Abstract
The need for a new antibiotic pipeline to confront threat imposed by resistant pathogens has become a major global concern for human health. To confront the challenge there is a need for discovery and development of new class of antibiotics. Nature which is considered treasure trove, there is re-emerged interest in exploring untapped microbial to yield novel molecules, due to their wide array of negative effects associated with synthetic drugs. Natural product researchers have developed many new techniques over the past few years for developing diverse compounds of biopotential. Taking edge in the advancement of genomics, genetic engineering, in silico drug design, surface modification, scaffolds, pharmacophores and target-based approach is necessary. These techniques have been economically sustainable and also proven efficient in natural product discovery. This review will focus on recent advances in diverse discipline approach from integrated Bioinformatics predictions, genetic engineering and medicinal chemistry for the synthesis of natural products vital for the discovery of novel antibiotics having potential application.
Keywords: Genome mining, Genome shuffling, Metabolomics, Combinatorial synthesis and antibiotics
1. Introduction
Microbial natural products (MNPs) have been a prominent source for drug discovery since the discovery of the penicillin by Alexander Fleming. MNPs has been critical importance in therapeutically viable drugs against infectious diseases, antioxidants, cancer, diabetes, etc. [1] During the age of antibiotic discovery 1940–1970 has been accounted for major antibiotics known today as a result of vast screening efforts by natural product researchers. FDA assessment of natural products reveals that nearly one-quarter of these has been derived from microbial sources [1], [2]. The long-established approach of ‘herbal remedy’ has been substituted with precise compounds emerging from the gene and molecular altered microbial products. Hence, microbial natural products have replaced plant-derived compounds as a source of therapeutic agents and has been a potential source for the discovery of novel therapeutics in the past decades. In recent years, there has been declination of natural products but an increase in their synthetic derivatives. This has greatly impacted therapeutic pipeline from the exploration of novel natural products being abandoned in the last decade by pharmaceutical industries. The perception of all the major classes of antibiotics had been found to be the main reason for the abandonment. Synthetic drugs have been preferred as screening methods for natural products, resulted in redundant isolates than novel compounds, also due to large-scale amount and time required for discovery [3]. Nowadays, the funding opportunities have been provided through academia, charities and governmental institutions for the quest. Due to the emergence of antimicrobial resistance (AMR), the antibiotic pipeline for combating serious life-threatening infections is almost empty against drug-resistant pathogens [1], [2]. But, the diversity of chemical skeletons provided by MNPs has been a ray of hope. To tackle the unmet clinical challenge posed by multidrug resistance and also to cure ailments there is a need to readdress the issue with a new class of antibiotics through innovative approaches with completely different new framework using a directed synthesis of MNPs [2]. Earlier studies state natural products, as a result of evolutionary modification by mutation or through variation leading to change in the biosynthetic pathways for emergence bioactive molecules with the potency and selectivity to bind to biological targets [3], [4]. This has attracted natural product researchers to explore the enzymology and biosynthesis for modulation possibilities for new molecular entities. There have been several new techniques developed for the natural product discovery in the recent years, initiating a ‘new era in natural therapeutic discovery’.
This review is an effort to examine the current scenario, new approaches for exploration of natural therapeutics and also highlights recent reports of novel natural products derived from recent drug discovery technologies.
2. Microbial natural products as therapeutics
Threat imposed by drug-resistant pathogens has been a global concern. The inefficacies of available antibiotic pipeline in combating existing multidrug-resistant pathogens has been a critical issue in recent years. This has been attributed due to overexploitation antibiotics resulting in resistance to these pathogens. MNPs could be a prominent strategy for meeting the present preclinical requisite exerted by drug-resistant pathogens. MNPs have unusual chemical skeletons and diversification which could target varied regions leading to the potential for drug discovery. The regained interest of natural products with the amalgamation of cutting-edge developments in drug discovery has gained utmost importance for a targeted approach. MNPs have offered an efficient source of new drug leads, however, it is attributed to evolutionary advantage by selective mutation for the organism. In this regard concoction of untapped sources with recent advances in drug discovery techniques would enable to provide a new class of antibiotics. MNP analogue libraries could provide a base for medicinal chemistry for pharmaceutical profiling for enhancing the desired phenotype as well as providing insights on target and pathways [5], [6].
3. Antibiotic resistance
Antibiotics had been effective against prevalence and prevention of infections in the yesteryears. But, recently there has been a complication of resistance adopted by most of the pathogens in the pipeline of available antibiotics. The resistance has been linked to overexploitation of antibiotics leading to molecular, gene, cellular and community level resistance. Antibiotics have high affinity to the targets if there is a single point of mutation could trigger the surface resulting in resistance and proliferation as well transfer of resistance gene by transformation. Enzyme catalysed inactivation of antibiotics by hydrolysis has been crucial reason for failure β lactam antibiotics preventing cell penetration, alteration of targets and resisting the action of antibiotics [7]. Formation of biofilms has been one of the major reasons for antibiotic failure. Microbes conglomerate to form biofilms leading to irreversible attachment to the biotic/abiotic surface. Biofilms aid consortia of microbes against oxidative stress, increase in specific efflux pumps and also protection by providing an outer layer of extracellular polysaccharide layer. In addition, the transfer of antibiotic resistance genes by horizontal gene transfer is found to be in increased levels [8], [9]. Hence, the concentration of antibiotic to inhibit these planktonic cells is much higher when compared to sessile cells.
4. Regulation of antibiotic biosynthesis
Antibiotics are secondary metabolites without a functionalised role in cell growth or reproduction. Antibiotics inherit specialised functions, which trigger intra/intracellular signalling (such as Carbon Catabolite Regulation, Nitrogen Metabolite Regulation, Phosphate Regulation), either for communication, nutrition and defence. The major diversity of antibiotics is attributed to combination synthesis of polyketides (PKS), non-ribosomal peptides (NRPS) pathways [10], [11].
