Abstract
In present study seven RAPD primers were used to access the diversity within and among twelve populations of three mushroom species Ganoderma lucidum, leucoagaricus sp. and Lentinus sp. Total of 111 bands were scored by 7 RAPD primers in 30 accessions of three mushroom species collected from different sampling sites of central India. Total 111 bands were generated using seven primers which were F-1, OPG-06, OPC-07, OPD-08, OPA-02, OPD-02, OPB-10. All 111 bands were polymorphic in nature (100%). Therefore, it revealed that the used primers had sufficient potency for population studies and 30 accessions had higher genetic differences among each other. In best of the knowledge, this is the first report, which accesses the genetic diversity between three mushroom species (Gd Ganoderma lucidum, Lg Leucoagaricus sp., Ls Lentinus). The polymorphic percentage ranged from 3.60 to 23% within twelve populations, while polymorphic percentage among group was 40.56, among population within groups was 41.12 and within population was 18.32. This indicated that the genetic diversity within the population was very low, but slightly higher in the populations of three species. Among three groups representing Gd., Lg and Ls, Among populations within groups shown highest percentage of variation (Pv = 41.12) while within populations, the lowest percentage of variation (18.32) was observed. This result also support that the highest genetic variation was present among groups in comparison to among the population within a species and lowest genetic variation was observed within the population.
Keywords: Genetic diversity, Polymorphic, Population, Primer, Variation
1. Introduction
Utilization of wild mushrooms as a food source started with prehistoric man. Human as a hunter collected wild fungi of forest that serve as important source of nutrition during the long period. There are many edible mushrooms i.e. Agarics, Volvarias, Polypores and tubers fungi that have been used as ethno-botanical food by the tribal of forest regions. These are obviously non toxic as these have been in intimate human consumption by native and tribal, since ancient past [1]. Mushrooms offer significant vital health benefits, including antioxidants, cholesterol-lowering properties, anti-hypertensive, anti-inflammatory, liver protection, as well as anti-diabetic, anti-viral, and anti-microbial properties [2]
Lentinus tigrinus and G. lucidium are proved anticholesterolmic [3]. Lentinus edodes has been used to enhance vigour, sexuality, energy and as an anti aging agent. Lentinan sulphate obtained from Lentinus species inhibits HIV [4]. Lentinus sajor-caju can easily be recognized in the field by its large, thin, whitish to cream pileus with pale brown disc, lacking or with small squamules at the center, short stipe with annulus or annular ridge [5] Lentinus sp. is high source of protein, carbohydrate and low amounts of fat and possess high quantities of micronutrients (vitamins and carotenoids) and minerals (P, K, Mn, Ni, and Fe) with strong antioxidant properties [6].
Ganoderma lucidum is considered to be a natural medicine that promotes longevity and maintains the vitality of human beings. G. lucidum is well known as traditional medicine used against cancer, viral and bacterial infection, diabetes, and liver injury. Among its activities, its anticancer properties have been the most interesting studies. It revealed cytotoxic activity of suppressed inflammatory breast cancer [7], [8], [9], [10]. The genus Leucoagaricus has been well studied in Europe. However, species diversity of Leucoagaricus in Asia remains poorly known.
Now a day’s molecular techniques are becoming very important for the taxonomic and phylogenetic relationship studies among different fungi [11]. By the use of DNA based techniques like Random Amplified Polymorphic DNA Polymorphism (RAPD), Amplified Fragments length Polymorphism (AFLP), Restriction Fragment Length Polymorphism (RFLP) or DNA sequence analysis (nSSU and mtSSU), limitation of identification of mushroom strains based on a few morphological characters can be overcome.
Any of the molecular methods mentioned above could be combined with morphological methods to make identification of fungal species reliable [12]. The random amplified polymorphic DNA (RAPD) is a convenient method to detect genetic diversity [13]. This method has been particularly successful when applied to check the strains of mushrooms with different origins [12]. RAPD have been used to examine material from the genera Agaricus, Coprinus and Lentinula [14]. Genetic Diversity Characterization of Pleurotus strains by Random Amplified Polymorphic DNA Fingerprinting has been performed by various workers [15], [16]. Fruiting body observations provide information about the fungi on the surface. In addition, evolutionary relationships cannot be determined accurately through morphology alone [17].
2. Materials and methods
2.1. Study sites
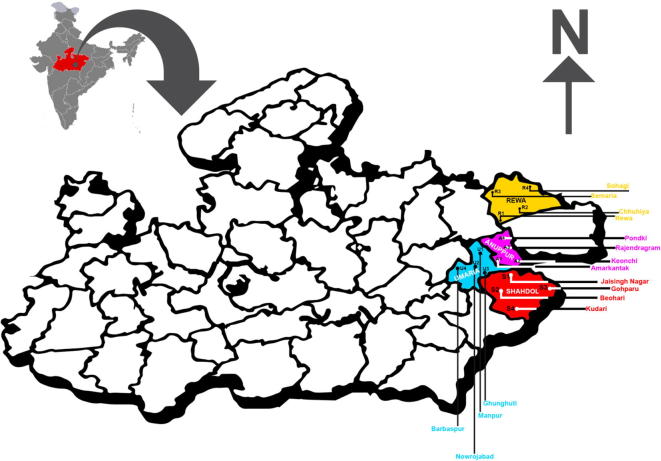
Forest areas of four districts Anuppur, Rewa, Shahdol and Umaria of Central India were main study sites of present study (Fig. 1). Sampling sites for mushroom collection of these four study sites are shown in the map.
Fig. 1.
Map showing sampling sites of Central India for study of mushroom diversity.
2.2. Samples collection and identification
Extensive survey of the sampling sites of forest areas of four districts Anuppur, Rewa, Shahdol and Umaria was done in rainy season (2012–13). Ecological features, macroscopic studies and mushroom field test were performed. Specimens were preserved in dried as well as in wet form. Samples were identified to their respective families, genera and species by consulting literature [18], [19], [20], [21], [22]. Help of mushroom guide “The great encyclopedia of mushrooms” [23] “Eye Witness Handbooks Mushroom” [24] was also taken. Help of experts in taxonomy of mushroom was also taken when ever required.
2.3. Genomic DNA isolation
DNA isolation and RAPD Analysis of Mushrooms was done by the following protocol [25], [26]. Total three species, 12 populations and 30 accessions were used for genetic diversity study for RAPD analysis 0.50 g of dried fruiting bodies of all 30 mushroom samples were cut into small pieces and were soaked in 1 ml buffer. After that all pieces were incubated at 65 °C for 2 h. After incubation the samples were homogenised using pestle and mortar.15 ml of Lysis buffer was added [25]. The tubes were incubated at 65 °C for 1 h in a water bath with intermittent mixing. Centrifuge at 1000 rpm for 15 min to separate out the unlysed cells. Supernatant was transferred to a fresh 30 ml centrifuge tube carefully. Equal volumes of chloroform was added and mixed well. Centrifuge this at 10,000 rpm for 15 min. The aqueous layer was pipette out into the fresh 30 ml centrifuge tube without taking the interface. Equal volumes of isopropanol and 1/10th volumes of 3 M sodium acetate was added and mixed well. Then left at room temperature to stand for 5–10 min. Centrifuge at 10,000 rpm for 15 min and the supernatant was discarded. The pellet was washed with 500 µl of 70% ethanol. The pellet air dried and suspended in 500 µ1 of 1X Tris–EDTA buffer. To remove PCR inhibitors, further the DNA sample was purified by Column purification.
