Abstract
Cellulose is an abundant natural biopolymer on earth, found as a major constituent of plant cell wall in lignocellulosic form. Unlike other compounds cellulose is not easily soluble in water hence enzymatic conversion of cellulose has become a key technology for biodegradation of lignocellulosic materials. Microorganisms such as aerobic bacteria, fungi, yeast and actinomycetes produce cellulase that degrade cellulose by hydrolysing the β-1, 4-glycosidic linkages of cellulose. In contrast to aerobic bacteria, anaerobic bacteria lack the ability to effectively penetrate into the cellulosic material which leads to the development of complexed cellulase systems called cellulosome. Among the different environments, the sediments of mangrove forests are suitable for exploring cellulose degrading microorganisms because of continuous input of cellulosic carbon in the form of litter which then acts as a substrate for decomposition by microbe. Understanding the importance of cellulase, the present article overviews the diversity of cellulolytic microbes from different mangrove environments around the world. The molecular mechanism related to cellulase gene regulation, expression and various biotechnological application of cellulase is also discussed.
Keywords: Cellobiohydrolases, Endoglucanases, Exoglucanases, Mangrove ecosystem, Microorganisms
1. Introduction
Cellulose is the most abundant component of plant biomass on the earth [1]. In the terrestrial environments, it is the primary product of photosynthesis and the most abundant renewable bio-resource (100 billions dry tons/year) produced in the biosphere [2]. Although produced by some animals (e.g. tunicates) and few bacteria, it is found exclusively in all plant cell walls. Cellulose is present in pure state, in most cases and primarily associated with hemicelluloses and lignin. It comprises about 35–50% of plant dry weight while hemicelluloses and lignin comprise 20–35% and 5–30% of plant dry weight respectively. Cellulose is a linear polysaccharide composed of monomers of the glucose unit, bound together by β-1, 4-glycosidic linkage.
Mangrove communities are highly ecosystem which provides large quantities of organic matter to the adjacent coastal water in the form of detritus. Hence it is rich in energy and contains a large active microbial population both attached and living free [3]. Roughly 30–50% of the organic matters in mangrove leaves are leachable, water-soluble compounds, such as tannins and sugars, the remaining fraction of the organic matter presumably consists of plant structural polymers commonly referred to as lignocelluloses [4]. As cellulose is not soluble like other substance, bacterial and fungal degradation occurs exocellularly and the products of cellulose hydrolysis are available as carbon and energy sources for other microbes that inhabit in these environments. Mangrove is composed of thick organic matter mixed with sediment hence it is anaerobic except the sediment surface. In such anaerobic environments, decomposition of cellulose such as mangrove leaves and woods are brought about by complex communities of interacting microorganisms by hydrolysing the β-1, 4-glycosidic linkages of cellulose [5]. Biodegradation of β-1, 4-glycosidic linkages of cellulosic biomass is usually done by enzymes such as cellulase and cellulosome, produced by numerous microorganisms. Decomposition of cellulose produces large quantity of detritus thus makes a mangrove a sink of organic matter. Flow of carbon from this fixed cellulosic sinks to atmospheric CO2 is very important for waste treatment processes. Thus, cellulose availability forms the basis of many microbial interactions that occurs in the anaerobic environments like mangroves [6] and the cellulase enzyme produced by these microorganisms that degrade cellulose have attracted much interest because of the diversity of their application. The major industrial application of cellulases are bio-polishing, bio-stoning, bio-finishing, etc. in textile industry, starch processing, grain alcohol fermentation, malting and brewing in beer and wine industry, extraction and processing of fruit and vegetable juices, etc. in food industry, in pulp and paper industry [7], controlling plant pathogen and disease in agriculture, as well as in house hold laundry detergents for improving fabrics softness and brightness, etc. [8]. Considering the importance of bioconversion of cellulosic biomass and role of cellulases in this bioconversion process, the present review highlights the diversity of cellulose degrading microorganisms from mangrove environment and their mode of action in degrading cellulosic biomass. Further the review also addresses genetics and the molecular mechanisms involved in cellulase gene expression along with various biotechnological applications of cellulase enzyme.
2. Screening methods for cellulase producing microorganisms
Cellulose degrading microorganisms from soil are generally isolated on CMC agar medium [9] containing: (g/l): Carboxymethylcellulose (CMC), 10; KH2PO4, 4; Na2HPO4, 4; Tryptone, 2; MgSO4·7H2O, 0.2; CaCl2·2H2O, 0.001; FeSO4·7H2O, 0.004; Agar,15 and pH adjusted to 7.0. The plates are incubated at 37 °C for 24–48 h. Zone of hydrolysis are visualised by flooding the plates with 0.1% Congo red solution and washed the plate with 1 M NaCl.
Another method is also available for screening of cellulose degrading microorganisms, in which strains are grown on ISP-4 media (carboxymethyl cellulose-10 g, K2HPO4-10 g, MgSO4-10 g, NaCl-3 or 4%, CaCO3-a pinch, (NH4)2SO4-2 g, MnCl2-1 mg, ZnSO4-1 mg, FeSO4-1 mg, agar-20 g) for 3–4 days and then flooded with 5 ml of 1.0% iodine solution along with 1 ml mercuric iodide. Colony produced clear yellow zones are considered to be cellulase positive [10]. Carboxymethyl cellulose hydrolysis capacity (HC value) of the isolates can be estimated by calculating the ratio of diameter of clearing zone and colony following the method of Lu et al. [11].
3. Cellulase activity assay
Several methods are standardised to determine the activity of cellulase [12], [13], [14], [15], [16], [17]. The CMCase or Endoglucanase enzyme production activities are assayed using method described by Mandels and Weber [13]. Endoglucanase activities are estimated using 1% solution of carboxymethyl cellulose (CMC) as substrate in 0.05 M citrate buffer (pH 4.8). The reaction mixture contained 0.5 ml of suitably diluted enzyme solution and 0.5 ml of substrate solution. The reaction mixtures are then incubated at 50 °C for 30 min. After 30 min the reactions are stopped by adding 3 ml of Dinitrosalicylic acid reagent solution to the reaction mixture. The reaction mixtures are then boiled at 100 °C for 5 min. Cooling down after 5 min of boiling, O.D. are taken with spectrophotometer at 540 nm [12]. One unit of CMCase or Endoglucanase activity is expressed as 1 μ mol of glucose liberated per ml of enzyme per minute. The values obtained are compared with glucose standard curve.
Filter paper activity (FPase) for total cellulase activity in the culture filtrates are determined by the standard method of Hankin and Anagnostakis [14]. The cultured filtrate are then diluted appropriately and used as enzyme source. To this culture filtrate, whatman No. 1 filter paper strip (1 × 6 cm; 50 mg) are added with one millilitre of 0.05 M Sodium citrate buffer (pH 5.0). After incubation at 50 ± 2 °C for 1 h, the reducing sugar released is estimated by dinitrosalicylic acid (DNS) method [12]. One unit of filter paper (FPU) activity is defined as the amount of enzyme releasing 1 μ mole of reducing sugar from filter paper per ml per min.
The β-glucosidase activities are determined using the method described by Wood & Bhat [16]. In this method, the p-nitrophenol released from p-nitrophenyl- β-d-glucoside is measured using spectrophotometers. One unit of β-glucosidase activity is defined as that releasing 1 μ mol of p-nitrophenol/min. But according to Zaldívar et al. [17], β-glucosidase activities are determined using one-tenth ml of the culture supernatant, incubated with 0.5 ml of 0.05 M acetate buffer (pH 5) containing 2.5 mg cellobiose. After incubation at 50 °C for 10 min, the amount of glucose released is measured by the glucose oxidase peroxidase method [17]. Hence many variations in experimental conditions from the enzymes to substrates, the complicated relationship between physical heterogeneity of the cellulosic materials, and the complexity of cellulase enzyme systems (synergy and/or competition) result in great challenges in cellulase activity assays [15], [16].
4. Cellulose degrading microorganisms in mangrove environments
The search for potential cellulolytic enzyme is continuing in the interest of successful bioconversion of lignocellulosic biomass. Several reports have been published of microorganisms degrading lignocellulosic biomass in mangrove environments around the world. These microorganisms include bacteria, fungi, yeast and actinomycetes.
Cellulose degrading bacterial isolates from decaying S. alterniflora plants collected from salt marshes on Sapelo Island, Ga were reported by Benner et al. [18]. Ramanathan et al. [19] reported comparatively a large number of cellulose degrading bacteria from Sundarban mangroves of West Bengal, India. Tabao and Monsalud [20] reported cellulase producing five bacterial strains such as Bacillus cereus, Bacillus licheniformis, two Bacillus pumilus and Bacillus sp. isolated from Philippines mangrove. Gao et al. [21] reported a cellulolytic bacterium, Vibrio xiamenensis isolated from mangrove soil in Xiamen, Fujian province of China. Thatoi et al. [22] reported seven cellulose degrading bacterial sp. such as Pseudomonas sp., Bacillus polymyxa, B. mycoides, B. brevis from mangrove soil of Bhitarkanika, Odisha, India. Bacterial species showed endo- and exoglucanase activities in the sediment from a mangrove located in Ilha do Cardoso, SP, Brazil were reported by Soares junior et al. [23]. Two cellulolytic bacteria, Paenibacillus sp. (IARI-S-1) and Bacillus sp. (IARI-S-3) were isolated from Sundarban mangrove India [24]. Similarly two cellulolytic bacteria Bacillus cereus (IARI-B-1) and Lysinibacillus sp. (IARI-B-3) isolated from Bhitarkanika mangrove soil were reported by Pandey et al. [24]. Kalaiselvi et al. [25] isolated 35 cellulose degrading bacteria from water and sediment samples of Uppanar estuary, India. Out of the 35 isolates Klebsiella ozaenae, and Pseudomonas aeruginosa created the largest zone of lyses. Reka and Ananthi [26] reported cellulase producing Bacillus subtilis from Indian mangrove soil. Fifteen cellulase degrading bacterial sp. such as Micrococcus sp., Bacillus sp., Pseudomonas sp., Xanthomonas sp., and Brucella sp., were isolated from mangrove soil of Mahanadi river delta Odisha, India [27]. Castro et al. [28] reported endophytic bacteria associated with plants of Brazilian mangrove ecosystem. Among these bacterial isolates 62% showed endoglucanase activity and of the isolates, Bacillus showed the highest activity. Jose et al. [29] reported Bacillus sp isolated from Indian mangrove species Rhizophora mucronata for the production of cellulase enzyme.