5. Conventional discovery and development of antibiotics
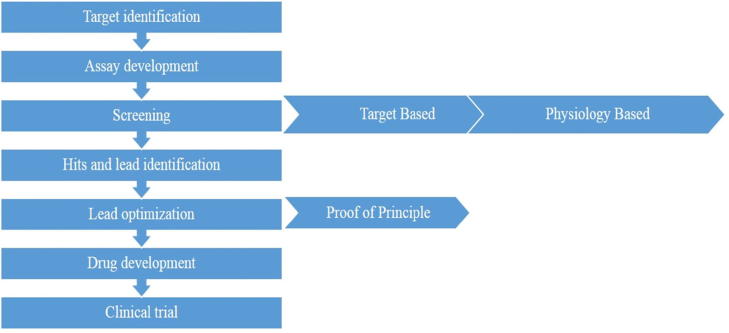
Drug discovery involves identification of compounds with potential for blocking a single gene, gene product or molecular mechanism determined by genetic analysis or biological observations associated with the disorder [12]. Drug discovery involves multistep process Fig. 1. Target identification, assay development, screening, Hits and lead identification, lead optimization, drug development and clinical trial [13].
Fig. 1.
Multi step process of conventional drug discovery.
Identification of target is a crucial preliminary step in drug discovery. The target could be an enzyme or protein which needs to be either blocked or activated to induce response for the cure of abnormality. Assay development involves a quest for bioactive compounds e.g., bioautography, paper disk method inhibiting the target to restore homeostasis. Screening enables high-throughput data for analysing a multitude of compounds. Hits and leads stage, a variety of parameters are examined such as the specificity, toxicity, and molecular structure of the bioactive molecule. Lead optimization is employed to ameliorate specificity, reduce toxicity, and also to enable a wide antibiotic spectrum of lead compound [14]. Once the criteria are achieved according to standards, drug development and clinical trials are followed. The drug is tested for safety and efficacy, in animal models preliminarily, followed in three stages on humans in clinical settings.
6. Omics based approach for biodiscovery of microbial natural products
6.1. Potential sources of microbial antibiotics for drug discovery
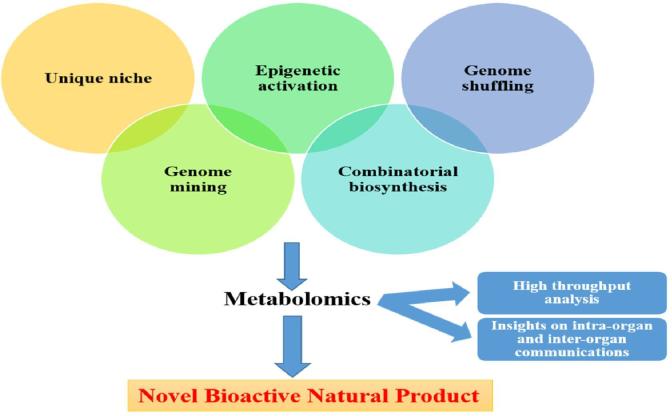
Unique niche in extreme climatic conditions, unusual landscapes and geographical location leading to the multiplex of adaptations (physiological and behavioural) could result in unusual biology for inhabiting species. These unique reservoirs are a potential source to isolate broad assemblage of microbes having pharmaceutical value. Advancement in metagenomics has resulted in the knowledge base of diverse and complex microbial communities surviving in lakes, rivers, sediments and marine environments, also in extreme conditions like ice cores, hydrothermal vents, sub-seafloor sites and caves. These bio-diverse populations could serve as an antibiotic mining source, having diverged and variable chemical structures of pharmaceutical interest [15], [16] (Fig. 2).
Fig. 2.
Omics based approach for biodiscovery of microbial natural products.
6.2. Genome mining
In the past decade, genome mining as an alternate strategy which has been considered against traditional screening for drug discovery. The genome mining approach aid in identifying and prediction analysis of biosynthetic gene clusters from genes to molecules. With a large library of information of DNA existing with the computational approach, there is the probability of wide array of novel molecules encoded in genome providing a new class of antibiotics. Genome mining has proved to be fast, the user-friendly and reliable technique also cost effective. Mining of genome has been potential for previously unfeasible antibiotics. The mining approach would enable target directed discovery by predicting the chemical structure and class using bioinformatics in amalgamation with bioassay-guided isolation, along with silent gene activation and heterologous expression techniques for novel bioactive metabolites [17], [18]. For instance, polyketides (PKs) and non-ribosomal peptides (NRPs) represent abundances of molecules with biological diversity such as daptomycin, erythromycin, and many others. NRPs and PKs of microbial origin represents a significant proportion of natural bioactive products. The accumulated knowledge of the NRPSs or PKSs constituting biosynthetic pathways or another type of NPs gene clusters helps to predict and isolate new gene clusters, engineering or production of NPs in heterologous hosts. There has been a significant development in a number of algorithms developed based on probabilistic approach, evolutionary distances in the genome to understand the pathways for biosynthesis of natural products. Most of the algorithms presently employed are based on domains and Pfam regions in well-known secondary metabolite pathways such polyketide synthases (PKS) and non-ribosomal peptide synthatases (NRPS).