2.4. Column purification
The column was placed in collection tube, 400 µl of equilibration buffer was added to the column and centrifuged at 10,000 rpm for 1 min. Collected buffer was discarded. 400 µl of equilibration buffer was added to the DNA samples, mixed and loaded into the column (This step was repeated till the DNA sample was completed). Flow through was discarded. 500 µl of wash buffer 1 was added, centrifuged at 10,000 rpm for 1 min and buffer was discarded. 500 µl of wash buffer 2 was added, centrifuged at 10,000 rpm for 1 min and buffer was discarded. The column was centrifuged with empty collection tube to completely remove the wash buffer for 2 min. 50 µl of elution buffer was added to the column placed in new collection tube. Incubated at room temperature for 2 min and centrifuge at 10,000 rpm for 1 min and eluted sample was saved (elution 1). Previous step was repeated. (DNA may elute in this fraction also) (elution2) Quantization of eluted DNA samples was done by loading into the Agarose gel.
Genomic DNA was isolated from the dried fruiting bodies of mushrooms and quantity and quality was observed using UV spectrophotometer. The amount of isolated DNA was varied from 461.92 to 1980.38 ng/µl DNA and absorbance ratio of A260/280 was obtained in the range of 1.71–1.98.
2.5. Random Amplified Polymorphic DNA (RAPD)
Isolated DNA from all 30 accessions of 12 populations of mushroom species were amplified using 7 fungal RAPD Primer F-1, OPG-06, OPC-07, OPD-08, OPA-02, OPD-02, OPB-10 and observed under UV light after resolving on 1.5% agarose gel with mid range ruler ranging from 0.1, 0.2, 0.3, 0.6, 1.0, 1.5, 2.0, 2.5, 3.0 and 3.5 kb. Supplier name ARISTOGENE BIOSCIENCES PVT Ltd. Number of bands were 10. Sequence of primers, GC%, annealing temperature and length is shown (Table 1). All 30 samples were categorized into three groups; in each group four populations were taken which belongs to Anuppur, Rewa, Shahdol and Umaria districts of Vindhyan Region. And the first group consists 14 accessions, while the second and third group have 8 accessions each group.
Table 1.
Primers with their sequence, GC%, Annealing temperature and length.
| S. No. | Primers | Sequence | GC% | Annealing temperature | Length |
|---|---|---|---|---|---|
| 1. | F-1 Primer | CTGGACACAC | 60 | 45 °C throughout the experiment | 10 bp |
| 2. | OPC-06 | GAACGGACTC | 60 | ||
| 3. | OPC-07 | GTCCCGACGA | 70 | ||
| 4. | OPD-08 | GTGTGCCCCA | 70 | ||
| 5. | OPA-02 | TGCCGAGCTG | 70 | ||
| 6. | OPD-02 | GGACCCAACC | 70 | ||
| 7 | OPB-10 | CTGCTGGGAC | 70 |
The amplification of genomic DNA was done by 7 random decamer nucleotide primers which were OPA-02 (TGCCGAGCTG), OPB-10 (CTGCTGGGAC), OPD-02 (GGACCCAACC), OPC-06 (GAACGGACTC), OPD-08 (GTGTGCCCCA), OPC-07 (GTCCCGACGA), F-1 Primer (CTGGACACAC).
2.5.1. Polymerase chain reaction
Reaction recipe for PCR amplification of genomic DNA was as follows. PCR master mix 20 µl, double distilled water17 µl, random primer 1 µl, template DNA 2 µl. Each reaction volume (40 µl) was pipetted into Eppondorf tube and placed in thermal cycler for amplification. The following thermal profile was applied for RAPD-PCR. Step 1 – Initial denaturation at 94 °C for 5 min, step 2 – Denaturation at 94 °C for 30 s, step 3 – annealing at 45 °C for 1 min, step 4 – extension at 72 °C for 1.30 min step 5 – final extension at 72 °C for 7 min.
2.5.2. Electectrophoretic analysis of RAPD products
RAPD-PCR product were analysed on 1.5% agarose gel to generate fragments and later stained with ethidium bromide, which were visualized with uv-transilluminater then documented by gel documentation system. The molecular weight of bands was estimated using a mid range ruler ranging from 0.1, 0.2, 0.3, 0.6, 1.0, 1.5, 2.0, 2.5, 3.0 and 3.5 kb.
2.6. Statistical analysis
The molecular weight of all bands was calculated and binary matrix was prepared by scoring as absence and presence. For each RAPD markers, polymorphic information content (PIC) and heterozygosity (H) were calculated using Molecular Kinships 3.0. Similarity indices, distance matrix and phylogenetic tree were prepared using the PAST software [27], using Jaccard’s coefficient and UPGMA, using binary matrix. Observed number of alleles (Na), effective number of alleles (Ne), gene diversity (H) described by [28], Shannon's information index (I), relative differentiation (GST), estimate of gene flow from [GST (Nm)], total heterozygosity (Ht), within population heterozygosity (Hs), genetic identity and genetic distance within population and group, number of polymorphic bands (NPB) and percentage of polymorphic bands (PPB) were calculated using Popgene version 1.31. Average genetic distances in the all populations and groups were calculated and Phylogenetic tree was analysed using MEGA software version 5 [29]. A matrix of pair wise FST (Genetic distances between populations), Average pair wise differences [30] AMOVA, sum of squares (SS), variance components (Vc), percentage of variation (Pv) and fixation index (FST) were calculated with significance level 0.005 using software Arlequin version 3.5 [31]. Input files were converted using Microsoft Excel based GenAlEx 6.5 software [32] from one form to another.
3. Results
The genetic structure among and within the 12 populations of three species was estimated with following parameters-
3.1. Genetic diversity according to primers
3.1.1. Percentage of polymorphism
Percentage of polymorphism was calculated according to the presence and absence of bands for all RAPD primers. All seven primers used for diversity study generate total 111 bands and average 15.86 ± 5.21 bands per primer. Maximum 26 bands were generated by F-1 and minimum 11 bands were generated by the OPB-10. All bands were polymorphic and 100% polymorphism was observed (Table 2)
Table 2.
