Abstract
The phosphatidylinositol 3-kinases (PIK3s) are lipid kinases. Mutation in the exon 9 and exon 20 determined as a predictive factor in anti-HER-2 therapy. In some countries, such as Singapore, China, and Peru, PIK3CA exon 9 E545A was reported to produce the highest rate of mutation. In this research, we developed and optimized PIK3CA exon 9 E545A detection methods with intercalating dye SYBR Green I based on the Tm Shift approach by using prepared recombinant plasmid pGEMT-easy PIK3CA exon 9 and PIK3CA exon 9 E545A. Recombinant plasmid was used due to the limited number of samples.
Methods
Recombinant plasmid was prepared based on manufactured procedures, and this process was then followed by Tm prediction with Poland software, Tm Shift SYBR Green I development, and its characterization (reproducibility, repeatability, sensitivity, qPCR efficiency, and qPCR amplification), respectively.
Result
A method for PIK3CA E545A detection based on TM shift SYBR Green I has been successfully developed. The melting temperature for PIK3CA exon 9 was 78.1 ± 0.1 °C, while that for PIK3CA exon E545A was 80.20 °C. The Tm of mutant was the same as that predicted using Polland Software. The reproducibility of the methods was high, with the coefficient values for inter and intra assays were below 10% with a high sensitivity at 1%, while R2 0.99 and PCR efficiency was 97.75%.
Conclusion
The results presented here demonstrate that the PIK3CA exon 9 E545A detection method has a good sensitivity and efficacy assay, which proves that the method has a high diagnostic accuracy in breast cancer.
Keywords: SYBR Green I, PIK3CA E545A, Breast cancer, Real time PCR, Recombinant plasmid
1. Introduction
Phosphatidylinositol-4.5-bisphosphate 3-kinase catalytic subunit α located at chromosome 3q26.3 consists of 20 exons with a total of 1068 amino acids. Normally, PIK3CA plays an important role in cell growth, motility, proliferation, survival, and differentiation. PIK3CA’s abnormality plays an important role in breast carcinogenesis and may serve as a potential target for therapy. The gene abnormality of PIK3CA was common to breast carcinoma which was responsible for around 8–40% of incidents [1], [2], [3], [4]. In 2014, the European Society for Medical Oncology (ESMO) recognized mutation in PIK3CA as a marker for anti-HER-2 predictive therapy. The hotspots of PIK3CA mutations were clustered at exons 9 at codon E542K&E545K in helical domain and exon 20 at codon H1047R in kinase domain [5], [6], [7], [8]. Some studies show that those mutants have gained a function which induces tumor formation and to increase cancer progression [9]. This somatic mutation of PIK3CA is not only common in breast cancer, but also exists in colorectal cancer, endometrial cancer, lung cancer, gastric cancer, ovary cancer, and skin cancer, among others [10]. Gymnopoulus et al. [11] reported fifteen rare mutants of p110α: R38H, K111N, N345K, C420R, P539R, E545A, E545G, Q564K, Q546P, H710P, T1025S, M1043I, M1043V, H1047L, and H1047Y. Those fifteen mutants also gained a function which induced oncogenic transformation, except H1047Y which showed no phenotype. However, studies of breast cancer in Singapore and colorectal cancer in Shanghai, China reported that the highest frequency of PIK3CA mutations in exon 9 was found in E545A. In a different continent, research on Peruvian women also showed the same trend as that prevailing in those countries, with PIK3CA E545A having the highest number of incidents found in PIK3CA exon 9 mutations [12], [13], [14].
PIK3CA E545A mutation is located at c. 1634A>C which substituted glutamic acid at 545 to Alanine (A). Only a limited number of methods for detecting PIK3CA E545A have been developed to date. Most of these methods were developed commercially and found in scientific reports on PIK3CA detection which focused on the hotspot position, E542K, E545K, and H1047R. Available PIK3CA methods were developed based on the probe approach [15] or using sophisticated equipment such as Pyrosequencing [16] and SNApshot assay [17]. Since the PIK3CA exon 9E545A has been proven to produce a gain-of-function effect, especially in some countries which showed the highest frequency of incidents, it is important to develop an effective method of detection at this position.
