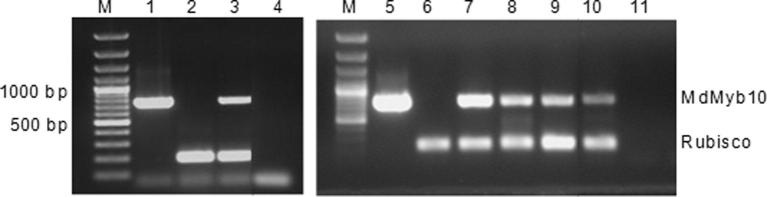
Multiplex RT-PCR was performed to confirm transcription of the transgene using Myb10-for specific primer and pJan-polyA-rev primer (the reverse primer located between stop codon and polyadenylation region of pJan vector) and Rubisco primers (Table 1). The expected fragments of 729 bp (MdMyb) and 200 bp (Rubisco) were amplified from selected transgenic lines (Fig. 5).
Figure 5.
Multiplex RT-PCR of different transgenic lines of Holsteiner Cox (HC) and Gala using Myb10-for specific and pJan-polyA-rev primers (PCR products of 729 bp) and Rubisco-for. and rev. primers (PCR products of 200 bp) as an internal control; M, 100 bp plus DNA molecular weight marker; Lanes 1 & 5, pJan-Myb10 plasmid as positive control; lane 2, HC negative control; Lane 3, Holsteiner Cox transgenic line HC-1; Lanes 4 & 11, water control; Lane 6, Gala negative control; Lanes 7, 8, 9 and 10, Gala transgenic lines Ga-1, Ga-2, Ga-3 and Ga-4, respectively.
3.3. Quantitative Real-time PCR
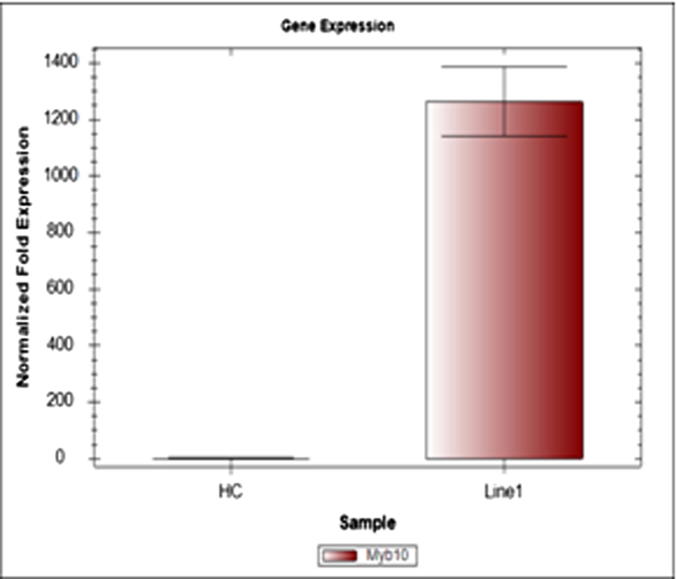
Real-time PCR was performed using MdMyb10 gene specific primers (Table 1) to obtain transcription profiles for the transgene MdMyb10. Expression levels of MdMyb10 gene were determined and expressed relatively to the Rubisco and RNA polymerase genes. The transcript of MdMyb10 was detected in both transgenic and non-transgenic control plants. The transcription level of MdMyb10 in the transgenic line of HC-1 was 1261-fold (Fig. 6).
Figure 6.
Expression of MdMyb10 gene in Holsteiner Cox (HC) transgenic line (transformed using pJan–MdMyb10 vector) and respective control. The values are expressed in relative to the level of mRNA transcript levels of the reference genes. Values are mean and standard error (error bars ± 1 SE) of three replicates. (HC, non transgenic plant; Line1, HC-1 transgenic plant).
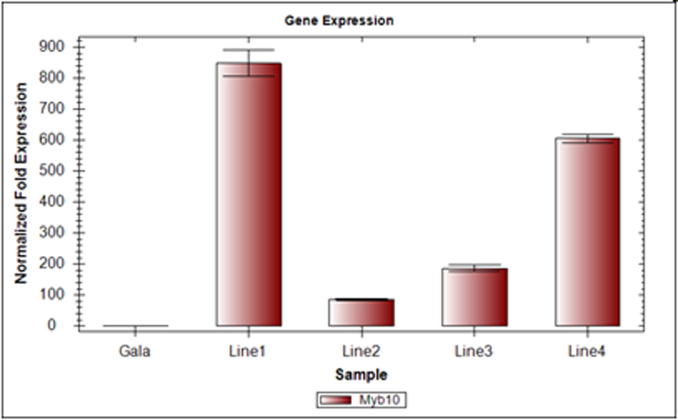
While transcription levels of the Gala transgenic lines were 847-fold (line Ga-1), 86- fold (line Ga-2), 190-fold (line Ga-3) and 610-fold (line Ga-4), (Fig. 7).