Total numbers of bands, percentage of polymorphic bands and their size generated by RAPD primers in all 30 accessions (mushroom samples collected from different sampling sites).
| S. No. | Primer Name | Total bands | NPB | PPB | Band size (Aprox.) |
|---|---|---|---|---|---|
| 1 | F-1 | 26 | 26 | 100% | 210-2600 |
| 2 | OPC- 06 | 18 | 18 | 100% | 060-3100 |
| 3 | OPC -07 | 12 | 12 | 100% | 180-2200 |
| 4 | OPD-08 | 12 | 12 | 100% | 270-2600 |
| 5 | OPA -02 | 15 | 15 | 100% | 240-2400 |
| 6 | OPD -02 | 17 | 17 | 100% | 230-3200 |
| 7 | OPB -10 | 11 | 11 | 100% | 320-2400 |
| Mean ± SD | 15.86 ± 5.21 | 15.86 ± 5.21 | 100% | ||
PPB = Percentage of polymorphic Band, NPB = No of polymorphic Band.
3.1.2. Polymorphic Information Content (PIC)
The program Molecular Kinships 3.0 was used for the simple calculation of polymorphic information content (PIC) and heterozygosity (Ho) for every primer between and within the group. Among the populations, mean of Ho observed was with 0.913 ± of mean PIC, while the maximum Ho (0.9412) and PIC (93.81) were observed in OPA-02 primer and minimum Ho (0.8409) and PIC (82.51) were recorded in OPB-10 primer. In the first group (GD), 0.811 ± 0.101 mean Ho with 78.820 ± 11.311 mean PIC was observed. Maximum Ho (0.9074) and PIC (90.01) was observed in primer OPC-06, while minimum Ho (0.6122) and PIC (56.98) was observed in primer OPB-10. The second group (Lg) had mean Ho and PIC of (0.820 ± 0.044) and (79.757 ± 5.344)respectively with maximum Ho (0.8571) and PIC (84.40) in primer OPC-07, while in primer OPD-08 had a minimum Ho (0.7959) and PIC (76.56). The third group (Ls) had mean Ho and PIC of 0.738 ± 0.083 and 70.077 ± 10.304 respectively with maximum Ho (0.8281) and PIC (80.55) in primer OPD-08, had a minimum Ho (0.6250) and PIC (55.47) in primer OPD-02 (Table 3).
Table 3.
Polymorphic Information Content (PIC) and Heterozygosity (Ho) for every primer and between and within Groups (Ganoderma lucidum, Leucoagaricus sp., Lentinus sp.)
| SN | Name of primer | All Group |
Group 1 Gd |
Group 2 Lg |
Group 3 Ls |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | HO | PIC | N | HO | PIC | n | HO | PIC | n | HO | PIC | ||
| 1 | F-1 | 28 | 0.9178 | 91.35 | 10 | 0.8878 | 87.70 | 10 | 0.8438 | 82.73 | 10 | 0.8125 | 79.75 |
| 2 | OPC- 06 | 26 | 0.9256 | 92.20 | 13 | 0.9074 | 90.01 | 5 | 0.7400 | 70.14 | 9 | 0.7656 | 74.77 |
| 3 | OPC -07 | 23 | 0.9339 | 93 | 7 | 0.7959 | 76.81 | 10 | 0.8571 | 84.40 | 7 | 0.7969 | 76.74 |
| 4 | OPD-08 | 17 | 0.9262 | 92.14 | 8 | 0.8438 | 82.53 | 6 | 0.7959 | 76.56 | 7 | 0.8281 | 80.55 |
| 5 | OPA -02 | 22 | 0.9412 | 93.81 | 9 | 0.8622 | 84.72 | 10 | 0.8594 | 84.51 | 4 | 0.6914 | 64.01 |
| 6 | OPD -02 | 14 | 0.9028 | 89.50 | 5 | 0.7653 | 72.99 | 7 | 0.7959 | 77.19 | 3 | 0.6250 | 55.47 |
| 7 | OPB -10 | 13 | 0.8409 | 82.51 | 4 | 0.6122 | 56.98 | 7 | 0.8469 | 82.77 | 4 | 0.6484 | 59.25 |
| Mean ± SD | 20.429 ± 5.855 | 0.913 ± 0.034 | 90.644 ± 3.834 | 8.000 ± 3.055 | 0.811 ± 0.101 | 78.820 ± 11.311 | 7.857 ± 2.116 | 0.820 ± 0.044 | 79.757 ± 5.344 | 6.286 ± 2.690 | 0.738 ± 0.083 | 70.077 ± 10.304 | |
where Gd = Ganoderma lucidum, Lg = Leucoagaricus sp., Ls = Lentinus sp.
Bold fonts indicate maximum and minimum values in given table.
3.1.3. Polymorphism and PIC of each primer
Total number of observed bands, percentage of polymorphic band, Ho and PIC of each primer is described. RAPD primer F-1 amplified total 26 bands among the range of 210–2600 bp. (Table 2). Polymorphic information content (PIC = 91.35) and heterozygosity (Ho = 0.9178) of this primer was recorded (Table 3) in all the groups. PIC 87.70, 82.73 and 79.75 and Ho 0.8878, 0.8438 and 0.8125 was observed in group 1, 2, and 3 respectively.
Total 18 bands were generated by fungal RAPD Primer OPC-06, with the range of 060 to 3100 bp. Thus 100 percent of polymorphic bands (PPB) were observed (Table 2). Polymorphic information content (PIC = 92.20) and heterozygosity (Ho = 0.9256) of this primer was observed (Table 3). Fungal RAPD Primer OPC-07 amplified total 12 bands between the range of 180 to 2200 bp. Polymorphic information content (PIC = 93) and heterozygosity (Ho = 0.9339) of this primer was observed. Total 12 bands were generated by RAPD primer OPD-08 between the range of 270–2600 bp. Polymorphic information content (PIC = 92.14) and heterozygosity (Ho = 0.9262) of this primer was detected (Table 3). RAPD primer OPA-02 produced total 15 bands between the range of 240 to 2400 bp. 100 percentage of polymorphic band (PPB) was observed (Table 2). Polymorphic information content (PIC = 93.81) and heterozygosity (Ho = 0.9412) of this primer was recorded (Table 3). Total 17 bands were generated by RAPD Primer OPD-02 with the range of 230 to 3200 bp (Table 2). Polymorphic information content (PIC = 89.50) and heterozygosity (Ho = 0.9028) of this primer was observed (Table 3) RAPD Primer OPB-10 produced total 11 bands between the range of 320 to 2400 bp. Out of 100.00 percentage of polymorphic band (PPB) was observed (Table 2). Polymorphic information content (PIC = 82.51) and heterozygosity (Ho = 0.8409) of this primer was recorded (Table 3).
3.2. Genetic variation
Observed number of alleles (Na), effective number of alleles (Ne), gene diversity (H), Shannon's information index (I), total heterozygosity (Ht), within population heterozygosity (Hs), number of polymorphic bands (NPB) and Percentage of polymorphic bands (PPB) were calculated for the estimation of genetic variation in the populations. The observed genetic variation among and within the populations and the species are described below.