In a previous report, we successfully developed a PIK3CA E545A detection method using the Tm Shift SYBR Green I approach [18]. Tm Shift SYBR Green I has been described as a low-cost, simple, and massive detection method. In this report, we seek to improve previous results by doing two sets of primer comparison and reformulation, followed by confirming and validating the used material and the developed method. Because of the limited number of samples we used plasmid recombinant as the material. Our results confirmed and validated recombinant plasmid as an effective material which can be further applied for the quality control of PIK3CA E545A detection. The utilization of plasmid as the material for developing a diagnostic real time PCR has been reported in many scientific papers. Chan et al. [19] utilized pooled recombinant plasmid as the material for developing methods, as well as the control material for developing a diagnostic real time PCR. Sims et al. [20] reported the utilization of DNA plasmid as the material for quality control and calibrator for a next generation sequence.
2. Materials and methods
2.1. Samples
DNA samples were obtained from breast cancer patients in West Sumatera province as the DNA source. This research had acquired an ethical approval from the Indonesian Ministry of Health. DNA genome extraction was performed according to the Purelink Invitrogen DNA kit and confirmed with A260/A280 values ranging between 1.8 and 2.0.
2.2. Plasmid recombinant preparation
DNA recombinant plasmid was prepared by using the TA cloning method inserted into pGEMT-easy. pGEMT-easy PIK3CA E545A was prepared with direct PCR. Mixed qPCR contains DNA plasmid 3 ng, 10 pmol primer for each, 5 µl KAPPA SYBR Green I fast master mix (2×) universal, and 3.23 µLMQ. The forward and reverse primers were (5′-gggccggatccccagaggggaaaatatgac-3′) and (5′-gggccggatccattttagcacttacctgtg-3′), respectively. The PCR condition was as follows: an initial denaturation 95 °C for 3 min, 35 cycle of 95 °C denaturation at 30 s, 57 °C annealing at 30 s, 72 °C extension at 30 s, and one cycle of 72 °C for post extension for 5 min. Meanwhile, pGEM-T easy PIK3CA exon 9 was developed with nested PCR, following Bachman et al. [21].
2.3. Plasmid melting curve prediction analysis with the Poland software
In order to predict the melt curve of the developed plasmid, especially for pGEM-T easy PIK3CA E545A isolated from pseudogene, we analyzed the prepared plasmid by comparing it with the real PIK3CA E545A using an in silico program, i.e. the Poland software (http://www.biophys.uni-duesseldorf.de/local/POLAND/poland.html).
2.4. Tm Shift SYB Green I assay
Tm Shift SYBR Green I assay for PIK3CA exon 9 E545A was developed by comparing two primer sets, with several trial-and-error primer formulations for each set because we had to look for the most specific primer which could recognize the target. Both of the primer sets contain two forward primers. The first forward primer recognized mutants which contained a long GC tail, while the second forward primer recognized the WT which contained a short GC tail. Both primer sets have the same forward primer that recognized WT, which is 5′-GCGGGCtcctctctctgaaatcactga-3′, and the same reverse primer, which is 5′-gggccggatccattttagcacttacctgtg-3′. The first forward primer for the first primer set was 5′-GCGGGCAGGGCGGCtcctctctctgaaatcaccgc-3′, which was the same as that reported in a previous study [18]. Meanwhile, the second primer set was 5′-GCGGGCAGGGCGGCtcctctctctgaaatcattgc. The difference between the first and the second forward primers lies in the mismatch position (shown in red colors). The qPCR conditions were the same as above, and the process was followed by melting it from 65 °C to 95 °C with a ramp rate of 0.2 °C per second. The process for producing the best primer design and formulation was then followed by the optimization of annealing temperature from 65 to 68 °C with no qPCR extension step.