Although many reports have been published on bacteria that degrade cellulose in mangrove, reports of fungi degrading cellulose are also well documented. El-Morsy [30] using mangrove roots of Avicennia marina, isolated several cellulose degrading fungi such as Aspergillus niger, Cladosporium cladosporioides, Cladosporium sphaerospermum, Penicillium chrysogenum, Scopulariopsis brevicaulis, Stachybotrys chartarum, Verticillium cycolsporum, and Chaetomium hamadae from the Red Sea Coast of Egypt. Luo et al. [31] screened twenty-nine fungal isolates from mangroves (and other marine sources) in Thailand, Hong Kong and Vietnam, for extracellular enzyme activity and found that most of these fungal isolates produced endoglucanases and were mostly ascomycetes, except for a basidiomycete, Calathella mangrovei, and a mitosporic fungus, Cirrenalia tropicalis. Seven fungal species such as Acremonium sp. Alternaria chlamydospora, Alternaria sp. Aspergillus sp. Aspergillus sp. Fusarium sp. and Pestalotiopsis sp. from Nethravathi mangrove, on the southwest coast of India, were reported to produce cellulase enzyme [32]. Kathiresan and manivannan [33] reported cellulose degrading activity of a fungal strain Penicillium fellutanum, isolated from coastal soil of the mangrove Rhizophora annamalayana in India. Devanathan et al. [34] reported the highest cellulase activity by Aspergillus niger isolated from Indian mangrove soil when wheat bran was used as a substrate. Ravindran et al. [35] screened for alkaline fungal cellulases from mangrove leaves (endophytes) and wood litter and identified alkaline tolerant fungi (Chaetomium sp. NIOCC 36) with high cellulolytic activity under a pH range 4–12 which are also stable up to 50 °C. Gilna and Khaleel [36] reported cellulase producing Aspergillus fumigates from mangrove rich sites of Kunhimangalam, Kannur districts, Kerala, India. Kathiresan et al. [37] showed that fungi belonging to Trichosporon sp., Aspergillus sp., and Fusarium sp. exhibited the maximum cellulases activity when mangrove leaves (Rhizophora mucronata and Avicennia marina) were used as a substrate. In Singapore, Ravindran et al. [38] isolated and characterised a new lignocellulose degrading fungus (Coniochaeta sp.) from rotten wood in mangroves. Arfi et al. [39] used transcriptomic and proteomic approaches on an isolated halotolerant lignocellulolytic mangrove fungus, Pestalotiopsis sp. NCi6, from Rhizophora stylosa trees in Saint Vincent Bay, New Caledonia. They found that this fungus holds over 400 lignocellulolytic enzymes using de novo transcriptomic assembly. Nathan et al. [40] isolated three fungal strains from Valanthakad mangroves, Kerala, India, which showed cellulose degrading activity and among the three strains Trichoderma viride showed a higher cellulase activity.
Besides bacteria and fungi, actinomycetes were also reported to degrade cellulose from mangrove environment. Gulve and Deshmukh [41] reported actinomycetes belong to Streptomyces sp., Micromonospora sp., Intrasporangium sp., Saccharopolyspora sp., Rhodococcus sp., Saccharomonospora sp. and Nocardia sp. possessing cellulase activity from five coastal sites of Konkan coast of Maharashtra. Several microbial species showing cellulase activity such as actinobacteria, fungi such as Trichosporon sp., Aspergillus sp., and Fusarium sp., Trichoderma asperellum, T. aggressivum, T. spirale, T. polysporum, yeasts such as, Pichia salicaria, Geotrichum sp., Pichia fermentans, Cryptococcus dimennae isolated from decomposing leaves of Rhizophora Mucronata and Avicennia marina trees in the mangrove forest of the Vellar estuary in southeast coast of India [37]. The actinomycetes, Streptomyces sp. from Coringa mangrove forest, Andhra Pradesh, India, reported to produce cellulase [10]. Mohanta [42] reported nine cellulose degrading actinomycetes from mangrove soil of Bhitarakanika, Odisha, India. Reyad [43] reported 58 cellulase producing actinomycetes associated with mangrove rhizosphere in Jazan Coast, Saudi Arabia.
5. Mechanism of cellulose hydrolysis by microorganisms
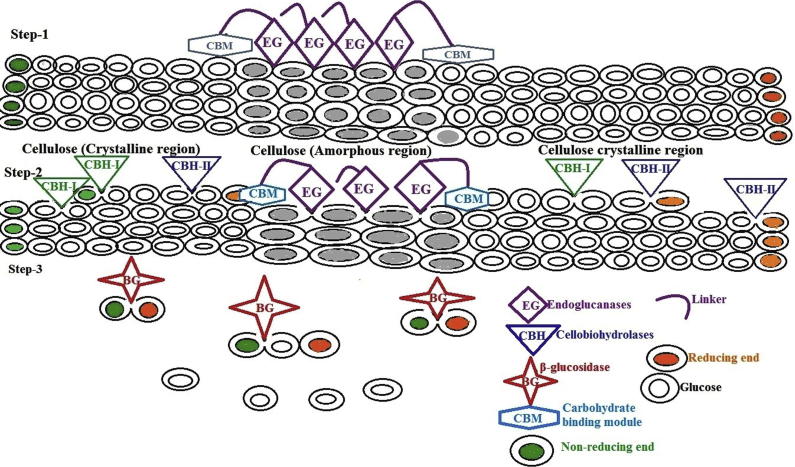
Cellulases are enzymes which able to break down cellulose by hydrolyse β-1-4 glycosidic bonds of cellulose polymer. The complete hydrolysis of cellulose into glucose requires the synergetic action of at least three enzymes, endoglucanases (EG; endo-1,4-β-d-glucanase, EC 3.2.1.4), preferably, attack amorphous regions and randomly cleave the internal bonds of the glycan chains, thus providing reducing or nonreducing ends of cellooligosaccharides for cellobiohydrolases (CBH; or exoglucanase, 1,4-β-d-glucan-cellobiohydrolase, EC 3.2.1.91) to attack (Fig. 1). CBH then hydrolyses those chain ends in the processive manner, yielding cellobiose as the major product. Lastly, β-glucosidase (BG; cellobiase, β-d-glucosideglucanohydrolase, EC 3.2.1.21) further hydrolyses cellobiose to glucose and also releases glucose from the nonreducing ends of soluble cellooligosaccharides [44]. There is a high degree of synergy between cellobiohydrolases (exoglucanases) and endoglucanases, which is required for the efficient hydrolysis of cellulose [45]. The products of endoglucanases and cellobiohydrolases, which are cellodextrans and cellobiose, respectively, are inhibitory to the enzyme’s activity. Thus, efficient cellulose hydrolysis requires the presence of β-glucosidases which cleaves the final glycosidic bonds producing glucose (end product) [46]. Typically, cellobiose and cellodextrins are taken up by the microorganism and internally cleaved via cellodextrin phosphorylases or cellobiose phosphorylases to create glucose monophosphate, which is energetically favoured [46]. Some bacteria also produce intra- or extracellular β-glucosidases to cleave cellobiose and cellodextrins and produce glucose to be taken up or assimilated by the cell [46].
Fig. 1.
Schematic representation of enzymatic hydrolysis of cellulose.
Mechanistically, the reactions catalysed by all cellulases are suggested to involve general acid–base catalysis by a carboxylate pair at the enzyme active site, though different in structure [46]. One residue acts as a general acid and protonates the oxygen of the o-glycosidic bond; at the same time, the other residue acts as a nucleophile. Depending on the distance between the two carboxylic groups, either inverting (∼10 Å distances) or retaining (∼5 Å-distances) mechanisms are observed in cellulases. Moreover, the involvement of multiple enzymes with a wide range of substrate specificities enables constant enzymatic actions on lignocellulosics [46].
Unlike soluble substrates that can diffuse the active sites of enzymes, cellulose is insoluble; thus, cellulases, on the contrary, have to diffuse, attach, and move the segment of the cellulose polymer to their active sites [47]. Most cellulases are modular proteins comprising discrete catalytic modules that typically appended one or more carbohydrate-binding modules (CBMs) joined by a flexible linker [48]. The CBM functions as a cellulose probe, in which the main responsibility is binding the enzyme to the cellulose and increasing the effective concentration of enzymes on the surface of the cellulose [49]. In addition, some CBMs are known to possess the ability to disrupt crystalline cellulose [48]. Therefore, the presence of CBMs appears to be important in enhancing the enzymatic activity towards insoluble polysaccharides, as well as crystalline cellulose.
6. Cellulosome, structure and function
In contrast to aerobic bacteria, anaerobic bacteria lack of the ability to effectively penetrate into the cellulosic material and perhaps had to find the alternative mechanisms for degrading cellulose and gaining access to products of cellulose hydrolysis in the presence of competition from other microorganisms and with limited ATP available for cellulose synthesis. This could have led to the development of “complexed” cellulase systems called cellulosome [50].
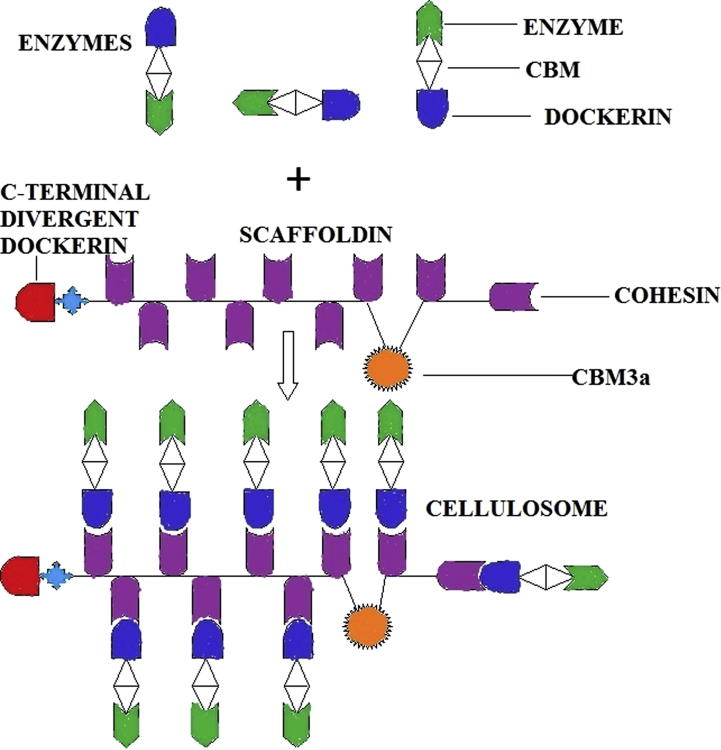
Cellulosomes were first observed by Bayer and Lamed and their colleagues as large protuberances on the surface of C. thermocellum [51].The complex was found to consist of a non enzymatic or non catalytic scaffolding protein to which attached a number of enzymatic or catalytic subunits [51], [52].