Figure 7.
Expression of MdMyb10 gene in Gala transgenic lines (transformed using pJan–MdMyb10 vector) and respective control. The values are expressed in relative to the level of mRNA transcript levels of the reference genes. Values are mean and standard error (error bars ± 1 SE) of three replicates (Gala: non-transgenic plant, Line1, Ga-1; Line2, Ga-2; Line3, Ga-3; Line4, Ga-4 transgenic plants).
3.4. Phenotype
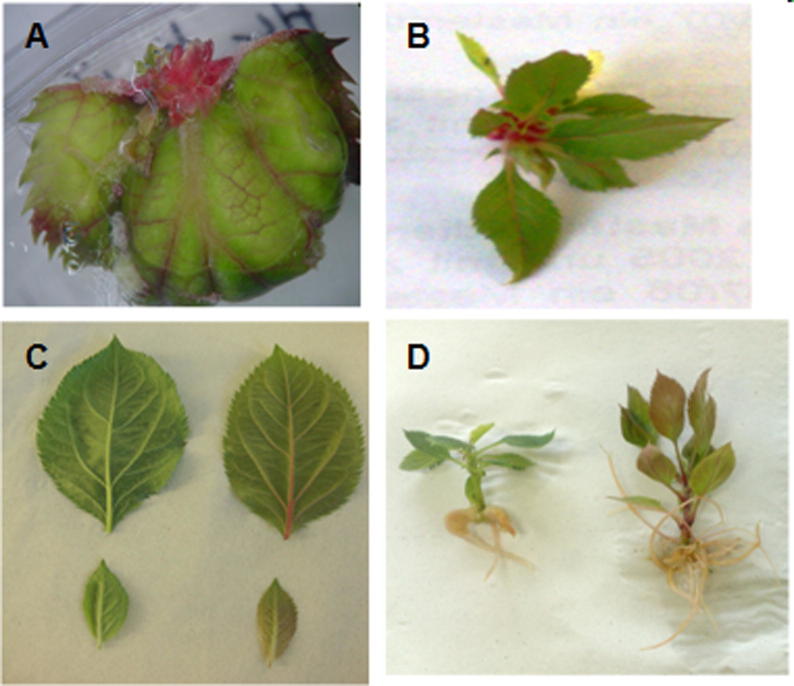
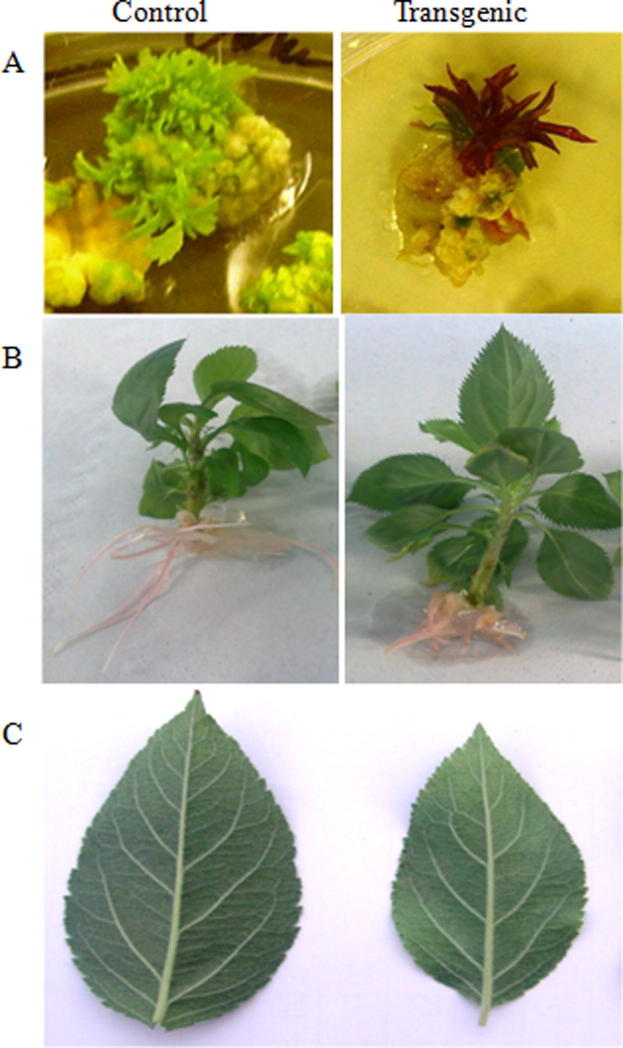
Phenotypical changes were observed in transgenic HC line. Starting with red calli and then red colored veins of leaves from regenerated shoots, as well as rooted and acclimatized plants in the greenhouse (Fig. 8). While in case of Gala, the change could be seen only in the early stages (calli and regenerated shoots) and subsequently disappeared (Fig. 9).