2.5. Sensitivity assay
To determine the sensitivity assay, we mixed and diluted pGEM-T easy PIK3CA E545A with pGEM-T easy PIK3CA exon 9, with E545A proportions of 100%, 50%, 25%, 10%, 5%, and 1% under optimized qPCR reaction.
2.6. Limit detection
Limit detection was performed starting from 3 ng-0.00003 ng with 10 times dilution.
2.7. Data analysis
Statistical analysis was performed for the calculation of %CV (coefficient of variation) intra and inter assay. Further analysis was done to calculate the percentage of R2 and PCR efficiency.
3. Results
3.1. Plasmid recombinant preparation
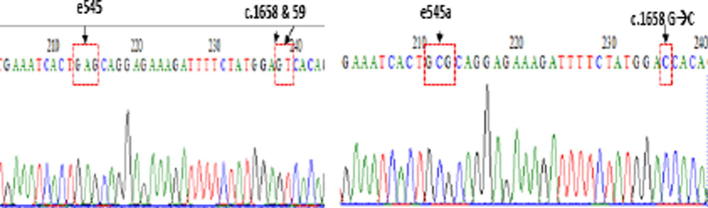
We found a pseudogene in exon 9 PIK3CA which showed a high similarity with chromosome 22. This pseudogene showed a base substitution which was identical with the mutation in PIK3CA exon 9, c. 1634A>C (E545A). The difference between this pseudogene and exon 9 PIK3CA was in the substitution and deletion mutation in the downstream area. We developed recombinant plasmid which contained a pseudogene identical with exon 9 PIK3CA E545, which was isolated using the direct PCR method. Meanwhile, PIK3CA exon 9 was isolated using the nested PCR method. The PCR fragments for both were inserted into pGEM-T easy, following the manufactured procedure. Then, the sequence of recombinant plasmid was confirmed (Fig. 1). Mutant preparation with the direct PCR of the template will produce more benefits compared with site-directed mutation because it is more cost-effective and has shorter preparation time.
Fig. 1.
DNA sequence of the prepared recombinant plasmid, mutant E545A (left) and WT PIK3CA (right).
The melt curve analysis was then performed using the Poland software to confirm and compare between the melt curve of the pseudogene and that of PIK3CA E545A fragment. The results show that the melt curve of PIK3CA E545A was similar to that of the PIK3CA pseudogene, which was at 80 °C for both (Fig. 2). Moreover, we can use the PIK3CA pseudogene as a template to develop PIK3CA E545A detection methods based on Tm Shift SYBR Green I.
Fig. 2.
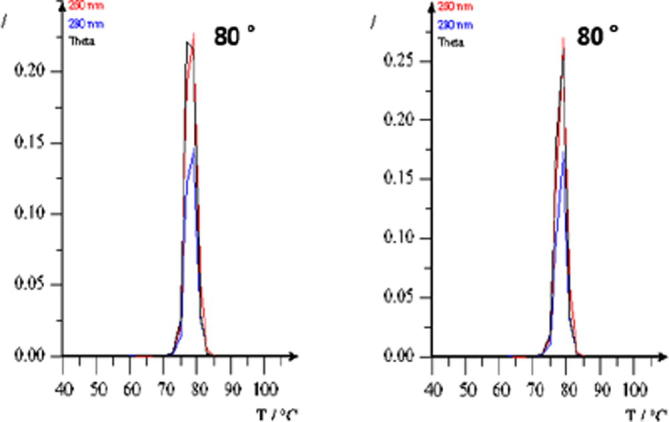
Melt curve analysis of the PIK3CA E545A (left) and PIK3CA pseudogene (right) in the same DNA fragment area.