Hence modular cellulases and hemicellulases produced by anaerobic microbes consist of a catalytic part which contains dockerin, enzyme and carbohydrate-binding modules (CBMs) [52]. Cellulosome assembly is completed when this Dockerin part binds to the cohesion part of the noncatalytic scaffolding (Fig. 2). Scaffoldings also contain a cellulose-specific family 3 CBM (CBM3a) and a C-terminal divergent dockerin that target the cellulosome to the plant cell wall and the bacterial cell envelope, respectively. The linkers join the modules in the scaffoldings and catalytic subunits [52].
Fig. 2.
Mechanism of cellulosome assembly.
The CBM is comprised of approximately 35 amino acid residues, and the linker region is a highly glycosylated region unusually consisting of threonine, serine and proline amino acid residues [46]. The cohesins are modules made up of ∼150 amino acid residues and usually present as tandem repeats in scaffoldings [46], while dockerins consist of approximately 70 amino acids containing two duplicated segment (∼22 amino acid residues). Dockerins are usually present in a single copy at the C terminus of cellulosomal enzymes. The first 12 amino acid residues in each segment resemble the calcium-binding loop of EF-hand motifs (helix-loop-helix motif) in which the calcium-binding residues, aspartate or asparagine, are highly conserved [52]. Interaction between dockerin and cohesion is specific in a species, i.e. the dockerins that are found in Clostridium cellulolyticum cellulosomal enzymes do not show interaction with the cohesins that are found in C. thermocellum and vice versa [53]. Both the free and cellulosomal enzymes contain very similar types of catalytic domains. But the major difference between fungal, aerobic bacterial enzymes and anaerobic cellulosomal enzymes is that the fungal and aerobic bacterial enzymes usually contain a CBM for guiding the catalytic domain to the substrate, whereas the anaerobic cellulosomal enzymes carry a dockerin domain that incorporates the enzyme into the cellulosome system [54].
7. Regulation of cellulase gene
The regulation of cellulase production is finely controlled by activation and repression mechanisms, hence cellulases are known as inducible enzymes [55]. Only in the presence of the substrate these enzymes are induced and repressed when easily utilisable sugars are available [55]. The most probable inducer of the Trichoderma cellulase system is considered to be the disaccharide cellobiose or sophorose [56]. But sophorose doesn’t induce cellulase expression in A. niger, P. chrysosporium and P. janthinellum [57]. Sophorose (two b-1,2-linked glucose units) is believed to be formed from cellobiose by the transglycosylating activity of β-glucosidase [58]. In accordance with this, deletion of the bgl1 gene in T. reesei encoding the major extracellular β-glucosidase led to a delay in induction of cellulase gene transcription during growth on cellulose [59].
Growth in the presence of cellobiose (two b-1, 4-linked glucose units), the end product of the action of cellobiohydrolases [60], has been shown to induce cellulase expression in many species of fungi, including for instance all the main cellulases of T. reesei, eg1of Volvariella volvacea, egl2 of P. janthinellum, and eglA of A. nidulans [60], [61]. However, the reports concerning the inducing effect of cellobiose have been somewhat controversial, most likely due to the various culture conditions and e.g. cellobiose concentrations used in the experiments. The β-glucosidase (s) can either cleave cellobiose, the end product of cellobiohydrolase action, into glucose, which may cause repression or transglycosylate it into e.g. sophorose, which would lead to activation of enzyme gene transcription. Thus the outcome in cellobiose cultures depends on the balance of hydrolysis and transglycosylation, and the subsequent uptake of the generated sugars and the intracellular signals they provoke [61].
Lactose (1, 4-O-β-d-galactopyranosyl-d-glucose) which is utilised in commercial production of the enzyme owing to economic considerations is known as another inducer of cellulase [62]. Though the mechanism of lactose induction is not fully understood, it is believed that the intracellular galactose-1-phosphate levels might control the signalling [62].
In addition, the presence of various other oligosaccharides in the T. reesei cultures is known to induce cellulase expression. These include laminaribiose, gentiobiose, xylobiose and the monosaccharide L-sorbose [61]. It is proposed that due to the trans-glycosylation activity of basally expressed β-glucosidase, the inducer is generated [63].
It was reported that slow feeding of cellobiose into growth media leads to the production of high level of cellulase [63]. Northern analysis of another report showed that cellobiose induced cellulase gene such as cbh 1, cbh 2 and egl to a moderate level, which was reduced at latter stages of cultivation, when glucose started to accumulate in the culture medium [60]. Expression of a large majority of the cellulase genes that have been studied in filamentous fungi is repressed during growth on glucose. Glucose repression of cellulase system overrides its induction and derepression is believed to occur by an induction mechanism mediated by transglycosylation of glucose [64]. Simple incubation in media lacking any carbon source did not induce cellulase transcription suggesting that an induction mechanism is involved in the observed derepression of cellulase gene expression in carbon depleted conditions [61].
8. Transcriptional factor regulating cellulase gene expression
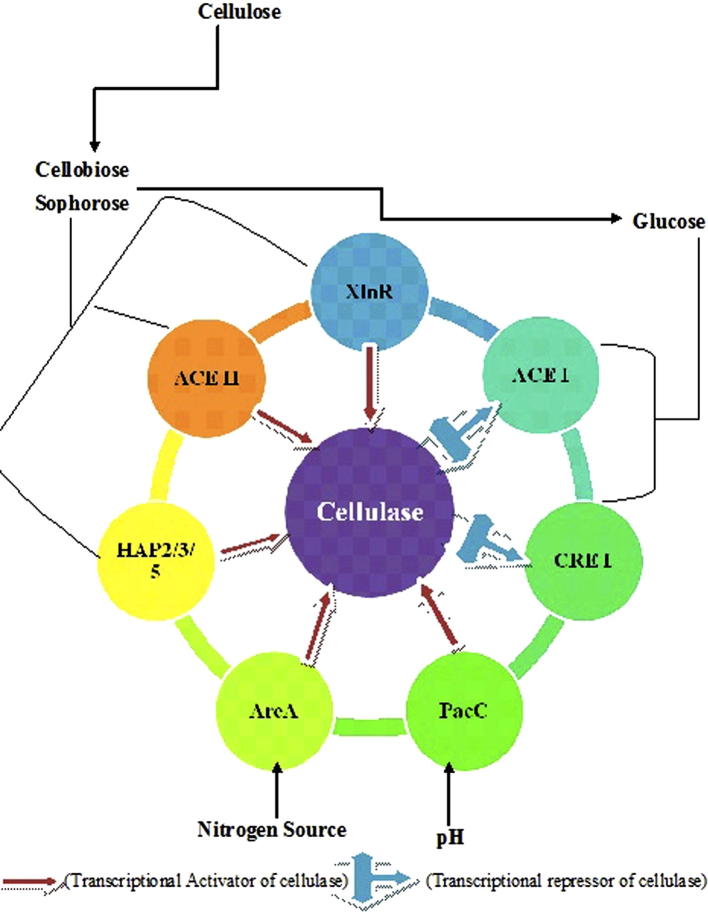
The enzymes responsible for cellulolytic activity of T. reesei include five endoglucanases- eg1/cel7b and eg2/cel5a, eg3/cel12a [65], [66] eg4/cel61a and eg5/cel45a genes [67] and two exoglucanaseses- the cellobiohydrolases cbh 1/cel7a and cbh 2/cel6a [68]. These enzymes act synergistically to convert cellulose into cellobiose, whose hydrolysis into glucose involves two β-glucosidases-bgl1/cel3a and bgl2/cel1a [69]. The promoter region of cbh 1, cbh 2, egl and eg2 genes has the binding sites for several transcriptional factors (activator/repressor) that regulate these cellulase gene expression.
XYR1 (Xylanase regulator 1) known as general and main activator of cellulase gene expression was found as an orthologue of the xlnR (Xylanase regulator) gene of A. niger [70]. The zinc binuclear cluster DNA-binding domain of XlnR was shown to bind in vitro to the 5′-GGCTAATAA sequence in the xylanase xlnA promoter. The binding site was proposed to contain a core of 5′-GGCTAR that is found in most hemicellulase and cellulase gene promoters of A. niger [70]. The expression of two endoglucanases (egl1 and egl2), two cellobiohydrolases (cbh1 and cbh2), two endoxylanases (xlnB and xlnC), a β-xylosidase (xlnD) and several genes encoding the side chain cleaving hemicellulases are co-ordinately regulated by this xylanolytic activator [70].
ACE II also identified as a transcriptional activator of cel7A and cel6A gene expression [71]. The ACE II gene was isolated in a yeast expression screening designed for isolation of factors binding to and activating the main cellulase promoter cbh1 of T. reesei. ACE II encodes a zinc binuclear cluster DNA-binding protein with no clear amino acid similarity to sequences in the databases. ACE II has been shown to bind in vitro to 5′ GGSTAA sequences in the promoters of cbh1 and xyn2 of T. reesei [71]. Deletion of the ACE II gene from the T. reesei genome resulted in the reduction of expression of all the main cellulase genes, cbh1, cbh2, egl1, egl2, and the xylanase gene xyn2, when the genes were induced upon growth on cellulose as the sole carbon. This indicates that ACE II acts as an activator but that in addition to ACE II, other factors also play a role in the cellulose-derived induction of these genes.
Zeilinger et al. [72] found HAP2/3/5 protein complex as an activator of cel6A gene transcription dependent on a CCAT box, which is a popular motif for transcription factor binding in eukaryotic gene. CCAAT sequences are found in the 5′ regions of approximately 30% of eukaryotic genes. A conserved multimeric protein complex recognises and binds to this sequence. The first CCAAT-binding complex described, designated as the Hap complex, consisting of Hap2, Hap3, Hap4 and Hap5 proteins, was identified from S. cerevisiae [73]. CCAAT sequences have been found in the promoters of many cellulase and hemicellulase genes as well as in the promoters of the ligninolytic genes mnp1 and mnp2 of P. chrysosporium and mnp1 of Ceriporiopsis subvermispora [74].