Figure 8.
Phenotypic changes in different stages of regenerated explants of Holsteiner Cox (HC) transformed with pJan-MdMyb10 binary vector showing red color compared to control plant. A, explant on regeneration medium; B, in vitro regenerated shoots; C, leaves of acclimatized plants (left, control; right, transgenic plant); D, rooted plantlets (left, control; right, transgenic plant).
Figure 9.
Change of phenotype in the first stages of transformation of Gala using pJan-MdMyb10 binary vector. A, explant on regeneration medium; B, in vitro rooted plants; C, leaves of acclimatized plants (left, control; right, transgenic plant).
3.5. Quantitative analysis of flavonoid levels in MdMyb10 transgenic plants
In MdMyb10 ‘HC-1’ transgenic plants, the concentration of some of the polyphenol classes analyzed were enhanced, while others were reduced (Table 2). The levels of total flavan-3-ols were increased to 1.4 times, with the strongest rises were observed for (epi) catechin-hexoside amounted to 1.44 times in comparison to their levels in the non-transgenic plants. The level of cyaniding-galactoside and dihydrochalcones were nearly the same in MdMyb10 ‘HC’ transgenic and non-transgenic plants. The total levels of flavonols were higher by 1.68 times in comparison to their levels in the non-transgenic plants. Quercetin-rhamnoside showed a rise of 2.7 times in comparison to the non-transgenic plants.
Table 2.
Content of individual phenolic compounds (mean ± SD, mg/100 g dry.wt., n = 5) and total phenolic compounds (mg/100 g dry.wt.) in leaves. HC, Holsteiner Cox; Ga, Gala; Ga-1, -2, -3, -4 different transgenic lines.
| HC control | HC-1 | Gala control | Ga-1 | Ga-2 | Ga-3 | Ga-4 | |
|---|---|---|---|---|---|---|---|
| Phenolic compounds | |||||||
| Catechin | 0.61 ± 0.43 | 0.47 ± 0.29 | 1.14 ± 0.5 | 1.26 ± 0.58 | 0.85 ± 0.38 | 2.03 ± 0.7 | 0.46 ± 0.1 |
| Epicatechin | 1.28 ± 0.61 | 1.35 ± 0.46 | 1.58 ± 0.5 | 3.01 ± 1.76 | 2.69 ± 1.09 | 3.65 ± 0.5 | 1.78 ± 0.5 |
| (epi)catechin-hexoside | 19.52 ± 6.48 | 28.1 ± 6.71 | 24.82 ± 14 | 60.93 ± 47.17 | 67.91 ± 29.22 | 45.28 ± 4.8 | 25.23 ± 1.22 |
| Total flavan-3-ols | 21.41 | 29.92 | 27.55 | 65.2 | 71.45 | 50.96 | 27.48 |
| Caffeic acid-glucose-ester | 0.36 ± 0.33 | 0.16 ± 0.05 | 1.25 ± 0.3 | 2.01 ± 0.98 | 0.74 ± 0.37 | 1.87 ± 0.7 | 0.43 ± 0.2 |
| Chlorogenic acid | 3.56 ± 1.09 | 4.35 ± 1.06 | 4.1 ± 4.2 | 2.62 ± 1.5 | 5.47 ± 3.04 | 3.38 ± 1.2 | 3.75 ± 1.9 |
| Coumaric acid-glucose-ester | 4.3 ± 2.96 | 2.04 ± 1.02 | 9.32 ± 2.5 | 9.94 ± 4.74 | 5.57 ± 1.7 | 8.48 ± 1 | 5.56 ± 2 |
| Ferulic acid-glucose-ester | 1.45 ± 0.78 | 0.78 ± 0.23 | 4.28 ± 1.3 | 4.76 ± 3.76 | 2.77 ± 1.25 | 7.67 ± 2.3 | 2.36 ± 1 |
| Coumaric acid-glucoside | 1.04 ± 0.78 | 0.76 ± 0.61 | 2.61 ± 1.2 | 5.64 ± 4.23 | 2.24 ± 0.82 | 4.16 ± 1.4 | 2 ± 0.8 |
| Total hydroxycinnamic acids | 10.71 | 8.1 | 20.56 | 24.97 | 16.8 | 25.56 | 14.1 |