3.2. Tm Shift SYBR Green I assay
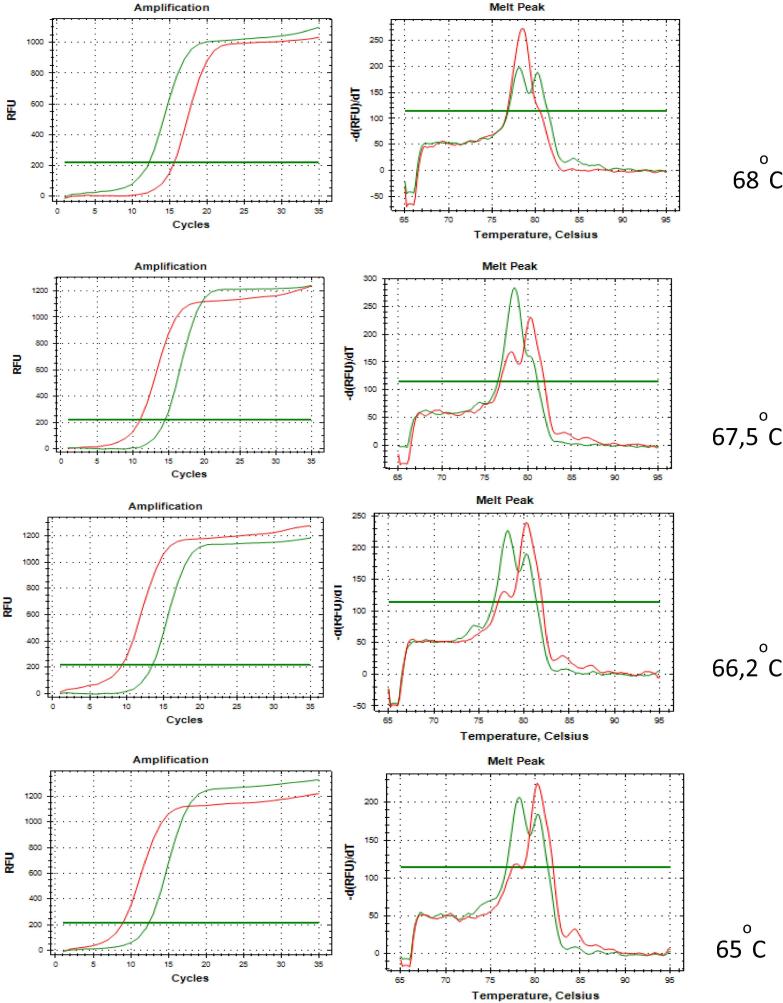
We have successfully developed Tm Shift SYBR Green I assay in one single closed tube with some trial-and-error primer proportion formulations (data are not shown). With this single tube approach, contamination problems could be prevented. The best primer proportions for PIK3CA E545A detection are 0.03 μM (long FW): 0.00175 μM and (short FW): 0.03 μM. This proportion formulation is better than that reported in a previous study [18]. In this experiment, we increased the primer annealing optimization to above 65 °C, namely 65 °C, 66.2 °C, 67.5 °C, and 68 °C, in order to increase the specificity primer to recognize the target. The result of this is the best annealing temperature for PIK3CA genotyping at 66.2 °C by deleting qPCR extension reaction (Fig. 3). We tried another set primer that had a different mismatch position from that of the first set. The results of the trial-and-error primer formulation at several annealing temperatures were not as specific as expected, because the set primer could not discriminate between WT exon 9 and PIK3CA exon 9 E545A (data are not shown). Based on this finding, we used the first set of primer for further PIK3CA exon 9 E545A validation and characterization assay, which consists of sensitivity assay, repeatability, reproducibility, and qPCR efficiency (see Fig. 4, Fig. 5).
Fig. 3.
PIK3CA E545A annealing temperature optimization from 65 to 68 °C.