In filamentous fungi the regulation of nitrogen catabolic enzymes during the use of secondary nitrogen sources such as nitrate, nitrite and purines involves GATA factors that are a class of eukaryotic transcriptional activators or repressors characterised by a highly conserved DNA binding motif comprising a Cys(4) zinc finger followed by a basic domain [61]. This motif mediates binding to DNA elements that have the core sequence 5′GATA. The major global positive-acting regulators, involved in nitrogen metabolism, are the GATA factors AreA and Nit-2 of A. nidulans and N. crassa respectively, that activate transcription of many structural genes encoding enzymes for nitrogen source catabolism under nitrogen limiting conditions [75]. Recently, A. nidulans areA mutants were used to study the role of AreA in the production of cellulases. The amount of total secreted cellulase activity was elevated in a strain containing a constitutively activating AreA allele whereas the cellulase amount was reduced in an AreA loss-of-function mutant in cellulose induced cultures where ammonium was used as a nitrogen source [61].
The enzymatic breakdown of cellulosic biomass in natural environment, can occur in different surrounding pH. Enzymes functional in a certain pH range are produced with respect to the ambient pH of the culture habitat. The regulatory mechanism controlling pH-dependent transcriptional regulation has been studied in detail in A. nidulans and a major role has been demonstrated for the zinc finger transcription factor PacC [76], which acts as an activator for alkaline-expressed genes and prevents expression of acid-expressed genes. PacC contains a DNA-binding domain with three Cys2His2 zinc fingers and it binds to promoter sites containing the core hexanucleotide sequence 5′GCCARG [76]. It acts as an activator of cellulase gene expression.
The ACE I gene was isolated in a similar screen as ACE II designed for the isolation of transcription factors binding and activating the T. reesei cbh1 promoter in S. cerevisiae. ACE I contains three Cys2His2-type zinc fingers and it was shown to bind in vitro to eight sites in the cbh1 promoter, all of which contain the core 5′ AGGCA followed by a sequence rich in A and T [77]. Although ACE I activated the cbh1 promoter in yeast, its deletion in T. reesei led to increased expression of all the main cellulase and xylanase genes studied in cellulose and sophorose induced cultures, indicating that ACE I has a negative effect on the induced expression of these genes. T. reesei strains deleted for ACE I expressed cellulases at 2–30-fold higher level than the host strain in the inducing conditions [78].
In fungi such as T. reesei, glucose repression of cellulase is supposed to be mediated through a carbon catabolite repressor protein CREA/CRE1 [79], [80]. The CREA/CRE1 contains two almost perfectly conserved C2H2 zinc fingers responsible for DNA binding [81]. CRE1 binding sites indicating fine control of these genes by carbon catabolite repression [82]. The CREI catabolite repressor protein usually binds to the promoter region of cellulases [83]. Numerous cellulase, hemicellulase and pectinase genes have been shown to be regulated by Cre proteins in T. reesei and Aspergillus species using cre mutant strains in the studies [84], [60]. In general, mutations of the cre gene lead to (partial) derepression of enzyme gene expression on glucose. For instance, a hypercellulolytic industrial mutant strain (Rut-C30) of T. reesei, in which the cellulase and most of the hemicellulase genes are expressed to some extent during growth on glucose, was shown to contain a truncated CRE I gene. Transformation of a full-length CRE I gene into this strain restored glucose repression of the studied cellulase and hemicellulase genes, indicating that glucose repression of cellulase and most of the hemicellulase genes is mediated by CRE I in T. reesei [84]. Hence mainly five positive transcriptional activators (XYR1, ACE II, HAP 2/3/5, PacC and AreA) as well as two repressor (ACE I and CRE I) have been involved in cellulase gene regulation and expression (Fig. 3).
Fig. 3.
Schematic representation of cellulolytic gene expression and repression.
9. Biotechnological application of cellulase
Cellulases have biotechnological potential in various industries, including food, brewery and wine, industrial waste to chemical feedstock, animal feed, textile and laundry, pulp and paper, agriculture, as well as in research and development of single-cell protein [85], [86]. Today, cellulases along with hemicellulases and pectinases account for about 20% of the world enzyme market [87]. Various application of cellulase is summarised in Table 1. Details of some of the most promising applications are described below.
Table 1.
Application of Cellulase in different industry.
| Industry Function | Function | Application | References |
|---|---|---|---|
| Food industry | Hydrolysis of cell wall components; decreasing the viscosity and maintaining the texture of fruit juice | Extraction of juice from fruits, food colouring agent, Alteration of the sensory properties of fruits and vegetables and oil from olives and soups, Controlling coronary heart disease and atherosclerosis; reducing food spoilage | [88] |
| Animal feed | Pretreatment of agricultural silage and grain feed for partial hydrolysis of lignocellulosic materials | Improvement in the nutritional quality of animal feed; weight gain by broiler chickens and hens; decreasing colonisation of pathogenic bacteria in large intestine | [97], [101] |
| Beer and wine | Hydrolysis of plant cell wall polysaccharides, Modification of aromatic residues | Improvement in skin maceration and colour extraction of grapes, quality, stability and clarification and aroma of wines | [103] |
| Textile and laundry | Act on the cotton fabric and break off the small fibre ends on the cotton fabric, thereby loosening the dye after washing, prevention or permanent removal of fuzz formation and pilling, Bio-polishing of cotton and non-denim fabrics, defibrillation of lyocell | Bio-stoning of denim fabrics: biopolishing of non-denim fabrics, defibrillation of lyocell containing fabrics and bio finishing, production of high quality and environmentally friendly washing powders. Production of high quality fabrics | [107] |
| Pulp and paper | Mechanical pulping, bio-modification of fibres, removing of ink coating and toners from paper | Increase tensile strength and high fibre qualities, energy consumption reduced, improving drainage of the paper mills, and manufacturing of soft papers like paper towels and sanitary papers | [116], [117], [118] |
| Agriculture industry | Solubilisation of plant or fungal cell walls, Inhibition of spore germination, germ tube elongation and fungal growth | Production of plant or fungal protoplasts, hybrid and mutant strains, hybrid and mutant strains, Soil fertility, plant growth | [131] |
| R&D industries | Affinity tag, affinity systems, conjugation and gene fusion; Expression of heterologous proteins and enzymes | Affinity purification, immobilisation and fusion of proteins, enzymes and antibodies; production of hybrid molecules for various applications, Production of high levels of proteins, enzymes and antibodies | [133] |
| Biofuel industry | Conversion of cellulosic material to glucose and other fermentable sugars | Production of single cell protein or fermentation products like ethanol | [55], [86] |
| Pharmacy industries | Digestion of cellulose fibre | Preparation of digestin, rapid hydrolysis of cellulose, hemicellulose and beta-glucan polymers in food | [46] |
| Waste management | Degradation of cellulosic wastes | Reduction of environmental pollution | [116], [138] |
9.1. Cellulase in food industry
Cellulases play a key role in food industry. [86], [88]. Macerating enzyme which mainly consists of cellulases with pectinases and hemicellulases are used to macerate fruit pulps to maximum possible liquefaction and results in more nutritive juice yield with better stability and reduction of processing time. Macerating enzymes are used to improve the cloud stability and texture of nectars and purees and decreasing their viscosity rapidly [89]. Another important role of macerating enzymes is in olive oil extraction with slower induction of rancidity and higher levels of antioxidants and vitamin E [88]. For the first time an enzyme named Olivex was prepared by mixing pectinase with cellulase and hemicellulase from Aspergillus aculeatus to improve the extraction of olive oil [90].
Cellulases are also used for food colouring agent production [46]. The main group of colouring substances in nature which are being responsible for many plant colours from red to yellow are Carotenoid [91]. Due to their natural origin, desirable properties, null toxicity, and high versatility, providing both lipo- and hydrosoluble colourants, carotenoids as food colourants has a continuous demand in market [91]. Enzymes such as cellulase and pectinase are used to disrupt the cell wall of sweet potato, carrot and orange peel and release the carotenoids [86].
Alteration of the sensory attributes of fruits, vegetables and other foods by enzymatic infusion has enormous demand in food biotechnology [88]. The aroma and volatile characteristics of specific fruits and vegetables can be increased by infusion of pectinases and β-glucosidases. In addition vacuum infusion of β-glucosidase has the potential to alter the texture, flavour and other sensory properties of foods [92].
Nutritive quality of fermented foods, homogeneous water absorption by cereals and dried vegetables and in the production of low-calorie food ingredient oligosaccharides also can be improved by the use of enzyme cellulases [93].
Cellulases in association with pectinases are used to release antioxidants from fruit and vegetable pomace, which helps in controlling coronary heart disease, atherosclerosis, and in reducing food spoilage [94].
9.2. Cellulase in animal feed industry
Important role of cellulase in animal feed industry, which is an important sector of agribusiness comprising of ruminants, poultry, pigs, pet foods and fish farming [88], [95]. The dietary fibre in animal food consists of non starch polysaccharides such as arabinoxylans, cellulose, many other plant components including resistant dextrins, inulin, lignin, waxes, chitins, pectins, β-glucan, and oligosaccharides, which can act as anti-nutritional factor for several animals. Cellulase improves the nutritional value of feed by eliminating these anti-nutritional factors present in raw materials by providing supplementary digestive enzymes such as proteases, amylases, and glucanases [46]. Cellulases also improve the nutritional value of feed or by degrading certain cereal components [96]. For instance, β-glucanases hydrolyse cereal β-glucans in the feed of monogastric animals which help in decreasing intestinal viscosity and releasing nutrients from grains, thereby markedly improving the digestion and absorption of feed components and weight gain by broiler chickens and hens [97], [98]. The addition of commercial enzyme preparations containing cellulase and xylanase to hay diet also increased the live weight gain of cattle [99]. Lewis et al. [100] reported 5–25% increase in milk yield in case of dairy cows. These cows were fed with forage, which were treated with commercial fibrolytic enzymes. Pre-treatment of agricultural silage and grain feed by cellulases or xylanases can also improve its nutritional value [101].
The undigested starch in the large intestine can act as a substrate for bacterial fermentation and helps in proliferation of some pathogenic bacteria [102]. Cellulases have a positive effect on the caecal fermentation processes by increasing the production of propionic acid, which act as a bacteriostatic material and thus can decrease the colonisation of pathogenic bacteria [102]. Cellulases have also been used to improve the feed utilisation, milk yield and body weight gain by ruminants [88].
9.3. Cellulase in beer and wine industry
Beer brewing involves malting of the barley in a malt house followed by the preparation and fermentation of the wort in the brewery. Malting depends mainly on germination of seed, which initiates the biosynthesis and activation of amylases, carboxypeptidase and cellulases which act in synergy under optimal conditions to produce high-quality malt [46]. In brewery, use of unmalted or poor quality barley during malting and fermentation, poor filtration of the wort, slow run-off times, low extract yields and development of haze in the final product lead to gel or precipitate formation and low extract yield of beers. To overcome this problem endoglucanases are used [88], [103]. Glucanases are added either during mashing or primary fermentation to hydrolyse glucan, reduce the viscosity of wort, and improve the filterability [104]. Therefore, the addition of cellulases is known to improve not only the beer qualities but also their overall production efficiency [105].