Cluster Assignment by Islands of Sites (CASSIS) was developed by Wolf et al. CASSIS scans the genome (FASTA format) for key enzymes prerequisite for secondary metabolite production such as polyketide synthases (PKS), non-ribosomal peptide synthetases (NRPS) and dimethylallyltryptophan synthases [19]. Another program antiSMASH 3.0 was developed by Weber et al. an updated version of the previous server-based program. The program uses the probabilistic approach and allows the user to input using multi-FASTA, GenBank and EMBL formats. The embedded modified Cluster Blast and ClusterFinder with better sensitivity and specificity to identify biosynthetic gene clusters has potential for discovering novel drugs [20]. A new PKS-NRPS hybrid product, haliamide was determined by in silico genome analysis using antiSMASH from myxobacterium Haliangium ochraceum. The biosynthetic gene cluster constituted one NRPS module with four PKS modules having 21.7 kbp. The putative gene cluster harboured in myxobacterium could lead to the isolation of additional compounds with biological potential [21]. Salinilactam, a polyene macrolactam was discovered along with 17 novel biosynthetic gene clusters coding for polyene derivatives from a marine actinomycete Salinispora tropica [22]. Genome mining and genetic manipulations strategies were employed on marine actinomycete Streptomyces olivaceus yielded two new naphthoquinone macrolides olimycins A and B [23]. A 92 kb cryptic biosynthetic gene comprising hybrid polyketide and nonribosomal peptide marine Streptomyces pactum, was activated by genetic modification resulted in a novel sulfonate containing analogue totopotensamide C [24]. Sweeney et al. reported the genome mining of Streptomyces nodosus, the organism constituted several biosynthetic gene clusters for polyketides, peptides, siderophores and terpenes synthesis. These compounds were found to be expressed in low quantities or silent in comparison with amphotericin expression [25]. The notable anti-infectives through mining approach include clarepoxcins and landepoxcin [26], cypemycin, [27] haloduracin [28], Lactocillin [29], microcyclamide [30], teixobactin [31] reistomycin [32], microviridin [33] and microbisporicin [34] to mention in few. Besides the above programs there are 2metDB, BAGEL, CLUSEAN, ClusterFinder, eSNaPD, EvoMining, FunGeneClusterS, MIDDAS-M and SMURF [35], [36], [37], [38], [39], [40], [41], [42], [43].
Beside these antibiotics, other novel bioactive natural products include EstDZ2 bacterial esterolytic enzyme possessing moderate thermostability which exhibited stability to withstand high concentrations of organic solvents [44], BP-M-CPF4 (PhaC) a polyhydroxyalkanoate (PHA) synthase possessing very wide substrate specificity [45], EM3L4 lipase retaining salt-resistance property [46], genome mining studies by Roopa et al. for taxol from endophytes of Salacia oblonga revealed a total of 8 mycoendophytes, seven coded for DBAT (deacetylbaccatin III-10-O-acetyl transferase) and one for BPAT (C-13 phenylpropanoid side chain-CoA acyltransferase) proving the mining strategies, as a potential tool for new taxol derivatives possessing anticancer activity [47].
Streptomyces coelicolor genome mining revealed locus SCO 7131 of estA gene suggested that it might differ in substrate specificity with respect to members of the hormone-sensitive lipase (HSL) family of lipases and esterases. The estA gene was expressed using His-tagged protein in Escherichia coli, enzyme purified was found to be an esterase, which hydrolyzed the acetate ester of p-nitrophenol, but possessed mild activity on esters with longer side chains and exhibited optimal activity against caproate (C6) esters. Modification using site-directed mutagenesis led to an increased activity against butyrate and caproate esters but affected by reducing the thermostability of the enzyme. Using the genome mining data, the changes in the amino acid in conserved regions to indicate active sites could be employed for characterization of novel enzymes [48]. Genome mining of Streptomyces coelicolor revealed beyond 50 presumptive genes for putative lipolytic enzymes. EstC, purified recombinant enzyme from the gene cluster exhibited cold-active esterase activity along with the production of valuable esters. The enzyme possessed alcohol and organic solvent tolerance, which could be employed for organic synthesis of short-chain esters such as flavours [49]. Genome mining is a powerful toolbox for novel natural products biosynthesis and also to decipher regulation, distribution as well as elucidating biosynthesis pathways.
6.3. Epigenetic activation
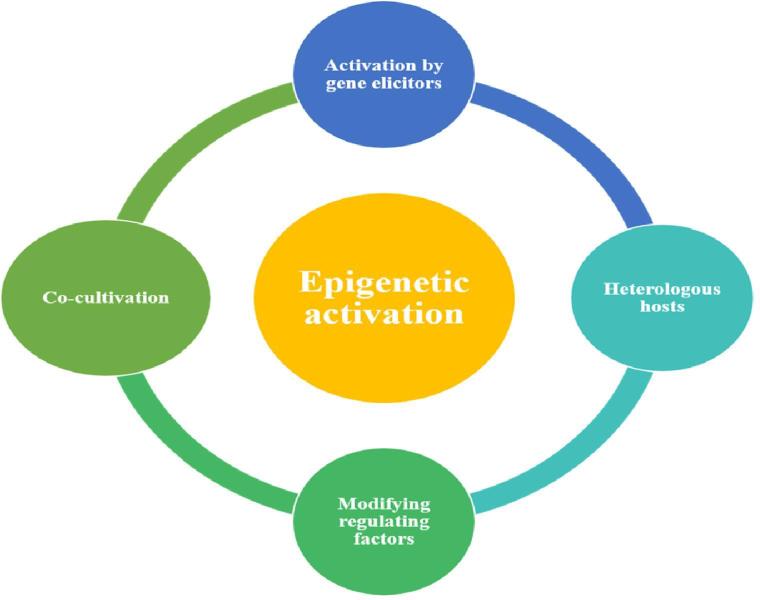
The metagenomics facilitated the next generation sequencing technology has provided insights towards the cryptic gene clusters. Various strategies have been put in effort past decade towards activating these silent gene clusters for new drugs. Many techniques have been put forward by natural product researchers in the past decade such as activation by gene elicitors, using heterologous hosts, regulating factors and co-cultivation for expression of gene clusters [50], [51].