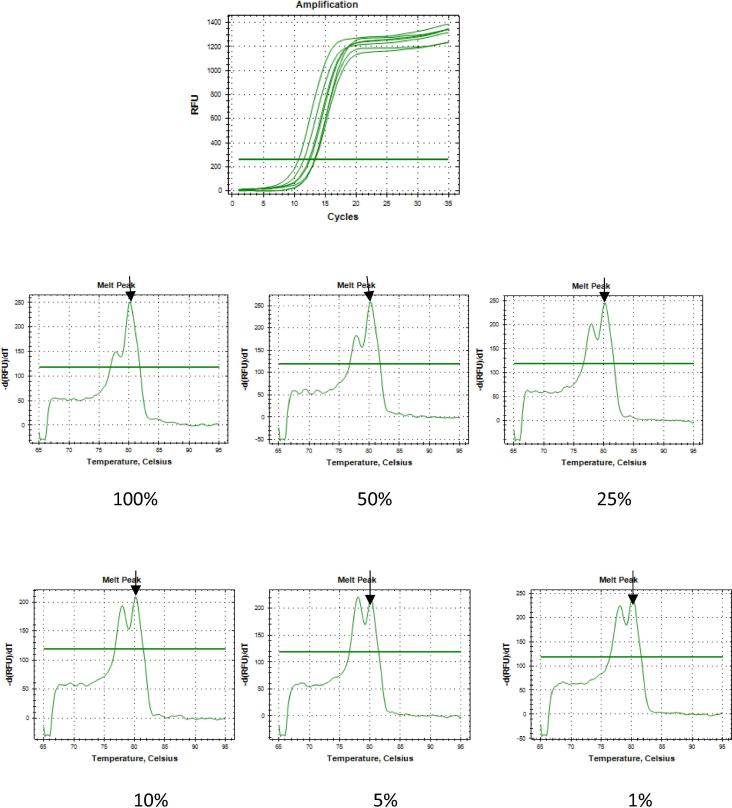
Fig. 4.
Sensitivity assay of PIK3CA E545A detection with the E545A proportions of 100%, 50%, 25%, 10%, 5%, and 1% under optimized qPCR reaction. A. Ct value for each proportion; B. Derived melt peak for each proportion.
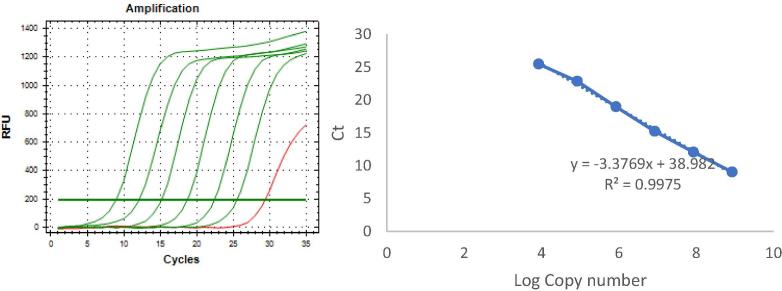
Fig. 5.
Amplification of Real Timer PCR for determining its efficiency and R2 value.
3.3. Sensitivity assay
In terms of sensitivity assay, the generated method has 1% sensitivity, which means that PIK3CA E545A could be recognized among 99% of the PIK3CA WT.
3.4. qPCR efficiency and amplification
The qPCR efficiency was high at 97.75%, with R2 was 0.9975. The reproducibility of the developed method was found to be below 10%, which means that it has satisfied the requirement for both intra and inter assays.
4. Discussion
Many diagnostic and detection methods are available commercially and reported in various scientific reports. Tm Shift SYBR Green I is considered as a rapid and low cost method for mutation detection [22]. The sensitivity of the developed targeted detection is 1%, which shows that it is more sensitive than that of the gold standard method PCR seq with sensitivity ranging from 15 to 25%. This sensitivity assay is an important finding because it is usually found in a low percentage.