In wine production, use of cellulases and other enzymes help in hydrolysing the plant cell wall polysaccharides, which considerably improves skin maceration, colour extraction of grapes, quality, stability, clarification and aroma of wines [103], [106]. β-Glucosidases can improve the aroma of wines by modifying glycosylated precursors [86]. Macerating enzymes also improve pressability, settling, and juice yields of grapes used for wine fermentation [86].
9.4. Cellulase in textile and laundry industry
Cellulases have now become the third largest group of enzymes used in textile and laundry [88]. The cellulases in textile industry are most commonly used to improve the fabric quality through controlled modification of cellulosic fibres by biostoning of denim garment, biopolishing of non-denim fabrics, defibrillation of lyocell containing fabrics [88] and bio finishing [107]. The aged or faded jeans were very popular in the late 1970s and early 1980s. Hence industrial laundries used pumice stones for washing the garments to produce faded jeans [88]. This washing is known as stone washing which partially removes the dye revealing the white interior of the yarn, which leads to the faded, worn and aged appearance [88]. But this stone washing caused several problems such as unsafe working conditions, rapid wear and tear of washing machines, large numbers of second class garments, environmental pollution, and the need for manual removal of pumice from pockets and folds of garments [88]. Hence, to overcome these problems, biotechnology provided a perfect alternative for stone-washing using microbial cellulases, later known as ‘bio-stoning’ [88]. In bio-stoning process, cellulases act on the cotton fabric and break off the small fibre ends on the cotton fabric, thereby loosening the dye after washing [86]. The advantages in bio-stoning treatment include less damage of fibres, less work-intensive, increased productivity of the machines and environmentally benign [55].
Cellulosic fibres, such as cotton, viscose, ramie, linen and lyocell (Lyocell is a pure cellulosic fibre from wood pulp obtained after the solvent spun with amino oxide which shows fibrillation on the surface) were used in fabric manufacturing. These fibres had a tendency for ‘fuzz’ formation (short fibres protruding from the surface) as well as ‘pilling’ (fluffy/loosened fuzz attached to the surface) which were considered as negative features of cellulosic fabrics [88]. Therefore, the defibrillation of lyocell fabrics and prevention of fuzz formation and pilling was carried out using cellulases in a process called ‘bio-polishing’ [88], [105]. Bio-polishing is usually accomplished during the wet processing stages by desizing, scouring, bleaching, dyeing, and finishing [86]. In bio-polishing, the cellulases act on small fibre ends that protrude from the fabric surface, where the mechanical action removes these fibres and polishes the fabrics which produces a smooth glossy appearance with improved colour brightness, hydrophilicity and moisture absorbance, environmentally friendly process and uniformly improved finishing [55], [105]. The acidic cellulases improve softness and water absorbance property of fibres, strongly reduce the tendency for pill formation, and provide a cleaner surface structure with less fuzz [108]. For treating pure lyocell garments acid cellulases are used while mixed lyocell garments are treated with neutral cellulases for better result [88]. Cellulase preparations rich in endoglucanases are best suited for biopolishing enhancing fabric look, feel, and colour without needing any chemical coating of fibres [103].
Cellulase rich in endoglucanase activity is proved better for biofinishing [86]. Due to the presence of partially detached micro fibrils on surface of most of the cotton garments, these become fluffy and dull and the use of cellulases can remove these microfibrils and restore a smooth surface and original colour to the garments [86], [109].
Cellulases are used in laundry to improve the production of high quality fabrics [106]. Use of cellulases along with protease and lipase in the detergents is a more recent innovation in this industry [110]. They are utilised in household washing powders to enhance the detergent performance by removing small fuzzy fibrils extruding from fabric surfaces. This removal of small fuzzy fibrils leads to an improved colour brightness along with the restoration of softness in cotton fabrics [101], [105]. Use of alkaline cellulase attracted much to the industrialist as a detergent additive for its potential ability to selectively contact the cellulose within the interior of fibres and remove soil in the inter fibril spaces [55], [110]. This is currently accomplished by adding a commercial cellulase preparation from H. insolens, active under mild alkaline conditions (pH 8.5–9.0), and at temperatures over 50 °C in washing powders [111].
9.5. Cellulase in pulp and paper industry
For bio-mechanical pulping, removing of ink coating and toners from paper, bio-modification of fibres, improving drainage of the paper mills, and manufacturing of soft papers like paper towels and sanitary papers, cellulases along with other enzymes have increasing demands in the paper and pulp industry [106], [112]. The mechanical pulping such as refining and grinding of the woody raw material lead to pulps with high content of fines, bulk, and stiffness [86]. Although these fibres are useful for producing different grades of papers, the main disadvantage of mechanical pulping is high-energy consumption [88], while in contrast, biomechanical pulping using cellulases from white-rot fungi resulted in substantial energy savings (20–40%) during refining and improvements in hand-sheet strength properties [88], [110]. Endoglucanase have been reported to decrease the pulp viscosity with a lower degree of hydrolysis [113]. In bio-modification of fibres, cellulase along with hemicellulase have been used to improve the pulp beatability, paper sheet density and runnability, leading to high productive and trouble free printing process [114]. Mansfield et al. [115] studied the action of a commercial cellulase preparation on different fractions of Douglas fir kraft pulp and observed that the cellulase treatment decreased the defibrillation, reducing the fibre coarseness. Cellulases have also been reported to enhance the bleachability of softwood kraft pulp producing a final brightness score comparable to that of xylanase treatment [110].
Cellulase have been observed to be the most effective for recycling the waste papers from books, magazines and newspaper which could have value addition via deinking and reuse of fibre either in manufacturing of newspaper or ethanol production [116], [117], [118]. Cellulase alone, or used in combination with xylanases, are beneficial for deinking of different types of paper wastes. The main advantage of enzymatic deinking is the avoidance of the use of alkali, improved fibre brightness, enhanced strength properties, higher pulp freeness and cleanliness, and reduced fine particles in the pulp [116], [117], [118]. Deinking, using cellulase at acidic pH, also prevents the alkaline yellowing, simplifies the deinking process, changes the ink particle size distribution and reduces the environmental pollution [107], [116], [117], [118], [119].
9.6. Cellulase in agriculture industry
For enhancing growth of crops and controlling plant diseases, various enzyme preparations consisting of different combinations of cellulases, hemicellulases, and pectinases have potential applications in agriculture [88], [120]. Enhanced seed germination, rapid plant growth and flowering and increased crop yields are known to be facilitated by many cellulolytic fungi such as Trichoderma sp., Geocladium sp., Chaetomium sp. and Penicillium sp. [121], [122], [88]. Induction of morphological changes in plants such as hyphal tip swelling, leakage of cytoplasm, formation of numerous septae and controlling of plant disease by inhibiting the growth of pathogenic Rhizopus solani and Fusarium sp. has been reported by β-1,3-glucanase isolated from T. harzianum CECT 2413 [123], [46]. It has been reported that the synergistic action of β-1,3-glucanase and N-acetylglucosaminidase from T. harzianum strain P1 inhibit the spore germination and germ tube elongation of B. cinerea [124]. The exoglucanase promoters of Trichoderma have been used for the expression of chymosin [125] and other proteins: glucoamylase, lignin peroxidase, and laccase [126], [127], [128]. Disease on cucumber seedlings caused by a plant pathogen, Pythium was reported to be reduced by hypercellulolytic mutant of T. longibrachiatum, which produced higher levels of β-1, 4-endoglucanase than wild type [120]. Thus, the use of cellulase could be useful as bio-control agents to protect the seeds and plants from plant pathogens [88].
Improvement of soil quality was also reported by the use of cellulase which reduces dependence of mineral fertilisers [129]. Decomposition of cellulose in soil was accelerated by addition of exogenous cellulase supplementation was reported by Fontaine et al. [130]. Therefore, using exogenous cellulase may be a potential means to accelerate straw decomposition and increase soil fertility [129].
9.7. Cellulase in R&D industries
For generating new strains capable of producing high levels of enzymes of commercial interest, cellulase and related enzymes can be used as potential tools [46]. To produce protoplast, mixture of cellulase and other enzymes can be used in the solubilisation of fungal or plant cell wall [131]. Cellulose-binding domains (CBD) of cellulase have been successfully used either as an affinity tag for the purification of proteins or immobilisation of fusion proteins because it functions normally when fused to heterologous proteins [132]. Similarly, using the scaffolding CBD of the C. thermocellum cellulosome, a novel affinity column was prepared for the purification of antibodies [133]. The biotinylated CBD bound to cellulose was attached to biotinylated protein A via avidin and used successfully for the purification of antibodies [133].
It has been reported that the cellobiohydrolase I gene possessed a strong promoter by which a number of heterologous proteins, enzymes and antibodies have been produced in mg and gram quantities [126]. The promotor of the A. niger galA gene triggered the synthesis and secretion of relatively high levels of glucoamylase and could be used for the production of heterologous proteins [126]. The first heterologous protein expressed in T. reesei and Aspergillus was calf chymosin [125] and was the first mammalian protein produced at the commercial level and used in the production of vegetarian cheese [134]. Using the cellobiohydrolase I promotor, other heterologous proteins were also expressed in T. reesei which include glucoamylase P from Hormoconis resinae [135], phytase and acid phosphatase from Aspergillus niger [136], lignin peroxidase and laccase from Phlebia radiata [127], [128], endochitinase from Trichoderma harzianum [84], antibody Fab fragments and single chain antibodies from murine [136] and interleukin-6 from mammalian origin [137]. Using cellobiohydrolase I from T. reesei, glucoamylase from A. niger and the bacterial phleomycin resistance protein, human lysozyme has been yielded of in a high amount [126].
9.8. Cellulase in bio fuel industry
A potential application of cellulases is the conversion of cellulosic material to glucose and other fermentable sugars, which in turn can be, used as microbial substrate for the production of single cell protein or fermentation products like ethanol [55], [116], [117], [118]. Production of ethanol from renewable resources via fermentation represents an important process for production of alternative fuels [55], [116], [117], [118].
9.9. Cellulase in pharmaceutical industries
Since cellulose fibres are not easily digested, people are using a digestive enzyme product, like digestin, that contains cellulase enzymes which is important for healthy cells. Fungal hemicellulase and cellulase enzyme system helps in rapid hydrolysis of cellulose, hemicellulose and beta-glucan polymers in food [46].