3.2.1. Within population genetic variation
Within populations, population Gd showed maximum Na (1.2342 ± 0.4254), Ne (1.2234 ± 0.4105), H (0.1137 ± 0. 2075), I (0.1588 ± 0.2892), NPB (26.) and PPB (23.42), while, the minimum Na (1.0360 ± 0.1872), Ne (1.0288 ± 0.1498), H (0.0160 ± 0.0832), I (0.0229 ± 0.1192), NPB (4) and PPB (3.60) were observed in population GdU (Table 4)
Table 4.
Genetic variation (Mean ± SD) within populations of Ganodermalucidum, Leucoagaricus sp., Lentinus sp.
| SL | Pop | N | Na | Ne | H | I | TNB | NPB | PPB |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GdA | 4 | 1.2342 ± 0.4254 | 1.2234 ± 0.4105 | 0.1137 ± 0. 2075 | 0.1588 ± 0.2892 | 111 | 26 | 23.42 |
| 2 | GdR | 3 | 1.0631 ± 0.2442 | 1.0505 ± 0.1953 | 0.0280 ± 0.1085 | 0.0401 ± 0.1554 | 111 | 7 | 6.31 |
| 3 | Gds | 4 | 1.1622 ± 0.3703 | 1.1261 ± 0.2990 | 0.0698 ± 0.1614 | 0.1006 ± 0.2312 | 111 | 18 | 16.22 |
| 4 | GdU | 3 | 1.0360 ± 0.1872 | 1.0288 ± 0.1498 | 0.0160 ± 0.0832 | 0.0229 ± 0.1192 | 111 | 4 | 3.60 |
| 5 | LgA | 2 | 1.0721 ± 0.2598 | 1.0721 ± 0.2598 | 0.0360 ± 0.1299 | 0.0500 ± 0.1801 | 111 | 8 | 7.21 |
| 6 | LgR | 2 | 1.0721 ± 0.2598 | 1.0721 ± 0.2598 | 0.0360 ± 0.1299 | 0.0500 ± 0.1801 | 111 | 8 | 7.21 |
| 7 | LgS | 2 | 1.0541 ± 0.2271 | 1.0541 ± 0.2271 | 0.0270 ± 0.1136 | 0.0375 ± 0.1574 | 111 | 6 | 5.41 |
| 8 | LgU | 2 | 1.0811 ± 0.2742 | 1.0811 ± 0.2742 | 0.0405 ± 0.1371 | 0.0562 ± 0.1901 | 111 | 9 | 8.11 |
| 9 | LsA | 2 | 1.0721 ± 0.2598 | 1.0721 ± 0.2598 | 0.0360 ± 0.1299 | 0.0500 ± 0.1801 | 111 | 8 | 7.21 |
| 10 | LsR | 2 | 1.1081 ± 0.3119 | 1.1081 ± 0.3119 | 0.0541 ± 0.1560 | 0.0749 ± 0.2162 | 111 | 12 | 10.81 |
| 11 | LsS | 2 | 1.0631 ± 0.2442 | 1.0631 ± 0.2442 | 0.0315 ± 0.1221 | 0.0437 ± 0.1693 | 111 | 7 | 6.31 |
| 12 | LsU | 2 | 1.0541 ± 0.2271 | 1.0541 ± 0.2271 | 0.027 ± 0.1136 | 0.0375 ± 0.1574 | 111 | 6 | 5.41 |
Na = Observed number of alleles.
Ne = Effective number of alleles.
H = Nei's gene diversity.
I = Shannon's Information index.
N = Sample size.
TNB = Total no of bands.
PPB = Percentage of polymorphic Band.
NPB = No of polymorphic Band.
Bold fonts indicate maximum and minimum values in given table.
3.2.2. Within group genetic variation-
Within populations of first group (Gd), the Na (1.5225 ± 0.5018), Ne (1.3292 ± 0.3649), H (0.1941 ± 0.1998), I (0.2891 ± 0.2894), Ht (0.1863 ± 0.0382) and Hs was (0.0569 ± 0.0059) while 58 NPB with 52.25 PPB was observed (Table 5).
Table 5.
Genetic Variation statistics among population within group (Ganoderma lucidum, Lecoagaricus sp. and Lentinus sp.) (Mean ± SD).
| S.No. | Group | Population Size | Sample Size | Na | Ne | H | I | Ht | Hs | TNB | NPB | PPB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Gd | 4 | 14 | 1.5225 ± 0.5018 | 1.3292 ± 0.3649 | 0.1941 ± 0.1998 | 0.2891 ± 0.2894 | 0.1863 ± 0.0382 | 0.0569 ± 0.0059 | 111 | 58 | 52.25 |
| 2 | Lg | 4 | 8 | 1.6577 ± 0.4766 | 1.4459 ± 0.3820 | 0.2565 ± 0.2016 | 0.3776 ± 0.2873 | 0.2565 ± 0.0406 | 0.0349 ± 0.0057 | 111 | 73 | 65.77 |
| 3 | Ls | 4 | 8 | 1.5045 ± 0.5022 | 1.3075 ± 0.3799 | 0.1788 ± 0.1990 | 0.2681 ± 0.2852 | 0.1788 ± 0.0396 | 0.0372 ± 0.0039 | 111 | 56 | 50.45 |
| All groups | 12 | 30 | 1.9640 ± 0.1872 | 1.6089 ± 0.2946 | 0.3545 ± 0.1343 | 0.5268 ± 0.1705 | 0.3610 ± 0.0187 | 0.2072 ± 0.0130 | 111 | 107 | 96.40 | |
Na = Observed number of alleles.
Ne = Effective number of alleles.
H = Nei's gene diversity.
I = Shannon's Information index.
TNB = Total no of bands.
NPB = No. of polymorphic site.
PPB = Percentage of polymorphic bands.
Within populations of Second group (Lg), the Na (1.6577 ± 0.4766), Ne (1.4459 ± 0.3820), H (0.2565 ± 0.2016), I (0.3776 ± 0.2873), Ht (0.2565 ± 0.0406) and Hs was (0.0349 ± 0.0057) while 73 NPB with 65.77 PPB was observed (Table 5).
Within populations of Third group (Ls) the Na (1.5045 ± 0.5022), Ne (1.3075 ± 0.3799), H (0.1788 ± 0.1990), I (0.2681 ± 0.2852), Ht (0.1788 ± 0.0396) and Hs was (0.0372 ± 0.0039) while 56 NPB with 50.45 PPB was observed (Table 5).
3.2.3. Among populations genetic variation (All group)
Within populations of All Group the Na (1.9640 ± 0.1872), Ne (1.6089 ± 0.2946), H (0.3545 ± 0.1343), I (0.5268 ± 0.1705), Ht (0.3610 ± 0.0187) and Hs was (0.2072 ± 0.0130) while 107 NPB with 96.40 PPB was observed (Table 5).