The utilization of recombinant plasmid is very useful for developing diagnostic or detection methods, especially with a limited number of samples. In this research, we took some benefits from using the pseudogene of PIK3CA exon 9 as the first step for developing a PIK3CA E545A detection method based on Tm Shift SYBR Green I method. Compared with the site-directed mutagenesis in the preparation of mutant DNA plasmid, the developed strategy was more cost-effective and worked faster. The mutant plasmid was prepared using the single step PCR, while PIK3CA exon 9 plasmid was prepared using the nested PCR. After the DNA plasmid was prepared, it could be further used in sensitivity and specificity assessment. In order to strengthen and increase the confidence of the prepared recombinant plasmid being used, we tested it by generating a melt curve prediction using the Polland software. According to Ramussen et al. [23], the melt peak prediction generated by this software approaches the real value. Furthermore, the Polland software is easy to operate. The Polland software analysis demonstrates that the melt curve of the PIK3CA from pseudogene produces a similar pattern, while the melt temperature value also shows the real PIK3CA E545A temperature at 80 °C. The presence of the deletion and substitution mutation in the downstream did not affect the melt peak temperature. Therefore, we can apply this plasmid as a material to develop targeted detection methods.
We have developed a PIK3CA E545A detection method using Tm Shift SYBR Green I with the first primer set design. In order to increase the specificity, we improved the available primer formulation, which was then followed by the optimization of annealing temperature starting from 65 °C to 68 °C. In this experiment,
additional mismatches at the 3rd site from the 3′ end (first primer sets), and this success gave destabilization effects which could further discriminate the E545A, while additional mismatches found at the 4th site did not affect the discrimination of the E545A. The selection of the right mismatches to increase primer specificity is still a challenging task. Different sites of mismatches have different primer ability to recognize the target. According to Liu et al. [24], mismatches only at 3′ terminal which correspond to a specific mutation site are not enough to discriminate between the target and the non-target. An additional mismatch site at 3rd nucleotide from the 3′ end has been statistically proven as having the highest allele specificity, compared with that of other mismatched sites, namely 2′ and 4′ from the 3′ end. Furthermore, the removal of the extension step could also be considered for increasing the primer specificity. The presence of the extension step in E545A development methods will decrease primer specificity [18].
The melting temperature in this developed method was 80.2 °C. This value was not significantly different from the predicted value of melting temperature generated by the Polland software, which is at 80 °C. Furthermore, based on the melt curve analysis, we can clearly see and differentiate between PIK3CA and PIK3CA E545A, although there was an unspecific peak. As shown in Fig. 3, the two annealing temperatures of 65 °C and 66.2 °C produced similar results. When higher annealing temperatures were used, the primer tended to interact with the wild type. The next step was selecting 66.2 °C as the annealing temperature. According to our observation, 66.2 °C could recognize the target more specifically.
As reported by Bustin et al. [25] in the MIQE guideline, there were some important indicators of a robust and precise qPCR, such as the limit of detection (LOD), accuracy, repeatability, and reproducibility which is expressed as CV for both intra and inter assays. Moreover, high PCR efficiency and PCR amplification also indicate a robust and precise qPCR assay [26]. All of these indicators are important, since they are usually found in a low percentage when used against tumor cells, but in a high percentage when used against non-tumor cells. In this present work, we performed sensitivity quantification by using mixed PIK3CA E545A with WT DNA plasmid in order to mimic the heterogeneity of a tumor. The mutant proportions were at 100%, 50%, 25%, 10%, 5%, and 1%, respectively among the WT. The developed method was able to detect up to 1% of the mutants. This suggests the high sensitivity of Tm Shift SYBR Green I because it could detect up to 1% of all DNAs contained in the mutation. This value indicates a higher sensitivity compared with that of PCR seq ranging from 5% to 25%. Furthermore, the slope of the standard curve represents the qPCR efficiency. When we used a triplicate for each concentration, the slope was −3.3769, which fell between −3.3 and −3.8. This indicated the ideal standards of efficiency at 97.75% and of R2 at 0.9975. Since the value was still within the required range, it means that there was no pipetting error or presence of the inhibitor in the reaction. Then, the CV value was 0% or below 10% as required, and this indicates the high accuracy and reproducibility of the developed Tm Shift SYBR Green I methods.