9.10. Cellulase in waste management
The wastes generated from forests, agricultural fields, and agro industries contain a large amount of unutilised or underutilised cellulose, causing environmental pollution [138]. Nowadays, by the use of enzyme like cellulase, amylase, lipase, protease etc, these so-called wastes are judiciously utilised to produce valuable products such as enzymes, sugars, biofuels, chemicals, cheap energy sources for fermentation, improved animal feeds, and human nutrients [117], [139].
9.11. Cellulase in recombinant DNA technology
By fusion of different lignocellulolytic genes or sections of genes from different organisms to give rise to novel chimeric gene-products with altered properties will be only possible by recombinant DNA technology [140]. A few examples will briefly be discussed to demonstrate the value and potential of these approaches in lignocellulolytic enzyme improvement. From Bacillus amyloliquefaciens and Bacillus macerans a novel thermostable hybrid (1, 3-1, 4)-β-glucanase encoding gene was expressed by gene fusion [141]. By artificial gene-fusion a unique lignocellulolytic enzymes with multiple activities can be created. A chimeric xylanases-endoglucanase [142] and a bifunctional exoglucanase-endoglucanase [143] were successfully constructed by this process. In T. reesei endoglucanase, the active carboxylic residues was identified by site-directed mutagenesis [144], opening up future research prospects for modifying these to improve the enzyme activity. A South African research team led by Pretorius has for years been active in engineering Saccharomycetes cerevisiae for efficient cellulose hydrolysis [145]. A heterologously expressed Neocallimastix patriciarum CelD encoding a multi-domain, multi-functional enzyme possessing endoglucanase, cellobiohydrolase and xylanase activity exhibited higher specific activities on avicel than two T. reesei cellobiohydrolase and a T. reesei endoglucanase [146]. Attempts have also been made to create a bifunctional hybrid of manganese-lignin peroxidase [147]. A genetically improved T. reesei RUT-P 37 showed a doubling of specific activity in both filter paper and endoglucanase units compared with the wild type QM 6a [148]. A genetically improved T. reesei mutant strain CL 847, resistant to catabolite repression exhibited a fourfold increase in cellulase productivity in compared to wild type T. reesei QM 9414 and increased β-d-glucosidase specific activity [149]. The mutant strains of T. harzianum, produced higher levels of β-1,3- and β-1,6-glucanases than the wild type strain, which have been shown to possess high anti-fungal activity for the control of plant pathogen [88]. A few strains with increased specific activities have been reported using a combination of genetic and DNA technology approaches, [148], [150]. A heterologous expressed Cellulomonas fimi exoglucanase with increased specific activity from E. coli was overproduced [151]. Attempts have also been made to clone cellulase and xylanase genes in order to produce transgenic animals, which would secrete the required enzyme into the gastrointestinal tract of the animal to facilitate its feed digestion efficiency [96].
10. Conclusion
Mangrove forests are an untapped resource for cellulosic biomass which provide opportunity for identifying novel microbial species that carry cellulase enzyme with novel biotechnological potential. Many findings listed in this present review update the evidence of various cellulose degrading microbial species from different mangrove reservoirs. To meet the growing demand for cellulases and to realise their full potential in biotechnology and research, continued multidisciplinary research on basic and applied aspects is vital. Microbial cellulases are now commercially produced by several industries globally and are being widely used in food, animal feed, fuel, paper industry, textile industry and also various chemical industries. The development of rapid and reliable methods for the screening of cellulases from microorganisms within inhospitable environments like mangrove will allow a greater number of novel bacterial cellulases to be isolated with purpose for various industrial uses. Moreover, detailed knowledge about the mechanism of cellulase degradation and variation of cellulase system in different microbes provides a detailed framework for further engineering using available knowledge of enzyme structure and function through rational design. Hence, recent development in cellulases including cellulosomes from microorganism has led to speculation and anticipation of their enormous commercial potential in biotechnology and will undoubtedly play a key role in future biotechnological research in the 21st century.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Tomme P., Driver D.P., Amandoron E.A., Miller J.R.C., Warren R.A.J., Kilburn D.G. J. Bacteriol. 1995;177:4356–4363. doi: 10.1128/jb.177.15.4356-4363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y.-H.P., Lynd L.R. Biotechnol. Bioeng. 2004;88:797–824. doi: 10.1002/bit.20282. [DOI] [PubMed] [Google Scholar]
- 3.Bano N., Nisa M.U., Khan N., Saleem M., Harrison P.J., Ahmed S.I., Azam F. Mar. Ecol. Prog. Ser. 1997;157:1–12. [Google Scholar]
- 4.Cundell A.M., Brown M.S., Stanford R., Mitchell R. Estuar. Coast. Mar. Sci. 1979;9:281–286. [Google Scholar]
- 5.Alongi D.M. Rev. Aquat. Sci. 1989;1:243–280. [Google Scholar]
- 6.Holguin G., Vazquez P., Bashan Y. Biol. Fertil. Soil. 2001;33:265–278. [Google Scholar]
- 7.Gao J.H., Zhu Weng D., Yuan M., Guan F., Xi Y. Bioresour. Technol. 2008;99:7623–7629. doi: 10.1016/j.biortech.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Cavaco paulo A. Carbohydr. Polym. 1998;37:273–277. [Google Scholar]
- 9.Ray A.K., Bairagi A., Ghosh S., Sen S.K. Acta. Ichthyol. Piscator. 2007;37:47–53. [Google Scholar]
- 10.Kavya D.M., Solomon S.M., Nagalakshmi D.M. Int. J. Pharm. Chem. Biol. Sci. 2012;2:110–116. [Google Scholar]
- 11.Lu W.J., Wang H.T., Yang S.J., Wang Z.C., Nie Y.F. J. Gen. Appl. Microbiol. 2005;51:353–360. doi: 10.2323/jgam.51.353. [DOI] [PubMed] [Google Scholar]
- 12.Miller G.L. Analytic. Chem. 1959;31:426–428. [Google Scholar]
- 13.M. Mandels, J. Weber, in cellulases and their applications, in: Hajny, G.J. & E.T. Reese (eds.), American Chemical Society, Washington DC, 1969, pp. 391–414.
- 14.Hankin L., Anagnostakis S.L. J. Gen. Microbiol. 1977;98:109–115. doi: 10.1099/00221287-98-1-109. [DOI] [PubMed] [Google Scholar]
- 15.Ghose T.K. Pure Appl. Chem. 1987;59:257–268. [Google Scholar]
- 16.Wood T.M., Bhat K.M. Meth. Enzymol. 1988;160:87–112. [Google Scholar]
- 17.Zaldívar M., Velásquez J.C., Contreras I., Pérez L.M. Elect. J. Biotechnol. 2001;4:1–9. [Google Scholar]
- 18.Benner R., Newell S.Y., Maccubbin A.E., Hodson R.E. Appl. Environ. Microbiol. 1984;48:36. doi: 10.1128/aem.48.1.36-40.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramanathan A.L., Singh G., Majumdar J., Samal A.C., Chauhan R., Ranjan R.K., Rajkumar K., Santra S.C. Ind. J. Marine Sci. 2008;37:159–165. [Google Scholar]
- 20.Tabao N.C., Moasalud R.G., Philip J. Syst. Biol. 2010;4:13–20. [Google Scholar]
- 21.Gao Z.M., Xiao J., Wang X.N., Ruan L.W., Chen X.L., Zhang Y.Z. Int. J. Syst. Evol. Microbiol. 2011;62:1958–1962. doi: 10.1099/ijs.0.033597-0. [DOI] [PubMed] [Google Scholar]
- 22.Thatoi H.N., Behera B.C., Dangar T.K., Mishra R.R. Int. J. Environ. Biol. 2012;2:50–58. [Google Scholar]
- 23.Soares F.L., Jr., Dias A.C.F., Fasanella C.C., Taketani R.G., de Souza Lima A.O., Melo I.S. Brazil J. Microbiol. 2013;44:969–976. doi: 10.1590/s1517-83822013000300048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey S., Singh S., Yadav A.N., Nain L., Saxena A.K. Biosci. Biotechnol. Biochem. 2013;77:1474–1480. doi: 10.1271/bbb.130121. [DOI] [PubMed] [Google Scholar]
- 25.Kalaiselvi V., Jayalakshmi S., Lakshmi narayanan R. Int. J. Curr. Microbiol. Appl. Sci. 2013;2:67–74. [Google Scholar]
- 26.Reka V., Ananthi T. Asian J. Environment. Sci. 2013;8:67–71. [Google Scholar]
- 27.Behera B.C., Parida S., Dutta S.K., Thatoi H.N., Americ J. Microbiol. Res. 2014;2:41–46. [Google Scholar]
- 28.Castro R.A., Quecine M.C., Lacava P.T., Batista B.D., Luvizotto D.M., Marcon J., Ferreira A., Melo I.S., Azevedo J.L. Sprin. Plus. 2014;3:382. doi: 10.1186/2193-1801-3-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jose A.C., Helen C.P., Jijo C.J. Int. J Agric. Environ. Biotechnol. 2014;7:367–370. [Google Scholar]
- 30.El-Morsy E. Fung. Divers. 2000;5:43–54. [Google Scholar]
- 31.Luo W., Vrijmoed Lilian L.P., Jones E.B.G. Bot. Mar. 2005;48:379. [Google Scholar]
- 32.Maria G.L., Sridhar K.R., Raviraja N.S. J. Agric. Technol. 2005;1:67–80. [Google Scholar]
- 33.Kathiresan K., Manivannan S. Afric. J. Biotechnol. 2006;5:829–832. [Google Scholar]
- 34.Devanathan G., Shanmugam A., Balasubramanian T., Manivannan S. Trend. Appl. Sci. Res. 2007;2:23–27. [Google Scholar]
- 35.Ravindran C., Naveenan T., Varatharajan G.R. Bot. Mar. 2010;53:275. [Google Scholar]
- 36.Gilna V.V., Khaleel K. Rece. Res. Sci. Technol. 2011;3:132–134. [Google Scholar]
- 37.Kathiresan K., Saravanakumar K., Anburaj R., Gomathi V., Abirami G., Sahu S.K., Anandhan S. Int. J. Adv. Biotechnol. Res. 2011;2:382–389. [Google Scholar]
- 38.Ravindran A., Adav S.S., Sze S.K. Proc. Biochem. 2012;47:2440–2448. [Google Scholar]
- 39.Arfi Y., Chevret D., Henrissat B., Berrin J.G., Levasseur A., Record E. Nat. Commun. 2013;4:1810. doi: 10.1038/ncomms2850. [DOI] [PubMed] [Google Scholar]
- 40.Nathan V.K., Rani M.E., Rathinasamy G., Dhiraviam K.N., Jayavel S. Spring. plus. 2014;3:1–12. doi: 10.1186/2193-1801-3-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gulve R.M., Deshmukh A.M. Rece. Res. Sci. Technol. 2011;3:80–83. [Google Scholar]
- 42.Mohanta Y.K. Bioeng. Biosci. 2014;2:1–5. [Google Scholar]
- 43.Reyad A.M. Annal. Biol. Res. 2013;4:100–108. [Google Scholar]
- 44.Jørgensen H., Kristensen J.B., Felby C. Biofuels. Bioprod. Biorefin. 2007;1:119–134. [Google Scholar]
- 45.Gupta R., Sharma K.K., Kuhad R.C. Bioresour. Technol. 2009;100:1214–1220. doi: 10.1016/j.biortech.2008.08.033. [DOI] [PubMed] [Google Scholar]
- 46.R. Gupta, G. Meheta, D. Deswal, S. Sharma, K.K. Jain, A. Singh, R.C. Kuhad, in: R.C. Kuhad and A. Singh (eds.), Biotechnology for Environmental Management and Resource Recovery, 2013, pp. 89–106. doi. 10.1007/978-81-322-0876-1_6.