| Cyanidin-galactoside | 1.5 ± 0.94 | 1.46 ± 0.82 | 0.98 ± 0.7 | 4.31 ± 4.31 | 1.66 ± 04 | 1.9 ± 0.8 | 0.93 ± 0.5 |
| Phloridzin | 127.8 ± 57.99 | 126.7 ± 43.07 | 102.5 ± 30.2 | 148.1 ± 61.56 | 148.9 ± 4215 | 157.3 ± 13.9 | 107 ± 33.5 |
| Phloretin-2′-O-xyloglucoside | 64.84 ± 25.9 | 56.07 ± 23.77 | 44.19 ± 13.3 | 50.08 ± 21.04 | 52.51 ± 17.46 | 61.52 ± 7.4 | 42.35 ± 15 |
| Phloretin | 55.43 ± 34.86 | 74.14 ± 37.41 | 48.83 ± 30.6 | 133.0 ± 698 | 125.6 ± 5454 | 120.4 ± 241 | 45.82 ± 23.7 |
| Total dihydrochalcones | 248.1 | 255.9 | 194.5 | 331.1 | 327 | 339.2 | 195.2 |
| Q-arabinoside | 11.29 ± 5.37 | 15.51 ± 5.22 | 15.53 ± 5.3 | 20.72 ± 7.67 | 25.23 ± 7.92 | 19.47 ± 2.5 | 14.94 ± 4.3 |
| Q-rutinoside | 1.17 ± 0.67 | 1.63 ± 1.14 | 1.4 ± 0.8 | 2.55 ± 1.65 | 2.73 ± 1 | 3.08 ± 0.6 | 1.34 ± 0.6 |
| Q-xyloside | 0.44 ± 0.7 | 0.23 ± 0.27 | 0.91 ± 0.7 | 1.7 ± 1.56 | 1.96 ± 1.08 | 1.1 ± 0.7 | 0.31 ± 0.4 |
| Q-galactoside | 11.55 ± 9.44 | 18.17 ± 7.81 | 22.71 ± 9.1 | 29.75 ± 16.28 | 41.14 ± 12.71 | 4.127 ± 4.8 | 21.57 ± 6.8 |
| Q-rhamnoside | 5.46 ± 2.38 | 14.65 ± 6.39 | 13.02 ± 5.7 | 21.56 ± 8.23 | 21.46 ± 7.59 | 16.8 ± 2.5 | 12.97 ± 3.5 |
| Total flavonols | 29.91 | 50.2 | 53.56 | 76.28 | 91.51 | 71.71 | 51.13 |
| Total phenolic content | 311.63 | 345.58 | 297.15 | 518.6 | 585.2 | 489.33 | 288.84 |
In MdMyb10 ‘Gala’ transgenic plants, the levels of most of the analyzed polyphenol classes were enhanced (Table 2) or remained the same (line Ga-4). The total content levels of flavon 3-ols were increased 2.3, 2.5 and 1.85 times for the lines Ga-1, Ga-2 and Ga-3, respectively. The strongest rises were observed for (epi) catechin-hexoside, which was 2.45, 2.74 and 1.83 times for the lines Ga-1, Ga-2 and Ga-3, respectively. The total levels of hydroxycinnamic acids were increased to 1.2 and 1.24 times for the lines Ga-1 and Ga-3, respectively. In contrary they were decreased to 0.81 and 0.69 times for the lines Ga-2 and Ga-4, respectively. The level of cyanidin-galactoside was increased by 5.3, 1.7 and 1.95 times for the lines Ga-1, Ga-2 and Ga-3, respectively, in comparison to the non-transgenic plants. Line Ga-4 didn’t show clear difference compared to the non-transgenic plants.
4. Discussion
4.1. Regeneration and transformation
In the present study, two different combinations of plant growth regulators for regeneration of the studied two cultivars were used. Since the optimum levels of plant growth regulators for shoot organogenesis are influenced by cultivar [30]. Similar results were reported by Yepes and Aldwinckle [31] who tested 13 genotypes under different growth regulators, while Puite and Schaart [32] tested different shoot regeneration and induction media as well callus induction media. For transformation, they used the gus gene and the transformation efficiency ranged between 0.2% and 8% depending on the variety. Szankowski et al. [33] reported the combination of BAP and IBA for HC and BAP alone for Elstar in propagation medium.