In conclusion, based on the results presented here, our research has successfully produced a good, sensitive, and easy method for the operation and interpretation of PIK3CA exon 9 E545A detection. In order to avoid the interference of pseudogene when applying the method on breast cancer DNA as a target, we recommend small modifications, especially to the primer reverse orientation, in future studies. We suggest redesigning the reverse primer, taking a nucleotide which has incompatible bases between pseudogene and the PIK3CA exon 9. This will increase the probability that it will be able to recognize the targeted nucleotide specifically, as achieved by Bachman et al. [21].
Acknowledgment
This research was funded by Kegiatan Unggulan dan Bahan Baku Obat LIPI, P.I Dr. Eng. Desriani, grant number SP DIPA-079.01.2.450083/2015.3403.002.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.J. Dunlap, C. Le, A. Shukla, J. Patterson, A. Presnell, M.C. Heinrich, L. Christipher, L. Corless, Megan L. Troxell. Phosphatiylinositol-3-kinase and AKT1 mutations occur early in breast cancer carcinoma. Breast Cancer Treat 2010;120:409–418. [DOI] [PubMed]
- 2.Dieci M.V., Guarneri V. PIK3CA: a Target or a marker in breast cancers. Curr Breast Cancer Rep. 2015;7:1–9. [Google Scholar]
- 3.Arthur L.M., Turnbull A.K., Renshaw L., Keysm J., Thomas J.S., Wilson T.R., Lackner M.R., Sims A.H., Dixon J.M. Changes in PIK3CA mutation status are not associated with recurrence, metastasic disease or progression in endocrine trated breast cancer. Breast Cancer Res Treat. 2014;147:211–219. doi: 10.1007/s10549-014-3080-x. [DOI] [PubMed] [Google Scholar]
- 4.Miller T.W., Rexer B.N., Garret J.T., Arteaga C.L. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13:224–236. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukohara T. PI3K mutations in breast cancer: prognostic and therapeutic implications. Breast Cancer (Dove Med Press) 2015;7:111–112. doi: 10.2147/BCTT.S60696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuels Y., Wang Z., Bardelli A., Sillimian N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S.M., Riggins G.J., Wilson J.K.V., Markowitz S., Kinzler K.W., Vogelstein B., Velculescul V.E. High frequency of mutation of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 7.Samuels Y., Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. 2010;347:21–41. doi: 10.1007/82_2010_68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang B.H., Liu L.Z. PI3K/PTEN signaling in angiogenesis and tumorigenesis. Adv Cancer Res. 2010;102:19–65. doi: 10.1016/S0065-230X(09)02002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer D.S., Brinkhaus H., Muller U., Muller M., Cardiff R.D., Bentire-Alj M. Luminal expression of pIK3CA mutant H1047R in the mammary gland induces heterogenous tumors. Can Res. 2011;71:4344–4351. doi: 10.1158/0008-5472.CAN-10-3827. [DOI] [PubMed] [Google Scholar]
- 10.Mayer I.A. Clinical implications of mutations in the PIK3 pathway in HER-2+ breast cancer: prognostic of predictive? Curr Breast Cancer Rep. 2015;7:210–214. doi: 10.1007/s12609-015-0197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gymnopoulos M, Elsiger MA, Vogt PK. Rare cancer -specific mutations in PIK3CA Show gain of function 2007;104:5569–74. [DOI] [PMC free article] [PubMed]
- 12.Liang X., Lau Q.C., Tellez M.S., Putti T.C., Loh M., Sukumar S. Mutational hotspot in exon 20 of PIK3CA in breast cancer among Singapore Chinese. Cancer Biol Ther. 2006;5:544–548. doi: 10.4161/cbt.5.5.2656. [DOI] [PubMed] [Google Scholar]