- 47.Wilson D.B. Curr. Opin. Microbiol. 2011;14:259–263. doi: 10.1016/j.mib.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 48.Shoseyov O., Shani Z., Levy I. Microbiol. Mol. Biol. Rev. 2006;70:283–295. doi: 10.1128/MMBR.00028-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Araki Y., Karita S., Tsuchiya T., Kondo M., Goto M. Biosci. Biotechnol. Biochem. 2010;74:802–805. doi: 10.1271/bbb.90845. [DOI] [PubMed] [Google Scholar]
- 50.Demain A.L., Newcomb M., Wu J.H.D. Microbiol. Mol. Biol. Rev. 2005;69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bayer E.A., Shimon L.J., Shoham Y., Lamed R. J. Struct. Biol. 1998;124:134–221. doi: 10.1006/jsbi.1998.4065. [DOI] [PubMed] [Google Scholar]
- 52.Fontes C.M.G.A., Gilbert H.J. Ann. Rev. Biochem. 2010;79:655–681. doi: 10.1146/annurev-biochem-091208-085603. [DOI] [PubMed] [Google Scholar]
- 53.Pages S., Belaich A., Belaich J.P., Morag E., Lamed R., Shoham Y., Bayer E.A. Proteins. 1997;29:517–527. [PubMed] [Google Scholar]
- 54.Bayer E.A., Belaich J.P., Shoham Y., Lamed R. Annu. Rev. Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 55.Sukumaran R.K., Singhania R.R., Pandey A. J. Sci. Ind. Res. 2005;64:832–844. [Google Scholar]
- 56.Mandels M., Parrish F.W., Reese E.T. J. Bacteriol. 1962;83:400–408. doi: 10.1128/jb.83.2.400-408.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bisaria V.S., Mishra S., Crit C.R.C. Rev. Biotechnol. 1989;9:61–103. doi: 10.3109/07388558909040616. [DOI] [PubMed] [Google Scholar]
- 58.Gritzali M., Brown R.D.J. Adv. Chem. Ser. 1979;181:237–260. [Google Scholar]
- 59.Fowler T., Brown R.D., Jr Mol. Microbiol. 1992;6:3225–3235. doi: 10.1111/j.1365-2958.1992.tb01777.x. [DOI] [PubMed] [Google Scholar]
- 60.Ilmen M., Saloheimo A., Onnela M.L., Penttila M.E. Appl. Environ. Microbiol. 1997;63:1298–1306. doi: 10.1128/aem.63.4.1298-1306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aro N., Pakula T., Penttila M., Microbiol F.E.M.S. Rev. 2005;29:719–739. doi: 10.1016/j.femsre.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Seiboth B., Hartl L., Pail M., Fekete E., Karaffa L., Kubicek C.P. Mol. Microbial. 2004;51:1015–1025. doi: 10.1046/j.1365-2958.2003.03901.x. [DOI] [PubMed] [Google Scholar]
- 63.Vaheri M., Leisola M., Kaupinnen M. Biotechnol. Lett. 1979;1:41–46. [Google Scholar]
- 64.Sternberg D., Mandels G.R. J. Bacteriol. 1979;139:761–769. doi: 10.1128/jb.139.3.761-769.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okada H., Tada K., Sekiya T., Yokoyama K., Takahashi A., Tohda H., Kumagai H., Morikawa Y. Appl. Environ. Microbiol. 1998;64:555–563. doi: 10.1128/aem.64.2.555-563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saloheimo M., Lehtovaara P., Penttila M., Teeri T.T., Stahlberg J., Johansson G., Pettersson G., Claeyssens M., Tomme P., Knowles J.K. Gene. 1988;63:11–22. doi: 10.1016/0378-1119(88)90541-0. [DOI] [PubMed] [Google Scholar]
- 67.Saloheimo M., Nakari-Setala T., Tenkanen M., Penttila M. Eur. J. Biochem. 1997;249:584–591. doi: 10.1111/j.1432-1033.1997.00584.x. [DOI] [PubMed] [Google Scholar]
- 68.Teeri T.T., Lehtovaara P., Kauppinen S., Salovuori I., Knowles J. Gene. 1987;51:43–52. doi: 10.1016/0378-1119(87)90472-0. [DOI] [PubMed] [Google Scholar]
- 69.Barnett C.C., Berka R.M., Fowler T. Biotechnology. 1991;9:562–567. doi: 10.1038/nbt0691-562. [DOI] [PubMed] [Google Scholar]
- 70.Van peij N.N., Visser J., de Graaff L.H. Mol. Microbiol. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- 71.Aro N., Saloheimo A., Ilmen M., Penttila M. J. Biol. Chem. 2001;276:24309–24314. doi: 10.1074/jbc.M003624200. [DOI] [PubMed] [Google Scholar]
- 72.Zeilinger S., Ebner A., Marosits T., Mach R., Kubicek C.P. Mol. Gene. Genom. 2001;266:56–63. doi: 10.1007/s004380100518. [DOI] [PubMed] [Google Scholar]
- 73.Olesen J.T., Guarente L. Genes Dev. 1990;4:1714–1729. doi: 10.1101/gad.4.10.1714. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y.Z., Reddy C.A., Rasooly A. Gene. 1991;97:191–198. doi: 10.1016/0378-1119(91)90051-c. [DOI] [PubMed] [Google Scholar]
- 75.Marzluf G.A. Microbiol. Mol. Biol. Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tilburn J., Sarkar S., Widdick D.A., Espeso E.A., Orejas M., Mungroo J., Penalva M.A., Arst H.N., Jr. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saloheimo A., Aro N., Ilmen M., Penttila M. J. Biol. Chem. 2000;275:5817–5825. doi: 10.1074/jbc.275.8.5817. [DOI] [PubMed] [Google Scholar]
- 78.Aro N., Ilmen M., Saloheimo A., Penttila M. Appl. Environ. Microbiol. 2002;69:56–65. doi: 10.1128/AEM.69.1.56-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strauss J., Mach R.L., Zeilinger S., Hartler G., Stoffler G., Wolschek M., Kubicek C.P. FEBS Lett. 1995;376:103–107. doi: 10.1016/0014-5793(95)01255-5. [DOI] [PubMed] [Google Scholar]
- 80.Suto M., Tomita F. J. Biosci. Bioeng. 2001;92:305–311. doi: 10.1263/jbb.92.305. [DOI] [PubMed] [Google Scholar]
- 81.Cubero B., Scazzocchio C. EMBO. J. 1994;13:407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.C.P. Kubicek, M.E. Penttila, In: E Harman and C P Kubicek (Eds), Taylor & Francis Ltd., London, 1998, 49–72.
- 83.Narendja F.M., Davis M.A., Hynes M.J. Mol. Cell. Biol. 1999;19:6523–6531. doi: 10.1128/mcb.19.10.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Margolles-Clark M., Ilmen M., Penttila M. J. Biotechnol. 1997;57:167–179. [Google Scholar]
- 85.Bajpai P. Biotechnol. Prog. 1999;15:147–157. doi: 10.1021/bp990013k. [DOI] [PubMed] [Google Scholar]
- 86.Kuhad R.C., Gupta R., Singh A. Enzyme Res. 2011:10. doi: 10.4061/2011/280696. (Article ID 280696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mantyla A., Paloheimo M., Suominen P. In: Trichoderma & Gliocladium- Enzymes, Biological Control and Commercial Applications. Harman G.F., Kubicek C.P., editors. Taylor & Francis; London: 1998. pp. 291–309. [Google Scholar]
- 88.Bhat M.K. Biotechnol. Adv. 2000;18:355–383. doi: 10.1016/s0734-9750(00)00041-0. [DOI] [PubMed] [Google Scholar]
- 89.Grassin C., Fauquembergue P. In: Industrial Enzymology. second ed. Godfrey T., West S., editors. Macmillan; UK: 1996. 226–4. [Google Scholar]
- 90.Fantozzi P., Petruccioli G., Montedoro G. Grasse. 1977;54:381–388. [Google Scholar]
- 91.Cinar I. Proc. Biochem. 2005;40:945–949. [Google Scholar]
- 92.Marlatt C., Ho C.-T., Chien M. J. Agric. Food. Chem. 1992;40:249–252. [Google Scholar]
- 93.Bhat M.K., Bhat S. Biotechnol. Adv. 1997;15:583–620. doi: 10.1016/s0734-9750(97)00006-2. [DOI] [PubMed] [Google Scholar]
- 94.Meyer A.S., Jepsen S.M., Sorensen N.S. J. Agric. Food. Chem. 1998;46:2439–2446. [Google Scholar]
- 95.Chesson A. In: Recent Advances in Animal Nutrition. Haresign W., Cole D.J.A., editors. Butterworths; London: 1987. pp. 71–89. [Google Scholar]
- 96.Ali S., Hall J., Soole K.L., Fontes C.M.C.A., Hazlewood G.P., Hirst B.H., Gilbert H.J. In: Carbohydrate Bioengineering Progress in Biotechnology. Petersen S.B., Svensson B., Pedersen S., editors. Elsevier; Amsterdam: 1995. pp. 279–293. [Google Scholar]
- 97.Cowan W.D. In: Industrial Enzymology. second ed. Godfrey T., West S., editors. Macmillan Press; London: 1996. pp. 360–371. [Google Scholar]
- 98.Hesselman K., Elwinger K., Thomke S. Anim. Feed Sci. Technol. 1982;7:351–358. [Google Scholar]
- 99.Beauchemin K.A., Rode L.M., Sewalt V.J.H. Can. J. Anim. Sci. 1995;75:641–644. [Google Scholar]
- 100.Lewis G.E., Hunt C.W., Sanchez W.K., Treacher R., Pritchard G.T., Feng P. J. Anim. Sci. 1996;74:3020–3028. doi: 10.2527/1996.74123020x. [DOI] [PubMed] [Google Scholar]
- 101.Godfrey T. In: Industrial enzymology. 2nd edition. Godfrey T., West S., editors. Macmillan Press; London: 1996. pp. 360–371. [Google Scholar]
- 102.Pazarlioglu N.K., Sariisik M., Telefoncu A. Proc. Biochem. 2005;40:767–771. [Google Scholar]
- 103.Galante Y.M. A. De Conti R. In: Harman G.F., Kubicek C.P., editors. Trichoderma & Gliocladium—Enzymes, Biological Control and Commercial Applications. Taylor & Francis; London: 1998. pp. 327–342. [Google Scholar]
- 104.Bamforth C.W. J. Cer. Sci. 2009;50:353–357. [Google Scholar]
- 105.Galante Y.M. A. De Conti R, Monteverdi. In: Harman G.F., Kubicek C.P., editors. Trichoderma & Gliocladium—Enzymes, Biological Control and Commercial Applications. Taylor & Francis; London: 1998. pp. 311–326. [Google Scholar]
- 106.Thomas L., Ram H., Kumar A., Singh V.P. J. Plant Develop. Sci. 2013;5:237–247. [Google Scholar]
- 107.Kirk O., Borchert T.V., Fuglsang C.C. Curr. Opin. Biotechnol. 2002;13:345–351. doi: 10.1016/s0958-1669(02)00328-2. [DOI] [PubMed] [Google Scholar]
- 108.Sreenath H.K., Shah A.B., Yang V.W., Gharia M.M., Jeffries T.W. J. Ferment. Bioengineer. 1996;81:18–20. [Google Scholar]
- 109.Hebeish A., Ibrahim N.A. Colourage. 2007;54:41–55. [Google Scholar]
- 110.A. Singh, R. C. Kuhad, O. P. Ward, in: R. C. Kuhad and A. Singh, (eds.), Lignocellulose Biotechnologgy: Future Prospects, I.K. International Publishing House, New Delhi, India, 2007, pp. 345–358.