4.2. Phenotype
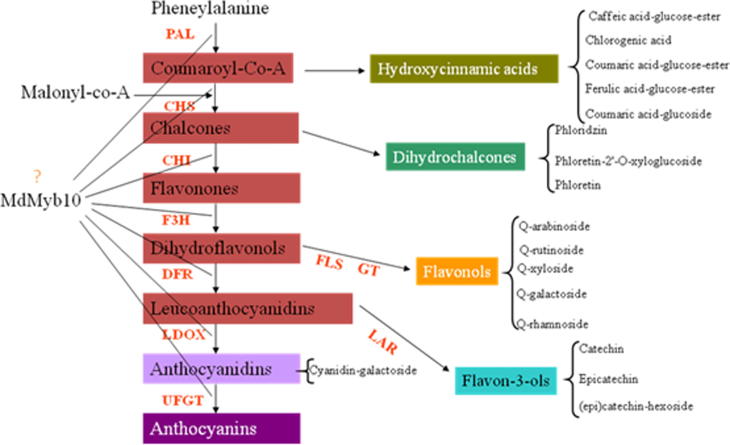
The biosynthetic pathway of flavonoids in apple and the expected roles of the transcription factor Myb10 are shown in Fig. 10 [15]. Transcription factors regulate the intensity and mode of anthocyanin accumulation and control transcription of different biosynthetic genes. One of these factors is Myb, which is involved in the regulation of anthocyanin synthesis [34].
Figure 10.
The biosynthetic pathway of flavonoids in apple and the expected role for MdMyb10 gene in regulation of this pathway. Enzymes required for each step are PAL, phenylalanine ammonia lyase; CHS, chalcone synthase; CHI, chalcone isomerase; F3 H, flavanone-3β-hydroxylase; FLS, flavonol synthase; GT, unidentified enzyme encoding a glycosyl transferase for flavonol glycone synthesis; DFR, dihydroflavonol-4-reductase; LAR, leucoanthocyanidin reductase; LDOX, leucoanthocyanidin dioxygenase; ANR, anthocyanidin reductase; UFGT, UDP-glycose:flavonoid-3-O-glycosyltransferase.
The phenotype change was highly distinguishable between HC′MdMyb10 transgenic plants and non-transgenic control plants by red colored calli, red shoots and well growing red plants (Fig. 8). The phenotype change in HC could be correlated with the content of flavan-3-ols, which was higher in transgenic HC line (29.92 mg/100 g dry matter) compared to control plant (21.41 mg/100 g dry matter) (Table 2). This was in agreement with Espley et al. [16], [21], [35] who reported that overexpression of MdMyb10 in apple generated a strong phenotype, with highly pigmented plants due to enhanced levels of anthocyanin. Li et al. [36] showed a similar result on phenotypic change due the LC expression in ‘HC’ transgenic apple plants.
Kortstee et al. [37] suggested the use of MdMyb10 as a selectable marker gene in transformation of apple and other crops to replace antibiotic selectable markers.
On the other hand, ‘Gala’ MdMyb10 transgenic plants were highly phenotypically distinguishable from non-transgenic control plants but only during the first 2–3 weeks on regeneration medium (Fig. 9). The content of flavan-3-ols was much higher in transgenic lines Ga-1,-2 and -3 with 65.2, 71.45 and 50.96 mg/100 g dry matter, respectively comparing with control plant (27.55 mg/100 g dry matter). This is in contrast to the results obtained by Espley et al. [35]. This is probably due to variation in expression level of the introduced MdMyb10 gene caused by the copy number, position of integration or silencing of the transgene [38].
4.3. Gene expression
The expression of MdMyb10 gene led to transcript increase in ‘HC’ and ‘Gala’ transgenic plants in comparison to the non-transgenic control plant (Figure 6, Figure 7). There was a 1261- fold and 150-847-fold increase in relative transcript levels for HC and Gala, respectively. It was noticed that the higher expression was correlated with the copy number found in Southern blot, the one copy number lines (Ga-1 and Ga-4) showed higher expression (847 and 610 fold, respectively) compared with two copies lines (Ga-2 and Ga-3) which had 86 and 190 fold, respectively. The variation in expression can be of several reasons such as copy number, cis-acting elements, transcriptional interference and chromatin structure [39].