Four gene clusters involved in the synthesis of sterigmatocystin, penicillin, terrequinone and orsellinic acid by H4 acetylation targets was reported by Soukup et al. The histone acetyltransferase had a crucial role in the synthesis of secondary metabolites and epigenetic regulation in Saccharomyces cerevisiae [52]. Du et al. reported new chlorinated polyketide, daldinone E a potent antibacaterial from Daldinia sp. using epigenetic modifier suberoylanilide hydroxamic acid (SAHA) [53]. Expression and production of polyketides have been difficult due to imitating factors such as Posttranslational Modification, Substrate Availability, Intracellular Factors, Transmembrane Transporters Post-PKS Polyketide Modification, Self-Resistance exerted by parent strain. The following candidates have been utilised in recent year as heterologous hosts Streptomyces coelicolor, Streptomyces glaucescens, Saccharopolyspora erythraea, Saccharomyces cerevisiae [54], Aspergillus oryzae and Aspergillus nidulans [55] which could serve robust, economically feasible and efficient system for production of compounds with pharmacological interest. Kumpfmüller et al. recently reported 6-deoxyerythronolide B (6dEB) by Bacillus subtilis. To enhance the production gene knockout surfactin (26 kb), bacillaene (76 kb), and plipastatin (38 kb) resulting in deletion of the prpBD operon showed a significant increase of the 6dEB yield. The study indicated B. subtilis could serve as a candidate for heterologous expression for the synthesis of complex polyketide [56] (Fig. 3).
Fig. 3.
Epigenetic activation strategies.
6.4. Combinatorial biosynthesis
Combinatorial biosynthesis can be generally referred as “induced evolution” for designer antibiotics through genetics and/or medicinal chemistry. The engineering of novel chemical entities by combinatorial approach utilises the key enzymes involved in secondary metabolite of antibiotic synthesis pathways such as polyketide synthase (PKS), nonribosomal peptide synthase (NRPS), thioesterase (TE), acyl transferase (AT), 6-deoxyerythronolide B synthase (DEBS), dehydratase (DH), enoyl reductase (ER), ketoreductase (KR) and ketosynthase (KS). The combinatorial synthesis has been proven successful between antibiotic-producing organisms for producing hybrid gene products. The feasibility of technique has been efficient among organisms constituting PKS gene (PKS Type I and II) due to the fact PKS families consist of variable and multifunctional roles [57], [58], [59]. PKS and NRPS assemblies constitute large enzymes composed of polypeptides ranging from 100 kDa to MDa. The antibiotic synthesis involves gene to enzyme creating chemical triggers in assembly-line fashion to synthesize the final product. To achieve effective biosynthesis natural product researchers must balance the antibiotic synthesis pathway with host cell growth which could possibly by achieved genetic engineering in certain scenarios.
Nine xantholipin analogues were obtained using PKS tailoring modification from Streptomyces flavogriseus. Biosynthetic gene cluster constituted 52 kb encoding xantholipin gene with type II polyketide synthases and regulators. In-frame mutagenesis of five enzymes resulted in new derivatives which were catalysed by FAD binding monooxygenase XanO4, followed by bridge formation by P450 monooxygenase XanO2 and hydroxylation of the carbon skeleton by the FAD-binding monooxygenase XanO5. The polycyclic polyketide derivatives can be potent antibiotic as well as antitumour [60]. Post PKS one step tailoring in putative cytochrome P450 monooxygenases, MakC2 and MakC3 resulted in a new spirotetronate-class antibiotic 29- Deoxymaklamicin from Maklamicin biosynthesis pathway in Micromonospora sp. 29- Deoxymaklamicin was found to be inhibitory against gram-positive bacteria [61].
6.5. Genome shuffling
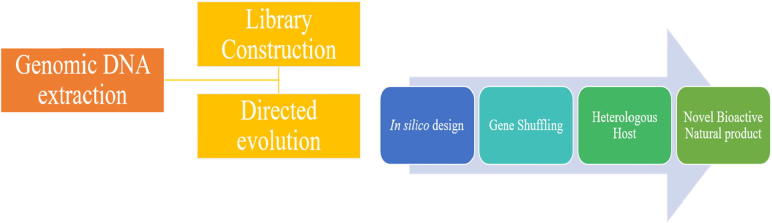
Conventionally the utilising random mutation and screening strategies had been used, although having some advantages it’s economically not feasible, labour and time intensive. With advancement in the ‘omics’ era, genome shuffling has been an alternate strategy which could even be employed in complex phenotypes which were difficult earlier. Genome shuffling with aid of advanced genetic tools has been able to assist in targeted genetic manipulation for a phenotype. The technique has been reliable for offering both strain improvement, also more providing insights into cellular information of the desired phenotype. Many molecules have been developed using directed evolution with DNA shuffling which has been a convenient strategy for whole cell and metabolic engineering. A concoction of high throughput analysis with in silico modelling/simulations has enabled to understand the global context of the metabolic system. Whole genome engineering could be considered as an inspiration from natural evolution which is a selection of desired phenotypic traits. With advancements in experimental and computational techniques genome shuffling has caught global consideration. The DNA shuffling allows accelerated directed evolution and pathway engineering, which is crucial for strain improvement of industrially important microorganisms [62], [63].
Increased production of avilamycin from Streptomyces viridochromogenes, from protoplast fusion, resulted in 36.8-fold in the recombinant. Mutation in ribosome protein S12 was considered the factor responsible for enhanced production as the well morphological difference in conidiospore, and hyphae pellets from the parent. The compound inhibits Staphylococcus pneumoniae, Streptococcus pyogenes, Enterococcus faecalis, Enterococcus faecium, methicillin-resistant Staphylococcus aureus (MRSA), and methicillin-resistant Staphylococcus epidermis (MRSE). The combination of genome shuffling and ribosome engineering was successful in improving the phenotype for yield production of avilomycin [64] (Fig. 4).