3.3. Phylogenetic tree (dendrogram)-
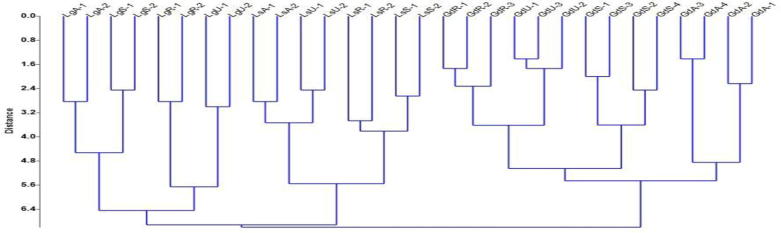
Phylogenetic tree were prepared using UPGMA (Unweighed Pair Group Method with Arithmetic Average) method. When dendrogram was prepared using a binary matrix of 30 accessions, three major clusters i.e. cluster I, cluster II and cluster III were generated, which represent the population of the species Ganoderma lucidum, Leucoagaricus sp. and Lentinus sp. respectively. Cluster I consist 3 sub cluster A, B and C while cluster II consist of 2 sub-clusters E and F and cluster III consist of 2 sub clusters G and H. All 7 sub-clusters represent 12 populations of three mushroom species (Table 7 and Fig. 2). In the cluster I, population of GdR was closely related to population GdU. In the cluster II, LsA was more similar to Ls U and Ls R was very close to Ls R. In cluster III, LgA was close to Lg S and Lg R was close to Lg U. Pair wise difference between and among Ganoderma lucidum, Leucoagaricus sp. and Lentinus sp. shown in Table 6.
Table 7.
Genetic Identity and Genetic distance among 30 accessions (Mushroom samples collected from 30 different sampling sites).
| 0.000 | GdA-1 | GdA-2 | GdA-3 | GdA-4 | GdR-1 | GdR-2 | GdR-3 | GdS-1 | GdS-2 | GdS-3 | GdS-4 | GdU-1 | GdU-2 | GdU-3 | LgA-1 | LgA-2 | LgR-1 | LgR-2 | LgS-1 | LgS-2 | LgU-1 | LgU-2 | LsA-1 | LsA-2 | LsR-1 | LsR-2 | LsS-1 | LsS-2 | LsU-1 | LsU-2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GdA-1 | 0.000 | 0.045 | 0.216 | 0.198 | 0.216 | 0.243 | 0.234 | 0.324 | 0.279 | 0.342 | 0.279 | 0.270 | 0.279 | 0.288 | 0.505 | 0.486 | 0.387 | 0.387 | 0.550 | 0.532 | 0.441 | 0.432 | 0.441 | 0.405 | 0.324 | 0.360 | 0.369 | 0.342 | 0.414 | 0.450 |
| GdA-2 | 0.045 | 0.000 | 0.207 | 0.225 | 0.189 | 0.216 | 0.189 | 0.297 | 0.270 | 0.333 | 0.270 | 0.225 | 0.234 | 0.243 | 0.496 | 0.477 | 0.378 | 0.378 | 0.541 | 0.523 | 0.432 | 0.423 | 0.450 | 0.414 | 0.333 | 0.351 | 0.360 | 0.333 | 0.423 | 0.423 |
| GdA-3 | 0.216 | 0.207 | 0.000 | 0.018 | 0.288 | 0.297 | 0.252 | 0.306 | 0.261 | 0.324 | 0.261 | 0.234 | 0.243 | 0.252 | 0.450 | 0.378 | 0.477 | 0.477 | 0.477 | 0.459 | 0.477 | 0.468 | 0.496 | 0.496 | 0.450 | 0.468 | 0.459 | 0.450 | 0.486 | 0.486 |
| GdA-4 | 0.198 | 0.225 | 0.018 | 0.000 | 0.288 | 0.297 | 0.270 | 0.324 | 0.261 | 0.342 | 0.261 | 0.252 | 0.261 | 0.270 | 0.450 | 0.378 | 0.477 | 0.477 | 0.477 | 0.459 | 0.477 | 0.468 | 0.496 | 0.496 | 0.432 | 0.468 | 0.459 | 0.450 | 0.468 | 0.505 |
| GdR-1 | 0.216 | 0.189 | 0.288 | 0.288 | 0.000 | 0.027 | 0.036 | 0.234 | 0.243 | 0.252 | 0.243 | 0.126 | 0.135 | 0.126 | 0.450 | 0.468 | 0.369 | 0.387 | 0.459 | 0.459 | 0.387 | 0.432 | 0.477 | 0.459 | 0.414 | 0.414 | 0.423 | 0.378 | 0.450 | 0.486 |
| GdR-2 | 0.243 | 0.216 | 0.297 | 0.297 | 0.027 | 0.000 | 0.063 | 0.225 | 0.234 | 0.243 | 0.234 | 0.099 | 0.108 | 0.099 | 0.441 | 0.459 | 0.360 | 0.378 | 0.450 | 0.450 | 0.396 | 0.441 | 0.486 | 0.486 | 0.405 | 0.423 | 0.414 | 0.387 | 0.459 | 0.496 |
| GdR-3 | 0.234 | 0.189 | 0.252 | 0.270 | 0.036 | 0.063 | 0.000 | 0.216 | 0.207 | 0.234 | 0.225 | 0.126 | 0.117 | 0.126 | 0.450 | 0.450 | 0.387 | 0.387 | 0.459 | 0.441 | 0.405 | 0.432 | 0.477 | 0.459 | 0.414 | 0.414 | 0.423 | 0.378 | 0.468 | 0.468 |
| GdS-1 | 0.324 | 0.297 | 0.306 | 0.324 | 0.234 | 0.225 | 0.216 | 0.000 | 0.117 | 0.036 | 0.117 | 0.234 | 0.225 | 0.234 | 0.432 | 0.450 | 0.405 | 0.423 | 0.441 | 0.441 | 0.423 | 0.450 | 0.514 | 0.532 | 0.414 | 0.432 | 0.459 | 0.450 | 0.505 | 0.486 |
| GdS-2 | 0.279 | 0.270 | 0.261 | 0.261 | 0.243 | 0.234 | 0.207 | 0.117 | 0.000 | 0.117 | 0.054 | 0.225 | 0.216 | 0.243 | 0.423 | 0.423 | 0.378 | 0.378 | 0.468 | 0.450 | 0.432 | 0.441 | 0.468 | 0.486 | 0.387 | 0.441 | 0.450 | 0.441 | 0.459 | 0.477 |
| GdS-3 | 0.342 | 0.333 | 0.324 | 0.342 | 0.252 | 0.243 | 0.234 | 0.036 | 0.117 | 0.000 | 0.117 | 0.234 | 0.225 | 0.234 | 0.450 | 0.468 | 0.405 | 0.423 | 0.459 | 0.459 | 0.441 | 0.468 | 0.532 | 0.550 | 0.432 | 0.450 | 0.459 | 0.450 | 0.523 | 0.505 |
| GdS-4 | 0.279 | 0.270 | 0.261 | 0.261 | 0.243 | 0.234 | 0.225 | 0.117 | 0.054 | 0.117 | 0.000 | 0.207 | 0.216 | 0.225 | 0.423 | 0.441 | 0.378 | 0.396 | 0.450 | 0.450 | 0.450 | 0.477 | 0.486 | 0.486 | 0.387 | 0.423 | 0.450 | 0.441 | 0.441 | 0.459 |
| GdU-1 | 0.270 | 0.225 | 0.234 | 0.252 | 0.126 | 0.099 | 0.126 | 0.234 | 0.225 | 0.234 | 0.207 | 0.000 | 0.027 | 0.018 | 0.396 | 0.414 | 0.423 | 0.441 | 0.423 | 0.423 | 0.405 | 0.432 | 0.496 | 0.496 | 0.396 | 0.432 | 0.405 | 0.378 | 0.468 | 0.486 |