- 13.Castaneda CA, Lopez-Ilasaca M, Pinto JA, Chirinos-Arias M, Doimi F, Neciosup SP, Rojas KI, Vidaurre T, Balko JM, Arteaga CL, Gomez HL. PIK3CA mutations in Peruvian Patients with HER-2 amplified and triple negative non mteatatic breast cancers. Hematol Oncol Stem Cell Ther 2014;7:142–8. [DOI] [PubMed]
- 14.Zhu Y.Z., Yu B., Li D., Ke H., Guo X., Xiao X. PI3K expression and PIK3CA mutation are related to colorectal cancer metastases. World J Gastroenterol. 2012;28:3745–3751. doi: 10.3748/wjg.v18.i28.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kompier L.C., Lurkin I., van der Aa M.N.M., van Thijn B.W.G., van der Kawast T.H., Warthoff E.C. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS ONE. 2010;5:1–13. doi: 10.1371/journal.pone.0013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker C.L., Vaughn C.P., Samowitz W.S. A PIK3CA Pyrosequencing based assay that excludes pseudogene interference. J Mol Diagn. 2011;14:56–60. doi: 10.1016/j.jmoldx.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Schneck H., Blassl C., Meier-Stiegen F., Neves R.P., Janni W., Fehm T., Neubauer H. Analysing the mutational status of PIK3CA in circulating tumor cells from metastatic breast cancer patients. Molecul Oncol. 2013;976–986 doi: 10.1016/j.molonc.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Ahwani F., Desriani D., Widyastuti U., Suharsono S. Method development for detection of E545A mutation PIK3CA gene in breast cancer patients using Tm Shift SYBR Green I qPCR. Indoens J Biotechnol. 2016;21:22–28. [Google Scholar]
- 19.Chan M., Jiang B., Tan T.Y. Using pooled recombinant plasmid as control materials for diagnostic real-time PCR. Clin Lab. 2016;61:1893–1901. doi: 10.7754/Clin.Lab.2016.160114. [DOI] [PubMed] [Google Scholar]
- 20.Sims D.J., Harrington R.D., Polley E.C., Forbes T.D., Mehaffey M.G., McGregor P.M., Camalier C.E., Harper K.N., Bouk C.H., Da B., Conley B.A., Doroshow J.H., Williams P.M., Lih C.J. Plasmid Based Material as multiplex quality controls and calibrators for clinical next generation sequencing assay. J Mol Diagn. 2015;18:335–349. doi: 10.1016/j.jmoldx.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachman K.E., Argani P., Samuels Y., Siliman N., Ptak J., Szabo S., Konishi H., Karakas B., Blair B.G., Lin C., Peters B.A., Velculescu V.E., Park B.H. The PIK3CA gene is mutated with high frequency in humans breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 22.Germer S., Higuchi R. Single-tube genotyping without oligonucleotide probes. Genome Res. 1999;9:72–78. [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen J.P., Saint C.P., Monis P.T. Use of DNA melting simulation software for in silico diagnostic assay design: targeting regions with complex melting curves and confirmation by real-time PCR using intercalating dyes. BMC Bioinformat. 2007;8:107–118. doi: 10.1186/1471-2105-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Huang S., Meiyu S., Liu S., Liu Y., Wang W., Zhang X., Wang H., Hua W. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Meth. 2012;8:1–9. doi: 10.1186/1746-4811-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustin S.A., Benes V., Garson J.A., Hellemans J., Hugget J., Kubsita M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:1–12. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 26.Dorak M.T. Taylor & Francis Group; New York (USA): 2006. Real-time PCR. [Google Scholar]