Fig. 4.
Genome shuffling for novel MNPs.
6.6. Metabolomics
Metabolomics provides high throughput analysis of metabolites and simultaneous comparison platform of biological samples providing insights on crucial roles in cellular processes, intra-organ and inter-organ communications. The analysis of the metabolome is especially arduous because of the varied chemical nature of metabolites. Metabolomics is an expeditious turning into one in all the platform sciences of the ‘omics’. The technique is an extension of identification that is predicated on the drug target identification and it's biological transformation sanctions, the drug replication data for a clinical disorder. This review of the utilisation of metabolomics in integrated applications wherever metabolomics data has been amalgamated with different ‘omic’ cognizance sets to transmute more immensely colossal understanding of a biological system. The potential of metabolomics for natural product drug revelation and practical analysis, primarily as incorporated into broader ‘omic’ erudition sets is enormous. Recent advances in metabolomics technologies have been extensively applied in biomedical applications. Especially, metabolomics is progressively being employed in disease diagnose, determining novel drug targets, custom designed drugs and supervise therapeutic consequence. The expeditiously developing field of metabolomics consolidates methodologies to apperceive and quantify cell metabolites utilising multi-variation strategies for data mining and information processing. Concurrently, substantial notional theorizations were made to investigative ways to decipher distinctive cell items, for example, those from quality articulation (transcripts), proteins, and metabolites. These alleged omics approaches, including genomics, transcriptomics, proteomics, and metabolomics, are viewed as vital contrivances to be connected and used to comprehend for drug discovery. A scope of analytical advances has been made to investigate metabolites in biological samples. It has been suggested that metabolomics greatest potential lies in disease marker revelation and detection. These metabolites are not only biomarkers for the disease but additionally accommodate as ‘biomarkers of efficacy’. This designates they sanction the utility of incipient drug which has to be rapidly assessed in cell-predicated or enzymatic assays. Furthermore, some metabolites may accommodate as potential targets for incipient drug therapies or as potential drugs, in and of themselves. Many genetic disorders arise from multiple gene defects and being able to distinguish between these disorders and to identify their root causes which are critical in finding out the drug targets, felicitous drugs or cumulations of dietary supplements to treat them [65], [66], [67]. Metabolomics has made an impact on drug revelation and development processes dramatically by quantifying accurately the spectrum of biochemical changes and mapping these vicissitudes to metabolic pathways. This technology provides data that is less intricate, precise, germane and quantitative than genomics, transcriptomics or proteomics. With this methodology, it is now possible to develop a construal of disease states and incipient treatment modalities much more expeditious and more accurately than before. Metabolomics has broad applications across the drug revelation and development processes. Metabolomics is a puissant and new scientific approach for the revelation and development of drugs and the early diagnosis of disease onset.
The integration of ‘omics’ knowledge has potential in natural product drug discovery [68]. One of the often-perceived hurdles committed natural product drug discovery is that the chance of redundancy. There are numerous ‘dereplication’ procedures, which have established to deal with this drawback, significantly employed by researchers focusing on microbial drug leads. NMR and MS-based platform give structural information of metabolites in an early stage, the comparison of structural information procured by NMR and high-resolution mass spectrometry with literature. If structures are reported, discriminators can be identified without the need for laborious isolation process. Thus, metabolomics serves as an approach for dereplication.
New prenylated Isatin antibiotic Streptomyces Species MBT28 was reported using NMR-based metabolomics. NMR-guided isolation was achieved by tracking the target proton signal which resulted in the characterization of a 7-prenylisatin antibiotic effective against Bacillus subtilis. NMR-based metabolomics with proteomic approach facilitated identification of gene cluster which deciphered an enzyme indole prenyltransferase that catalyzed the conversion of tryptophan into 7-prenylisatin [69]. Wu et al. reported novel C-glycosylpyranonaphthoquinones in Streptomyces sp. MBT76 utilising combined approach of NMR based metabolomics and mining strategy. The qin gene cluster of type II PKS pathway was activated leading to 8-C-glycosylation, 5,14-epoxidation, and 13-hydroxylation also fusing deoxy aminosugar to the pyranonaphthoquinone. The new C-glycosylpyranonaphthoquinones were effective against Gram-positive bacteria, which led insight to pyranonaphthoquinone antibiotics [70]. Metabolomic analysis using high-resolution LC-MS and NMR for dereplication purposes was employed in the study of 64 actinomycetes was isolated from 12 different marine sponge species. The LC-MS/NMR based detection aided in prioritizing two isolates belonging to the genera Streptomyces (SBT348) and Micromonospora (SBT687) which exhibited distinct chemistry profiles. The isolates possessed potent anti-trypanosomal activities. The chemically unique strains were profiled rapidly with utilisation hyphenation techniques of metabolomics [71]. Chemical communication in co-culture of Cladosporium sp. WUH1 and Bacillus subtilis CMCC (B) 63,501 resulted in the production of previously reported diphenyl ethers with polyhydroxy sidechains including a novel antibiotic. The metabolomic approach integrating LC-MS analysis, along with statistical tools and molecular networking revealed diphenyl ethers might be a defensive response against growth inhibition resulting from surfactins by B. subtilis [72]. A series of lipidic spirohemiaminals possessing broad antimicrobial activity were reported by surveying unique molecular signatures identified in the mass spectrometry data obtained by co-culture of Streptomyces nigrescens HEK616 and Tsukamurella pulmonis TPB0596 [73]. NMR-based metabolomics combined with multivariate data analysis was employed in the determination of secondary metabolites produced by co-culture of revealed A. niger and S. coelicolor. The biotransformation studies revealed synthesis of novel compounds (E)-2-(3-hydroxyprop-1-en-1-yl)-phenol and (2E, 4E)-3-(2-carboxy-1-hydroxyethyl)-2, 4-hexadienedioxic acid, respectively [74]. The comparison of the metabolome profile of the co-culture with that of the individual microorganisms would enlighten differentiation of induced metabolites present in co-culture but absent in monocultures, also these de novo-engineered molecules have high chances of being novel compounds or new derivatives with biological activity.