| GdU-2 | 0.279 | 0.234 | 0.243 | 0.261 | 0.135 | 0.108 | 0.117 | 0.225 | 0.216 | 0.225 | 0.216 | 0.027 | 0.000 | 0.027 | 0.423 | 0.423 | 0.432 | 0.432 | 0.432 | 0.414 | 0.414 | 0.423 | 0.505 | 0.505 | 0.405 | 0.441 | 0.414 | 0.387 | 0.477 | 0.477 |
| GdU-3 | 0.288 | 0.243 | 0.252 | 0.270 | 0.126 | 0.099 | 0.126 | 0.234 | 0.243 | 0.234 | 0.225 | 0.018 | 0.027 | 0.000 | 0.396 | 0.414 | 0.423 | 0.441 | 0.423 | 0.423 | 0.405 | 0.450 | 0.496 | 0.496 | 0.414 | 0.450 | 0.423 | 0.396 | 0.468 | 0.468 |
| LgA-1 | 0.505 | 0.496 | 0.450 | 0.450 | 0.450 | 0.441 | 0.450 | 0.432 | 0.423 | 0.450 | 0.423 | 0.396 | 0.423 | 0.396 | 0.000 | 0.072 | 0.459 | 0.459 | 0.189 | 0.207 | 0.315 | 0.306 | 0.514 | 0.477 | 0.505 | 0.523 | 0.459 | 0.486 | 0.486 | 0.505 |
| LgA-2 | 0.486 | 0.477 | 0.378 | 0.378 | 0.468 | 0.459 | 0.450 | 0.450 | 0.423 | 0.468 | 0.441 | 0.414 | 0.423 | 0.414 | 0.072 | 0.000 | 0.459 | 0.441 | 0.171 | 0.171 | 0.297 | 0.270 | 0.477 | 0.459 | 0.486 | 0.505 | 0.477 | 0.468 | 0.450 | 0.468 |
| LgR-1 | 0.387 | 0.378 | 0.477 | 0.477 | 0.369 | 0.360 | 0.387 | 0.405 | 0.378 | 0.405 | 0.378 | 0.423 | 0.432 | 0.423 | 0.459 | 0.459 | 0.000 | 0.072 | 0.450 | 0.450 | 0.288 | 0.315 | 0.360 | 0.378 | 0.405 | 0.387 | 0.378 | 0.387 | 0.369 | 0.351 |
| LgR-2 | 0.387 | 0.378 | 0.477 | 0.477 | 0.387 | 0.378 | 0.387 | 0.423 | 0.378 | 0.423 | 0.396 | 0.441 | 0.432 | 0.441 | 0.459 | 0.441 | 0.072 | 0.000 | 0.432 | 0.432 | 0.270 | 0.279 | 0.342 | 0.324 | 0.405 | 0.387 | 0.378 | 0.387 | 0.333 | 0.333 |
| LgS-1 | 0.550 | 0.541 | 0.477 | 0.477 | 0.459 | 0.450 | 0.459 | 0.441 | 0.468 | 0.459 | 0.450 | 0.423 | 0.432 | 0.423 | 0.189 | 0.171 | 0.450 | 0.432 | 0.000 | 0.054 | 0.306 | 0.315 | 0.414 | 0.432 | 0.477 | 0.459 | 0.450 | 0.459 | 0.441 | 0.459 |
| LgS-2 | 0.532 | 0.523 | 0.459 | 0.459 | 0.459 | 0.450 | 0.441 | 0.441 | 0.450 | 0.459 | 0.450 | 0.423 | 0.414 | 0.423 | 0.207 | 0.171 | 0.450 | 0.432 | 0.054 | 0.000 | 0.324 | 0.315 | 0.414 | 0.432 | 0.459 | 0.459 | 0.450 | 0.441 | 0.423 | 0.423 |
| LgU-1 | 0.441 | 0.432 | 0.477 | 0.477 | 0.387 | 0.396 | 0.405 | 0.423 | 0.432 | 0.441 | 0.450 | 0.405 | 0.414 | 0.405 | 0.315 | 0.297 | 0.288 | 0.270 | 0.306 | 0.324 | 0.000 | 0.081 | 0.432 | 0.414 | 0.477 | 0.459 | 0.450 | 0.441 | 0.423 | 0.441 |
| LgU-2 | 0.432 | 0.423 | 0.468 | 0.468 | 0.432 | 0.441 | 0.432 | 0.450 | 0.441 | 0.468 | 0.477 | 0.432 | 0.423 | 0.450 | 0.306 | 0.270 | 0.315 | 0.279 | 0.315 | 0.315 | 0.081 | 0.000 | 0.423 | 0.405 | 0.432 | 0.432 | 0.405 | 0.396 | 0.414 | 0.432 |
| LsA-1 | 0.441 | 0.450 | 0.496 | 0.496 | 0.477 | 0.486 | 0.477 | 0.514 | 0.468 | 0.532 | 0.486 | 0.496 | 0.505 | 0.496 | 0.514 | 0.477 | 0.360 | 0.342 | 0.414 | 0.414 | 0.432 | 0.423 | 0.000 | 0.072 | 0.279 | 0.243 | 0.270 | 0.261 | 0.117 | 0.117 |
| LsA-2 | 0.405 | 0.414 | 0.496 | 0.496 | 0.459 | 0.486 | 0.459 | 0.532 | 0.486 | 0.550 | 0.486 | 0.496 | 0.505 | 0.496 | 0.477 | 0.459 | 0.378 | 0.324 | 0.432 | 0.432 | 0.414 | 0.405 | 0.072 | 0.000 | 0.279 | 0.243 | 0.270 | 0.261 | 0.099 | 0.117 |
| LsR-1 | 0.324 | 0.333 | 0.450 | 0.432 | 0.414 | 0.405 | 0.414 | 0.414 | 0.387 | 0.432 | 0.387 | 0.396 | 0.405 | 0.414 | 0.505 | 0.486 | 0.405 | 0.405 | 0.477 | 0.459 | 0.477 | 0.432 | 0.279 | 0.279 | 0.000 | 0.108 | 0.135 | 0.126 | 0.306 | 0.324 |
| LsR-2 | 0.360 | 0.351 | 0.468 | 0.468 | 0.414 | 0.423 | 0.414 | 0.432 | 0.441 | 0.450 | 0.423 | 0.432 | 0.441 | 0.450 | 0.523 | 0.505 | 0.387 | 0.387 | 0.459 | 0.459 | 0.459 | 0.432 | 0.243 | 0.243 | 0.108 | 0.000 | 0.135 | 0.126 | 0.288 | 0.288 |
| LsS-1 | 0.369 | 0.360 | 0.459 | 0.459 | 0.423 | 0.414 | 0.423 | 0.459 | 0.450 | 0.459 | 0.450 | 0.405 | 0.414 | 0.423 | 0.459 | 0.477 | 0.378 | 0.378 | 0.450 | 0.450 | 0.450 | 0.405 | 0.270 | 0.270 | 0.135 | 0.135 | 0.000 | 0.063 | 0.297 | 0.297 |
| LsS-2 | 0.342 | 0.333 | 0.450 | 0.450 | 0.378 | 0.387 | 0.378 | 0.450 | 0.441 | 0.450 | 0.441 | 0.378 | 0.387 | 0.396 | 0.486 | 0.468 | 0.387 | 0.387 | 0.459 | 0.441 | 0.441 | 0.396 | 0.261 | 0.261 | 0.126 | 0.126 | 0.063 | 0.000 | 0.270 | 0.270 |
| LsU-1 | 0.414 | 0.423 | 0.486 | 0.468 | 0.450 | 0.459 | 0.468 | 0.505 | 0.459 | 0.523 | 0.441 | 0.468 | 0.477 | 0.468 | 0.486 | 0.450 | 0.369 | 0.333 | 0.441 | 0.423 | 0.423 | 0.414 | 0.117 | 0.099 | 0.306 | 0.288 | 0.297 | 0.270 | 0.000 | 0.054 |
| LsU-2 | 0.450 | 0.423 | 0.486 | 0.505 | 0.486 | 0.496 | 0.468 | 0.486 | 0.477 | 0.505 | 0.459 | 0.486 | 0.477 | 0.468 | 0.505 | 0.468 | 0.351 | 0.333 | 0.459 | 0.423 | 0.441 | 0.432 | 0.117 | 0.117 | 0.324 | 0.288 | 0.297 | 0.270 | 0.054 | 0.000 |