- 111.Uhlig H. John Wiley & Sons Inc; New York: 1998. Industrial Enzymes and their Applications; p. 435. [Google Scholar]
- 112.Saranraj P., Stella D., Reetha D. Int. J. Biochem. Biotech. Sci. 2012;1:1–12. [Google Scholar]
- 113.Pere J., Siika-aho M., Buchert J., Viikari L. Tappi J. 1995;78:71–78. [Google Scholar]
- 114.Noe P., ChevalierMora J.F., Comtat J. J. Wood Chem. Technol. 1986;6:167–184. [Google Scholar]
- 115.Mansfield S.D., Wong K.K.Y., De Jong E., Saddler J.N. Tappi J. 1996;79:125–132. [Google Scholar]
- 116.Kuhad R.C., Gupta R., Khasa Y.P. In: Wealth From Waste. Lal B., editor. Teri Press; New Delhi: 2010. pp. 53–106. [Google Scholar]
- 117.Kuhad R.C., Gupta R., Khasa Y.P., Singh A. Bioresour. Technol. 2010;101:8348–8354. doi: 10.1016/j.biortech.2010.06.043. [DOI] [PubMed] [Google Scholar]
- 118.Kuhad R.C., Mehta G., Gupta R., Sharma K.K. Biomass Bioenergy. 2010;34:1189–1194. [Google Scholar]
- 119.Liu R.H., Dai S.A., Chang F.J., Cheng W.T., Shih Y.F., Taiwan J. Inst. Chem. Eng. 2010;41:344–351. [Google Scholar]
- 120.Chet I., Benhamou N. S. Haran. In: Harman G.F., Kubicek C.P., editors. Trichoderma & Gliocladium-Enzymes, Biological Control and Commercial Applications. Taylor & Francis; London: 1998. pp. 327–342. [Google Scholar]
- 121.Bailey B.A., Lumsden R.D. In: Trichoderma & Gliocladium —Enzymes, Biological Control and Commercial Applications. Harman G.F., Kubicek C.P., editors. Taylor & Francis; London: 1998. pp. 327–342. [Google Scholar]
- 122.Harman G.E., Bjorkman T. vol. vol 2. Taylor & Francis Ltd; London: 1998. pp. 229–265. (Trichoderma and Gliocladium : enzymes, Biological Control and Commercial Applications). [Google Scholar]
- 123.Benitez T., Limon C., Delgado-Jarana J., Rey M. In: Trichoderma & Gliocladium —Enzymes, Biological Control and Commercial Applications. Harman G.F., Kubicek C.P., editors. Taylor & Francis; London: 1998. pp. 101–127. [Google Scholar]
- 124.Lorito M., Pietro Hayes C.K., Di A., Woo S.L., Harman G.E. Phytopathology. 1994;84:398–405. [Google Scholar]
- 125.Harkki A., Uusitalo J., Bailey M., Penttila M., Knowles J.K.C. Biotechnology. 1989;7:596–603. [Google Scholar]
- 126.Penttila M. In: Trichoderma & Gliocladium- Enzymes, Biological Control and Commercial Applications. Harman G.F., Kubicek C.P., editors. Taylor & Francis; London: 1998. pp. 365–382. [Google Scholar]
- 127.Saloheimo M., Bajaras V., Niku-Paavola M.-L., Knowles J.K.C. Gene. 1989;85:343–351. doi: 10.1016/0378-1119(89)90427-7. [DOI] [PubMed] [Google Scholar]
- 128.Saloheimo M., Niku-Paavola M.-L. Biotechnology. 1991;9:987–990. [Google Scholar]
- 129.Han W., He M. Bioresour. Technol. 2010;101:3724–3731. doi: 10.1016/j.biortech.2009.12.104. [DOI] [PubMed] [Google Scholar]
- 130.Fontaine S., Bardoux G., Benest D., Verdier B., Mariotti A., Abbadie L. Soil Sci. Soc. Am. J. 2004;68:125–131. [Google Scholar]
- 131.Beguin P., Aubert J.P. FEMS Microbiol. Rev. 1994;13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 132.Tomme P., Warren R.A.G., Gilkes N.R. Adv. Microb. Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 133.Bayer E.A., Morag E., Wilchek M., Lamed R., Yaron S., Shoham Y. In: Carbohydrate Bioengineering Progress in Biotechnology. Petersen S.B., Svensson B., Pedersen S., editors. Elsevier; Amsterdam: 1995. pp. 251–260. [Google Scholar]
- 134.Dunn-Coleman N.S., Bloebaum P., Berka R.M., Bodie E., Robinson N., Armstrong G., Ward M., Przetak M., Carter G.L., La Cost R., Wilson L.J., Kodama K.H., Baliu E.F., Bower B., Lamsa M., Heinsohn H. Biotechnology. 1991;9:976–981. doi: 10.1038/nbt1091-976. [DOI] [PubMed] [Google Scholar]
- 135.Joutsjoki V., Torkkeli T., Nevalainen H. Curr. Gent. 1993;24:223–229. doi: 10.1007/BF00351796. [DOI] [PubMed] [Google Scholar]
- 136.Nyyssonen E., Penttila M., Harkki A., Saloheimo A., Knowles J.K.C., Keranen S. Biotechnology. 1993;11:591–595. doi: 10.1038/nbt0593-591. [DOI] [PubMed] [Google Scholar]
- 137.J. Demolder, X. Saelens, M. Penttila, W. Fiers, R. Contreras, in: Second European Conference on Fungal Genetics, abstract no. B38, Lunteren, The Netherlands, 1994.
- 138.Milala M.A., Shugaba A., Gidado A., ENe A.C., Wafar J.A. Res. J. Agric. Biol. Sci. 2005;1:325–328. [Google Scholar]
- 139.Kuhad R.C., Singh A., Eriksson K.E. Adv. Biochem. Eng. Biotechnol. 1997;57:45–125. doi: 10.1007/BFb0102072. [DOI] [PubMed] [Google Scholar]
- 140.Howard R.L., Abotsi E., Jansen van Rensburg E.L., Howard S. Afric. J. Biotechnol. 2003;2:602–619. [Google Scholar]
- 141.R. Borris, J. Hofmeister, K.K. Thomsen, O. Olsen, D. Von Wettstein, Patent Abstracts 5470725, pp. 81.
- 142.Tomme P., Gilkes N.R., Miller R.C., Jr, Warren R.A.G., Kilburn D.G. Protein Eng. 1994;7:117–123. doi: 10.1093/protein/7.1.117. [DOI] [PubMed] [Google Scholar]
- 143.Warren R.A.J., Gerhard B., Gilkes N.R., Owolabi J.B., Kilburn D.G., Miller R.C., Jr. Gene. 1987;61:421–427. doi: 10.1016/0378-1119(87)90204-6. [DOI] [PubMed] [Google Scholar]
- 144.Okada H., Mori K., Tada K., Nogawa M., Morikawa Y. J. Mol. Catal. B: Enzymat. 2000;10:249–255. [Google Scholar]
- 145.Van Rensburg P., van Zyl W.H., Pretorius I.S. Yeast. 1998;14:67–76. doi: 10.1002/(SICI)1097-0061(19980115)14:1<67::AID-YEA200>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 146.Aylward J.H., Gobius K.S., Xue G.-P., Graham D.-S., Dalrymple B.P. Enzyme. Microb. Technol. 1999;24:609–614. [Google Scholar]
- 147.Timofeevski S.I., Nie G., Reading N.S., Aust S.D. Plant Peroxid. News lett. 1999;13:99–111. [Google Scholar]
- 148.Montenecourt B.S., Sheir-Neiss G.I., Gosh A., Gosh B.K. In: Proc. Duchworth H.E., Thompson E.A., editors. International Symposium on Ethanol from Biomass. Royal Society of Canada; Ottawa: 1983. pp. 309–414. [Google Scholar]
- 149.Durand H., Clanet M. Enzyme Microb. Technol. 1988;10:341–346. [Google Scholar]
- 150.Sheir-Neiss G., Montenecourt B.S. Appl. Microbiol. Biotechnol. 1984;20:46–53. [Google Scholar]
- 151.Lam T.-L., Wong R.S.C., Wong W.-K.R. Enzyme Microb. Technol. 1997;20:482–488. doi: 10.1016/s0141-0229(96)00203-7. [DOI] [PubMed] [Google Scholar]