Metabolomics approach for natural product discovery greatly depends on natural product libraries which enable rapid characterization of known compounds. Spectroscopic data libraries such as TCM Database@Taiwan, traditional Chinese medicine integrative database (TCMID), Chinese ethnic minority traditional drug database (CEMTDD), SuperToxic, NPACT, 3DMET DNP (Dictionary of Natural Products), Dictionary of Marine Natural Products, ACD/Labs NMR, ACD/Spectrus, UCSD Natural Products Database Napralert, NuBBE and SuperNatural have been developed over the recent years [75], [76].
Metabolomics, the study of the whole repertoire of tiny molecules in cells, tissues, organs, and biological fluids, which represents a significant and speedily evolving part of the new systems biology. Relative to different omics fields, metabolomics remains a comparatively young discipline. There are challenges in terms of technology, experimental design, information analysis, and information integration that may influence the sphere of metabolomics and its application to systems biology. However, many advances in metabolomics technologies have been developed. Instrumentation has been quicker, more sensitive with reliable and automated reducing of any error in constituent measurements, also applied statistical analyses for data validation. Given the potential of this technology in this field, there is a rise in the number of publications in the recent years. Metabolomics has real potential to facilitate the study of systems biology for predicated drug revelation by assessing drug response and its adverse reaction along with variation involved for natural product biodiscovery.
7. Conclusion
The regained interest in evaluating the efficacy of natural products from microbes as a source of new drugs is very crucial for antibiotic drug discovery. The conventional approach of antibiotic discovery should be re-evaluated with high-throughput screening, metagenomics and metabolomics for the pursuit of novel compounds. The amalgam of metagenomics with genomics, genetic engineering and medicinal chemistry would provide exciting novel antibiotic leads that could serve the issue of antibiotic resistance. The quest for novel bioactive natural products by the advancement of leading technologies has gained spotlight. Regardless of, amelioration in culturing techniques still render drawback of microbe uncultivable. With the advent of metagenomics have aided in isolation, identification and sequencing of these microbes. The boost in the knowledge base of microbe diversity in extreme environments with help of metagenomics has opened a new direction in the identification of novel isolates. Combination of metagenomics with recent developments of understanding in genome mining, genetic engineering, in silico design, metabolomics and medicinal chemistry could aid in the development of target-specific bioactive molecules for multifaceted necessities.
Acknowledgments
Acknowledgements
The authors would like to thank Department of Studies in Microbiology, University of Mysore, Manasagangotri, Mysore, Karnataka, India for providing facilities.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Naman C.B., Leber C.A., Gerwick W.H. 1st ed. Academic Press; 2017. Microbial resources: from functional existence in nature to applications; p. 103. [Google Scholar]
- 2.Patridge E., Gareiss P., Kinch M.S., Hoyer D. Drug Discovery Today. 2016;21(2):204–207. doi: 10.1016/j.drudis.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Harvey A.L., Edrada-Ebel R., Quinn R.J. Nat Rev Drug Discovery. 2015;14(2):111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 4.Abel-Santos E., Hedstrom L., Rotella D., Melander C., Thurston D., Fisher S., Khursigara C., Lazarides L., Garneau-Tsodikova S., Balibar C.J. Royal Society of Chemistry; 2017. Antibiotic Drug Discovery. [Google Scholar]
- 5.Katz L., Baltz R.H. J Ind Microbiol Biotechnol. 2016;43(2–3):155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 6.Huang T., Lin S. J Appl Microbiol Biochem. 2017;2:1. [Google Scholar]
- 7.Gabrani R., Sharma G., Dang S., Gupta S. Free Radicals Human Health Disease. 2015:369–379. [Google Scholar]
- 8.Blair J.M., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J. Nat Rev Microbiol. 2015;13(1):42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 9.Bazzini S., Buroni S., Udine C., Pasca M., Riccardi G. Caistee Academic Press; 2014. Burkholderia: from genomes to function; pp. 183–196. [Google Scholar]
- 10.Wang H., Fewer D.P., Holm L., Rouhiainen L., Sivonen K. Proc Natl Acad Sci. 2014;111(25):9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khosla C., Herschlag D., Cane D.E., Walsh C.T. Biochemistry. 2014;53(18):2875–2883. doi: 10.1021/bi500290t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman R.B., Holladay M.W. 3rd ed. Academic Press; 2014. The organic chemistry of drug design and drug action. [Google Scholar]
- 13.Bleicher K.H., Böhm H.J., Müller K., Alanine A.I. Nat Rev Drug Discovery. 2003;2(5):369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 14.Ratti E, Trist D. Pure and applied chemistry 73(1) (2001) 67–75 http://doi.org/10.1351/pac200173010067.