Gd = Ganoderma lucidum, Lg = Leucoagaricus sp., Ls = Lentinus sp., A = Anuppur, R = Rewa, S = Shahdol, U = Umaria, GdA 1–4 = Amarkantak, Keonchi, Rajendragram, Pondki, GdR 1–3 = Local Rewa ((University Campus), Chuhiya forest, Semariya Forest GdS 1–4 = Jaisinghnagar Beohari, Gohparu, Kudari, GdU 1–3 = Manpur, Nowrojabad, Ghunghuti, LgA-1and2 = Rajendragram Keonchi, LgR-1 and 2 = Local Rewa, Sohagi, LgS-1 and 2 = Kudari, Barbaspur, LgU-1 and 2 = Nowrojabaad, Ghunghuti.
LsA-1 and 2 = Amarkantak, Rajendragram, LsR-1 and 2 = Chuhiya Ghati, Sirmour, LsS-1 and 2 = Gohparu, Beohari, LsU-1 and 2 = Barbaspur, Ghunghuti
Fig. 2.
Dendrogram representing genetic identity and genetic distance among 30 accessions.
Table 6.
Pairwise Fst Distance method: Pairwise difference between and among Ganoderma lucidum, Leucoagaricus sp. and Lentinus sp.
| GdA | GdR | GdS | GdU | LgA | LgR | LgS | LgU | LsA | LsR | LsS | LsU | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GdA | 0.00000 | |||||||||||
| GdR | 0.57552 | 0.00000 | ||||||||||
| GdS | 0.58682 | 0.69113 | 0.00000 | |||||||||
| GdU | 0.61439 | 0.72034 | 0.71559 | 0.00000 | ||||||||
| LgA | 0.71367 | 0.88479 | 0.80089 | 0.90204 | 0.00000 | |||||||
| LgR | 0.69894 | 0.86182 | 0.78074 | 0.90683 | 0.84158 | 0.00000 | ||||||
| LgS | 0.75117 | 0.89831 | 0.81740 | 0.91930 | 0.65854 | 0.85714 | 0.00000 | |||||
| LgU | 0.70824 | 0.86694 | 0.79950 | 0.89721 | 0.74242 | 0.73438 | 0.78571 | 0.00000 | ||||
| LsA | 0.71917 | 0.88991 | 0.82734 | 0.91925 | 0.85047 | 0.79487 | 0.85106 | 0.81720 | 0.00000 | |||
| LsR | 0.65047 | 0.84403 | 0.76928 | 0.87570 | 0.82143 | 0.77273 | 0.82524 | 0.79000 | 0.6551 7 | 0.00000 | ||
| LsS | 0.68525 | 0.87730 | 0.81118 | 0.90711 | 0.85714 | 0.82353 | 0.87000 | 0.82979 | 0.74576 | 0.34483 | 0.00000 | |
| LsU | 0.72714 | 0.90220 | 0.82841 | 0.92802 | 0.86792 | 0.81818 | 0.87629 | 0.84211 | 0.44000 | 0.73134 | 0.79365 | 0.00000 |
where Gd = Ganoderma lucidum, Lg = Leucoagaricus sp., Ls = Lentinus sp. A = Anuppur, R = Rewa, S = Shahdol, U = Umaria.
Bold fonts indicate maximum and minimum values in given table.
4. Discussion
RAPD is still one of the cheapest and quickest method for accessing variability at the DNA level. Molecular markers were used for the genetic diversity analysis of various mushroom species such as discrimination of different strains of Agaricus bisporus [12], [33] Pleurotus sp. [34] Ganoderma lucidum [35], [25], [36] Lentinula edodes [37] RAPD marker is preferred over other molecular markers for the genetic diversity analysis because it is simple, easy to use, dominant and does not require any specific knowledge of the DNA sequence [26]. According to [13] phylogenetic analysis of RAPD profiles proved more useful in revealing both inter-generic and intra-species variability than ITS multiple sequence alignment alone. Inspite of many advantages there are certain limitations of RAPD. RAPD markers are dominant. Amplification either occurs at a locus or it does not, leading to scores based on band presence or absence. This means that homozygotes and heterozygotes cannot be distinguished. In addition, the absence of a band through lack of a target sequence cannot be distinguished from that occurring through the lack of amplification for other reasons (e.g., poor quality DNA), contributing to ambiguity in the interpretation of results.
[38], [25], [36] also used RAPD markers for studying genetic diversity among different isolates of Ganoderma lucidum. [31] Studied genetic diversity among six local isolates of Ganoderma lucidum. [39] reported that environmental factors, variability, inter hybridization and morphological propensity causes inaccurate identification of Ganoderma lucidum.
Polymorphism occurs when two or more different phenotypes exist in the same population of a species or the occurrence of more than one form or morph. Polymorphism is common in nature; it is related to biodiversity, genetic variation and adaptation; it usually functions to retain a variety of forms in a population living in a varied environment [40]. Polymorphism results from evolutionary processes, as does any aspect of a species. It is heritable and is modified by natural selection. The ability of a species to respond adaptively to environmental changes depends on the level of genetic variability it contains [41]. [42] have also studied genetic characterization of isolates of the basidiomycete Agaricus blazei by RAPD.