The increase in transcript is correlated with the red color appearance (change of phenotype) (Figure 8, Figure 9), in case of Gala transgenic plant the change of color was observed only in the first two weeks during the regeneration steps. Similar results were published with other species, indicating a positive correlation between high transcript levels of Myb10 and accumulation of anthocyanin. Expression of sweet cherry PavMyb10 gene transcript was analyzed using qPCR during fruit development in two cherry cultivars, ‘Rainier’ and ‘Stella’. Showing higher transcript accumulation of PavMyb10 in the last two stages of fruit development of cultivar ‘Stella’ resulted in a deep red color compared to the pink color of cultivar ‘Rainier’ [20]. The variation in phenotype response of HC and Gala can be explained by the fact that different cultivars show different transcript levels for the same gene which corresponds with the results of Lin-Wang et al. [20], who reported the expression level of the strawberry genes, FvMyb10 and FaMyb10 during fruit development of wild diploid strawberry Fragaria vesca and cultivated octaploid strawberry Fragaria ananassa. In F. ananassa, the transcript level of FaMyb10 in ripened fruits showed 40,000-fold increase in relative transcript level compared to FvMyb10 in F. vesca which showed a little change. Furthermore, Medina-Puche et al. [12] showed that FaMyb10 is receptacle-specific ripening-related TF that regulates EBG and LBG of the F/P pathway that are related to the biosynthesis of anthocyanins in the strawberry fruit receptacle.
Espley et al. [16] compared ‘Red Field’ and ‘Pacific Rose™’ red- and white-fleshed cultivars of apple respectively during fruit development using qPCR to analyze the expression of the MdMyb10 gene. The results showed increased transcript levels of MdMyb10 in the fruit tissues of ‘Red Field’ compared to ‘Pacific Rose™’.
The expression of MdMybA from apple skin was analyzed using northern blotting [40]. They found that the expression levels of MdMybA were higher in ‘Jonathan’ (a deep-red cultivar) than that in ‘Tsugaru’ (a pale-red cultivar) and the expression of MdMybA is correlated with amount of anthocyanins accumulated in the mature apple skin.
Similar data were also reported by Honda et al. [17], showing that the final anthocyanin concentrations in ‘Jonathan’ and ‘Fuji’ as red apple cultivar were much higher than in ‘Orin’ as yellow apple cultivar.
4.4. Metabolites
In the present study, MdMyb10 ‘HC’ and ‘Gala’ transgenic plants showed increases in the total contents of polyphenol compounds analyzed up to 1.1 and 1.96 times in comparison to non-transgenic ‘HC’ and ‘Gala’ plants, respectively.
The relation between polyphenol compounds levels and plant disease resistance was reported in many previous studies. Mikulic-Petkovsˇek et al. [41] reported that leaves infected with the V. inaequalis accumulated 6.2-fold higher levels of flavonols. Picinelli et al. [42] showed significant difference in flavanol contents and phloridzid/flavanol ratio between resistant and susceptible cultivars of apple, they suggested using these parameters to distinguish between resistant and susceptible cultivars.
Also, it was suggested that phloridzin (dihydrochalcon glucoside) is hydrolyzed in vivo by various fungi such as V. inaequalis to create phloretin, which, is degraded to phloroglucinol, phloretic acid and p-hydroxybenzoic acid, and inhibit the development of the fungus [43].
Further, Mikulic-Petkovšek et al. [44] found that the infected leaves with scab contained 1.2 to 2.8-fold more phenolic compounds i.e. chlorogenic acid, p-coumaric acid, catechin, epicatehin, phloridzin and TPC than the healthy ones. Santos and Furlan [13] observed that anthocyanins are over-accumulated under low temperatures and increased ozone concentration, suggesting that it can function as defenses against abiotic stresses.
In our case, we could demonstrate that ‘HC’ and ‘Gala’ transgenic plants, showed increases in the total content of dihydrochalcons up to 1.1 and 1.7 times in comparison to non–transgenic ‘HC’ and ‘Gala’ plants, respectively.
Furthermore, MdMyb10 ‘HC’ and ‘Gala’ transgenic plants showed increases in the total content of flavonols up to 1.6 and 1.7 times in comparison to ‘HC’ and ‘Gala’ non-transgenic plants, respectively.
To conclude, the role of flavonoids in plant disease resistance is well studied, and confirmed in some of previous studies. The present study provides a strategy to investigate the role of flavonoids in plant resistance through genetic modification by elevating their levels. Further studies are needed to challenge the transgenic apples with biotic and abiotic stresses to draw a final conclusion.