- 15.Cragg G.M., Newman D.J. Pharm Biol. 2001;39(sup1):8–17. doi: 10.1076/phbi.39.s1.8.0009. [DOI] [PubMed] [Google Scholar]
- 16.Berdy J. J Antibiot. 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 17.Zerikly M., Challis G.L. ChemBioChem. 2009;10(4):625–633. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
- 18.Li M.H., Ung P.M., Zajkowski J., Garneau-Tsodikova S., Sherman D.H. BMC Bioinf. 2009;10(1):185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf T., Shelest V., Nath N., Shelest E. Bioinformatics. 2015;32(8):1138–1143. doi: 10.1093/bioinformatics/btv713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Müller R., Wohlleben W., Breitling R. Nucleic Acids Res. 2015;43(W1):W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y., Tomura T., Sato J., Iizuka T., Fudou R., Ojika M. Molecules. 2016;21(1):59. doi: 10.3390/molecules21010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Udwary D.W., Zeigler L., Asolkar R.N., Singan V., Lapidus A., Fenical W., Jensen P.R., Moore B.S. Proc Natl Acad Sci. 2007;104(25):10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun C., Zhang C., Qin X., Wei X., Liu Q., Li Q., Ju J. Tetrahedron. 2018;74(1):199–203. [Google Scholar]
- 24.Chen R., Zhang Q., Tan B., Zheng L., Li H., Zhu Y., Zhang C. Org Lett. 2017;19(20):5697–5700. doi: 10.1021/acs.orglett.7b02878. [DOI] [PubMed] [Google Scholar]
- 25.Sweeney P., Murphy C.D., Caffrey P. Appl Microbiol Biotechnol. 2016;100(3):1285–1295. doi: 10.1007/s00253-015-7060-9. [DOI] [PubMed] [Google Scholar]
- 26.Owen J.G., Charlop-Powers Z., Smith A.G., Ternei M.a., Calle P.Y., Reddy B.V.B., Montiel D., Brady S.F. Proc Natl Acad Sci USA. 2015;112(14):4221–4226. doi: 10.1073/pnas.1501124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claesen J., Bibb M. Proc Natl Acad Sci. 2010;107(37):16297–16302. doi: 10.1073/pnas.1008608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClerren A.L., Cooper L.E., Quan C., Thomas P.M., Kelleher N.L., van der Donk W.A. Proc Natl Acad Sci USA. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donia M.S., Cimermancic P., Schulze C.J., Brown L.C., Martin J., Mitreva M., Clardy J., Linington R.G., Fischbach M.A. Cell. 2014;158(6):1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziemert N., Ishida K., Quillardet P., Bouchier C., Hertweck C., de Marsac N.T., Dittmann E. Appl Environ Microbiol. 2008;74(6):1791–1797. doi: 10.1128/AEM.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P., Mueller A., Schaberle T.F., Hughes D.E., Epstein S., Jones M., Lazarides L., Steadman V.A., Cohen D.R., Felix C.R., Fetterman K.A., Millett W.P., Nitti A.G., Zullo A.M., Chen C., Lewis K., Schaberle T.F., Hughes D.E., Epstein S., Jones M., Lazarides L., Steadman V.A., Cohen D.R., Felix C.R., Fetterman K.A., Millett W.P., Nitti A.G., Zullo A.M., Chen C., Lewis K. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spohn M., Kirchner N., Kulik A., Jochim A., Wolf F., Muenzer P., Borst O., Gross H., Wohlleben W., Stegmann E. Antimicrob Agents Chemother. 2014;58:6185–6196. doi: 10.1128/AAC.03512-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziemert N., Ishida K., Liaimer A., Hertweck C., Dittmann E. Angew Chem Int Ed Engl. 2008;47:7756–7759. doi: 10.1002/anie.200802730. [DOI] [PubMed] [Google Scholar]
- 34.Foulston L.C., Bibb M.J. Proc Natl Acad Sci USA. 2010;107:13461–13466. doi: 10.1073/pnas.1008285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bachmann B.O., Ravel J. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 36.van Heel A.J., de Jong A., Montalban-Lopez M., Kok J., Kuipers O.P. Nucleic Acids Res. 2013;41(W1):W448–W453. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber T., Rausch C., Lopez P., Hoof I., Gaykova V., Huson D.H., Wohlleben W. J Biotechnol. 2009;140(1):13–17. doi: 10.1016/j.jbiotec.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Cimermancic P., Medema M.H., Claesen J., Kurita K., Brown L.C., Mavrommatis K., Pati A., Godfrey P.A., Koehrsen M., Clardy J., Birren B.W. Cell. 2014;158(2):412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber T., Kim Hyun Uk. Synthetic and systems biotechnology. Synth Syst Biotechnol. 2016;1(2):69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruz-Morales P., Martínez-Guerrero C.E., Morales-Escalante M.A., Yáñez-Guerra L.A., Kopp J.F., Feldmann J., Ramos-Aboites H.E., Barona-Gómez F. BioRxiv. 2015:020503. [Google Scholar]
- 41.Andersen M.R., Nielsen J.B., Klitgaard A., Petersen L.M., Zachariasen M., Hansen T.J., Blicher L.H., Gotfredsen C.H., Larsen T.O., Nielsen K.F., Mortensen U.H. Proc Natl Acad Sci. 2013;110(1):E99–E107. doi: 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umemura M., Koike H., Nagano N., Ishii T., Kawano J., Yamane N., Kozone I., Horimoto K., Shin-ya K., Asai K., Yu J. PLoS ONE. 2013;8(12):e84028. doi: 10.1371/journal.pone.0084028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khaldi N., Seifuddin F.T., Turner G., Haft D., Nierman W.C., Wolfe K.H., Fedorova N.D. Fungal Genet Biol. 2010;47(9):736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zarafeta D., Moschidi D., Ladoukakis E., Gavrilov S., Chrysina E.D., Chatziioannou A., Kublanov I., Skretas G., Kolisis F.N. Sci Rep. 2016;6:38886. doi: 10.1038/srep38886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foong C.P., Lakshmanan M., Abe H., Taylor T.D., Foong S.Y., Sudesh K. J Polym Res. 2018;25(1):23. [Google Scholar]