Acknowledgements
The first author Kh. AL Rihani acknowledges the Islamic Development Bank-Merit Scholarship Program (Jeddah, Saudi Arabia) for the financial support of this study. The authors highly appreciate Prof. Iris Szankowski for her kind support for the work. The authors are grateful to Dr. M. Alkio and her group for their technical assistance.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.O'Rourke D.A. Haworth Press; Washington State, USA: 1994. The World Apple Market. [Google Scholar]
- 2.Hertog M.G.L., Hollman P.C.H., Katan M.B., Kromhout D. Nutr. Cancer. 1993;20:21–29. doi: 10.1080/01635589309514267. [DOI] [PubMed] [Google Scholar]
- 3.Treutter D., Feucht W. J. Hortic. Sci. 1990;65:511–517. [Google Scholar]
- 4.Dixon R.A. Comprehensive natural products chemistry. In: Sankawa U., editor. vol. 1. Elsevier; Amsterdam: 1999. pp. 773–823. (Polyketides and Other Secondary Metabolites Including Fatty Acids and their Derivatives). [Google Scholar]
- 5.Forkmann G., Martens S. Curr. Opin. Biotechnol. 2001;12:155–160. doi: 10.1016/s0958-1669(00)00192-0. [DOI] [PubMed] [Google Scholar]
- 6.Kreuzaler F., Ragg H., Fautz E., Kuhn D.N., Hahlbrock K. Proc. Natl. Acad. Sci. USA. 1983;80:2591–2593. doi: 10.1073/pnas.80.9.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto M., Nakatsuka S., Otani H., Kohmoto K., Nishimura S. Phytopathology. 2000;90:595–600. doi: 10.1094/PHYTO.2000.90.6.595. [DOI] [PubMed] [Google Scholar]
- 8.Treutter D. Plant Biol. 2005;7:581–591. doi: 10.1055/s-2005-873009. [DOI] [PubMed] [Google Scholar]
- 9.Treutter D. Environ. Chem. Lett. 2006;4:147–157. [Google Scholar]
- 10.Li C., Ng C.K.Y., Fan L.M. Exp. Bot. 2015;114:80–91. [Google Scholar]
- 11.Chaves N., Escudero J.C. In: Inderjit K., Dakshini M.M., Foy C.L., editors. CRC Press; Boca Raton: 1999. pp. 267–285. (Principles and Practices in Plant Ecology). [Google Scholar]
- 12.Medina-Puche L., Cumplido-Laso G., Amil-Ruiz F., Hoffmann Th., Ring L., Rodríguez-Franco A., Caballero J.L., Schwab W., Muñoz-Blanco J., Blanco-Portales R. J. Exp. Bot. 2013;65(2):401–417. doi: 10.1093/jxb/ert377. [DOI] [PubMed] [Google Scholar]
- 13.Santos A.C., Furlan C.M. Atmos. Pollut. Res. 2013;4(3):250–256. [Google Scholar]
- 14.Sandermann H., Ernst D., Heller W., Langebartels C. Trends Plant Sci. 1998;3:47–50. [Google Scholar]
- 15.Takos A.M., Ubi B.E., Robinson S.P., Walker A.R. Plant Sci. 2006;170:487–499. [Google Scholar]
- 16.Espley R.V., Hellens R.P., Putterill J., Stevenson D.E., Kutty-Amma S., Allan A.C. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda C., Kotoda N., Wada M., Kondo S., Kobayashi S., Soejima J., Zhang Z., Tsuda T., Moriguchi T. Plant Physiol. Biochem. (Paris) 2002;40:955–962. [Google Scholar]
- 18.Takos A.M., Jaffé F.W., Jacob S.R., Bogs J., Robinson S.P., Walker A.R. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin-Wang K., Bolitho K., Grafton K., Kortstee A., McGhie T., Espley R.V., Hellens R.P., Allan A.C. BMC Plant Biol. 2010;21:10–50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldoni E., Genga A., Cominelli E. Int. J. Mol. Sci. 2015;16:15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espley R.V., Bovy A., Bava Ch., Jaeger S.R., Tomes S., Norling C., Crawford J., Rowan D., McGhie T.K., Brendolise C., Putterill J., Schouten H.J., Hellens R.P., Allan A.C. Plant Biotechnol. J. 2013;11:408–419. doi: 10.1111/pbi.12017. [DOI] [PubMed] [Google Scholar]