- 46.Jeon Jeong Ho, Kim Jun Tae, Lee Hyun Sook. Evidence-Based Complement Altern Med. 2011;2011 [Google Scholar]
- 47.Roopa G., Madhusudhan M.C., Sunil K.C., Lisa N., Calvin R., Poornima R., Zeinab N., Kini K.R., Prakash H.S., Geetha N. J Genet Eng Biotechnol. 2015;13(2):119–127. doi: 10.1016/j.jgeb.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soror S.H., Rao R., Cullum J. Protein engineering. Des Select. 2009;22(6):333–339. doi: 10.1093/protein/gzp009. [DOI] [PubMed] [Google Scholar]
- 49.Brault G., Shareck F., Hurtubise Y., Lépine F., Doucet N. PLoS ONE. 2012;7(3):e32041. doi: 10.1371/journal.pone.0032041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu H., Sandiford S.K., van Wezel G.P. J Ind Microbiol Biotechnol. 2014;41(2):371. doi: 10.1007/s10295-013-1309-z. [DOI] [PubMed] [Google Scholar]
- 51.Rutledge P.J., Challis G.L. Nat Rev Microbiol. 2015;13(8):509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 52.Soukup A.A., Chiang Y.M., Bok J.W., Reyes-Dominguez Y., Oakley B.R., Wang C.C., Strauss J., Keller N.P. Mol Microbiol. 2012;86(2):314–330. doi: 10.1111/j.1365-2958.2012.08195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du L., King J.B., Cichewicz R.H. J Nat Prod. 2014;77(11):2454–2458. doi: 10.1021/np500522z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsunematsu Y., Ishiuchi K.I., Hotta K., Watanabe K. Nat Prod Rep. 2013;30(8):1139–1149. doi: 10.1039/c3np70037b. [DOI] [PubMed] [Google Scholar]
- 55.Anyaogu D.C., Mortensen U.H. Front Microbiol. 2015;6:77. doi: 10.3389/fmicb.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumpfmüller J., Methling K., Fang L., Pfeifer B.A., Lalk M., Schweder T. Appl Microbiol Biotechnol. 2016;100(3):1209–1220. doi: 10.1007/s00253-015-6990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walsh C.T. ChemBioChem. 2002;3(2–3):124–134. doi: 10.1002/1439-7633(20020301)3:2/3<124::AID-CBIC124>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen K.T., Ritz D., Gu J.Q., Alexander D., Chu M., Miao V., Brian P., Baltz R.H. Proc Natl Acad Sci. 2006;103(46):17462–17467. doi: 10.1073/pnas.0608589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baltz R.H., Brian P., Miao V., Wrigley S.K. J Ind Microbiol Biotechnol. 2006;33(2):66–74. doi: 10.1007/s10295-005-0030-y. [DOI] [PubMed] [Google Scholar]
- 60.Zhang W., Wang L., Kong L., Wang T., Chu Y., Deng Z., You D. Chem Biol. 2012;19(3):422–432. doi: 10.1016/j.chembiol.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 61.Daduang R., Kitani S., Sudoh Y., Pait I.G., Thamchaipenet A., Ikeda H., Igarashi Y., Nihira T. J Biosci Bioeng. 2015;120(6):608–613. doi: 10.1016/j.jbiosc.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 62.Stephanopoulos G. Nat Biotechnol. 2002;20(7):666–668. doi: 10.1038/nbt0702-666. [DOI] [PubMed] [Google Scholar]
- 63.Petri R., Schmidt-Dannert C. Curr Opin Biotechnol. 2004;15(4):298–304. doi: 10.1016/j.copbio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 64.Lv X.A., Jin Y.Y., Li Y.D., Zhang H., Liang X.L. Appl Microbiol Biotechnol. 2013;97(2):641–648. doi: 10.1007/s00253-012-4322-7. [DOI] [PubMed] [Google Scholar]
- 65.Kaddurah-Daouk R., Kristal B.S., Weinshilboum R.M. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 66.Wang L., McLeod H.L., Weinshilboum R.M. N Engl J Med. 2011;364(12):1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weinshilboum R. N Engl J Med. 2003;348:529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 68.Patti G.J., Yanes O., Siuzdak G. Mol Cell Biol. 2012;13(4):263. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu C., Du C., Gubbens J., Choi Y.H., van Wezel G.P. J Nat Prod. 2015;78(10):2355–2363. doi: 10.1021/acs.jnatprod.5b00276. [DOI] [PubMed] [Google Scholar]
- 70.Wu C., Du C., Ichinose K., Choi Y.H., van Wezel G.P. J Nat Prod. 2017;80(2):269–277. doi: 10.1021/acs.jnatprod.6b00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng C., MacIntyre L., Abdelmohsen U.R., Horn H., Polymenakou P.N., Edrada-Ebel R. PLoS ONE. 2015;10(9):e0138528. doi: 10.1371/journal.pone.0138528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi Y., Pan C., Wang K., Chen X., Wu X., Chen C.T., Wu B. Environ Microbiol. 2017;19:3606–3618. doi: 10.1111/1462-2920.13858. [DOI] [PubMed] [Google Scholar]
- 73.Sugiyama R., Nishimura S., Ozaki T., Asamizu S., Onaka H., Kakeya H. Angew Chem Int Ed. 2016;55(35):10278–10282. doi: 10.1002/anie.201604126. [DOI] [PubMed] [Google Scholar]
- 74.Wu C., Zacchetti B., Ram A.F., Van Wezel G.P., Claessen D., Choi Y.H. Sci Rep. 2015;5:10868. doi: 10.1038/srep10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie T., Song S., Li S., Ouyang L., Xia L., Huang J. Cell Prolif. 2015;48(4):398–404. doi: 10.1111/cpr.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marmann A., Aly A.H., Lin W., Wang B., Proksch P. Mar Drugs. 2014;12(2):1043–1065. doi: 10.3390/md12021043. [DOI] [PMC free article] [PubMed] [Google Scholar]