- 22.Chagné D., Carlisle C.M., Blond C., Volz R.K., Whitworth C.J., Oraguzie N.C., Crowhurst R.N., Allan A.C., Espley R.V., Hellens R.P., Gardiner S.E. BMC Genomics. 2007;8:212–223. doi: 10.1186/1471-2164-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J.J., Zhang Z., Peng R.H., Xiong A.S., Xu J., Zhu B., Yao Q.H. Mol. Biol. Rep. 2011;38(1):205–211. doi: 10.1007/s11033-010-0096-0. [DOI] [PubMed] [Google Scholar]
- 24.Murashige T., Skoog F. Plant Physiol. 1962;15:473–497. [Google Scholar]
- 25.Holsters M., Silva B., Van Vliet F., Genetello C., de Block M., Dhaese P., Depicker A., Inze D., Engler G., Villarroel R. Plasmid. 1980;3:212–230. doi: 10.1016/0147-619x(80)90110-9. [DOI] [PubMed] [Google Scholar]
- 26.De Greve H., Dhaese P., Seurinck J., Lemmers M., Van Montagu M., Schell J. J. Mol. Appl. Genetics. 1983;1:499–511. [PubMed] [Google Scholar]
- 27.Doyle J.J., Doyle J.L. Focus. 1990;12:13–15. [Google Scholar]
- 28.Southern E.M. J. Mol. Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J., Fritsch E.F., Maniatis T. Cold Sping Harbor Laboratory Press, Cold Spring Harbor; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 30.Korban S.S., Chen H. Biotechnology of apples. In: Hammerschlag F., Litz R., editors. CAB International; Oxford, UK: 1992. pp. 203–227. (Biotechnology of Fruit Tree Crops). [Google Scholar]
- 31.Yepes L.M., Aldwinckle H.S. Plant Growth Regul. 1994;15:55–67. [Google Scholar]
- 32.Puite K.J., Schaart J.G. Plant Sci. 1996;119:125–133. [Google Scholar]
- 33.Szankowski I., Briviba K., Fleschhut J., Schönherr J., Jacobsen H.J., Kiesecker H. Plant Cell Rep. 2003;22:141–149. doi: 10.1007/s00299-003-0668-8. [DOI] [PubMed] [Google Scholar]
- 34.Telias A., Lin-Wang K., Stevenson D.E., Cooney J.M., Hellens R.P., Allan A.C., Hoover E.E., Bradeen J.M. BMC Plant Biol. 2011;11:93–104. doi: 10.1186/1471-2229-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espley R.V., Brendolise C., Chagné D., Kutty-Amma S., Green S., Volz R., Putterill J., Schouten H.J., Gardiner S.E., Hellens R.P., Allan A.C. Plant Cell. 2009;21:168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Flachowsky H., Fischer T.C., Hanke M.V., Forkmann G., Treutter D., Schwab W., Hoffmann T., Szankowski I. Planta. 2007;226(5):1243–1254. doi: 10.1007/s00425-007-0573-4. [DOI] [PubMed] [Google Scholar]
- 37.Kortstee A.J., Khan S.A., Helderman C., Trindade L.M., Wu Y., Visser R.G., Brendolise C., Allan A., Schouten H.J., Jacobsen E. Transgenic Res. 2011;20(6):1253–1264. doi: 10.1007/s11248-011-9490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butaye K.M.J., Cammue B.P.A., Delauré S.L., De Bolle M.F.C. Mol. Breed. 2005;16(8):79–91. [Google Scholar]
- 39.Birch R.G. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:297–326. doi: 10.1146/annurev.arplant.48.1.297. [DOI] [PubMed] [Google Scholar]
- 40.Ban Y., Honda C., Hatsuyama Y., Igarashi M., Bessho H., Moriguchi T. Plant Cell Physiol. 2007;48(7):958–970. doi: 10.1093/pcp/pcm066. [DOI] [PubMed] [Google Scholar]
- 41.Mikulic-Petkovšek M., Stampar F., Veberic R. J. Plant Pathol. 2008;90:49–55. [Google Scholar]
- 42.Picinelli A., Dapena E., Mangas J.J. J. Agric. Food Chem. 1995;43:2273–2278. doi: 10.1021/jf9903197. [DOI] [PubMed] [Google Scholar]
- 43.Hamauzu Y. Stewart Postharvest Rev. 2006;2(2):1–7. [Google Scholar]
- 44.Mikulic-Petkovšek M., Stampar F., Veberic R. Can. J. Plant Sci. 2009;89:745–753. [Google Scholar]