In present study seven RAPD primers were used to access the diversity within and among twelve populations of three mushroom species Ganoderma lucidum, leucoagaricus sp. and Lentinus sp. Total of 111 bands were scored by 7 RAPD primers in 30 accessions of three mushroom species Ganoderma lucidum (Gd), Leucoagaricus sp (Lg), and Lentinus sp. (Ls). All 111 bands were polymorphic in nature (100%). Therefore, it revealed that the used primers had sufficient potency for population studies and 30 accessions had higher genetic differences among each other. In best of the knowledge, this is the first report, which accesses the genetic diversity between three mushroom species (Gd Ganoderma lucidum, Lg Leucoagaricus sp., Ls Lentinus).
The polymorphic percentage ranged from 3.60 to 23% within twelve populations, while polymorphic percentage among group was 40.56, among population within groups was 41.12 and within population was 18.32. This indicated that the genetic diversity within the population was very low, but slightly higher in the populations of three species.
Among seven populations, within the first group, within the second group and within the third group mean PIC = 90.644 ± 3.834, 78.820 ± 11.311, 79.757 ± 5.344 and 70.077 ± 10.304 respectively, showed high level of polymorphic informativeness of used markers and suggested that these markers were equally effective in determining polymorphisms. The PIC value of each RAPD primers was determined by both the number of alleles and their frequency distribution within a population and was used to assess their informativeness level (high PIC > 0.5, moderate 0.5 > PIC > 0.25 and low PIC < 0.25) [43], [44]. Therefore, in this study, polymorphic information content (PIC) of individual primers was calculated for among 12 populations and within the three groups. The PIC value has been used for evaluating genetic variation in many studies using RAPD markers [45], [46], [47], [48], [49], [50]. PIC was also evaluated in other molecular markers [51], [52], [53], [54].
4.1. Study of genetic variation
For the genetic diversity analysis, genetic variation in a diverse population was estimated through several parameters like observed number of alleles (Na), effective number of alleles (Ne), gene diversity (H), Shannon's Information index (I), observed and expected heterozygosity (Ht and Hs) [55], [56].
In the present study, within the population,very low value of the observed number of alleles (Na), effective number of alleles (Ne), gene diversity (H), Shannon's information index (I) was observed. Within each group (Gd Ganoderma lucidum, Lg Leucoagaricus sp., Ls Lentinus sp.) had slightly higher value of Na, Ne, H and I was observed as compared to within their population. Among 30 accession highest values of Na, Ne, H and I were recorded. Similarly the observed and expected heterozygosity (Ht and Hs) was higher in the 30 accessions in comparison to the groups. These informations further state that the genetic diversity was increasing in an order of within population, within the group, among the group and among accessions.
Study among population within group revealed that genetic variation among all groups was highest (96.40). Polymorphism percentage of first group was 52.2, second group 65.77 and third group was 50.45.
4.2. Relative differentiation and estimate of gene flow
Gene flow is a collective term that includes all the mechanism resulting in the movement of genes from one population to another. In this study, higher relative differentiation in within three groups (GST = 0.2198), (GST = 0.0788) and (GST = 0.1312) was observed in respect to the relative differentiation among 30 accessions (GST = 0.6737). On the other hand, gene flow was higher among the 30 accessions (Nm = 0.7396) in respect to the gene flow of within three groups (Nm = 0.198, 0.0788, 0.1312). These data revealed that the populations of three mushroom species (Ganoderma lucidum, Leucoagaricus sp. and Lentinus sp.) shown low genetic diversity (having a higher relative differentiation) and restricted gene flow (having a lower Nm value) as compare to the 30 accessions of mushroom species. Assessment of gene flow from one population to another is an important parameter for study of genetic diversity.
4.3. Analysis of Molecular Variants (AMOVA)
The significance of the covariance components was calculated with the different possible levels of genetic structure (among groups, among populations within groups and within populations). Among three groups representing Gd., Lg and Ls, Among populations within groups shown highest percentage of variation (Pv = 41.12) while within populations, the lowest percentage of variation (18.32) was observed. This result also support that the highest genetic variation was present among groups in comparison to among the population within a species and lowest genetic variation was observed within the population. Similar observations were also made by [25].
4.4. Genetic distance and phylogenetics
The phylogenetic tree developed through the distance matrix of 30 accessions of three mushroom species (Ganoderma lucidum, Leucoagaricus sp. and Lentinus sp.) also confirmed the findings of all the parameters like polymorphism, genetic variation, relative differentiation, gene flow and AMOVA. Because the phylogenetic tree consists three major clusters and population of this cluster represent their respective species and the genetic distance is lesser within the population and pretty higher within the species, while highest distance among population, which was in accordance with earlier reports. Phylogenetic tree also showed that the accessions of Ganoderma lucidum, Leucoagaricus sp, and Lentinus sp. collected from the same location were present at same sub-cluster in the tree, similarly, populations of all three species exist in the major clusters of the respective species. In addition to this, the phylogenetic tree also categorized the populations according to their geographical positions. These clustering system seem to be in agreement with [25]. He performed RAPD analysis to study inter species and intra species variations. He used 8 isolates of Agaricus bisporus, 16 isolates of Ganoderma lucidum and 22 isolates of Lactarious delicious. It clustered the 16 isolates of Ganoderma lucidum into three main groups. Genetic distance between the species showed that group I and group II were closely related to each other.
High levels of genetic diversity within populations are always desirable to ensure that they are genetically sustainable. Adaptability is correlated with diversity and should be an important driver for conservation in response to environmental change [57].
5. Conclusion
From this study it can be concluded that samples collected from same places show more similarity and less distance. Mushroom species collected from four different districts have shown good genetic distance and more percentage of variation (41.12%) which proved that these species are established species of this area and genetically adopted in the climatic conditions of this region. Dendrogram of 30 accessions showed that sampled species are having good genetic boundary according to the location.
This rich biodiversity of Vindhyan region needs further exploration to widen the nutritional and medicinal base of the rural population who depend on the mushrooms through conservation, cultivation and commercialization activities. The rich diversity of mushrooms in Umaria district of Vindhyan region, offers huge socio-economic potentials. However, they need to be properly documented for optimum application. Hence, this study is an important first step towards producing a checklist of mushrooms of Vindhyan region.
Acknowledgement
Authors are thankful to Dr. Pramod Sairkar Junior technical officer MPCST, Bhopal, MP for statistical analysis.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
Contributor Information
Sandhya Dwivedi, Email: sandhyabiotech2005@gmail.com.
Surendra Singh, Email: singhbiosci@yahoo.com.
U.K. Chauhan, Email: ukchauhan.dr@gmail.com.
Mahendra Kumar Tiwari, Email: mktiwari@aksuniversity.com.
References
- 1.Pandey VN, Srivastava AK. Fleshy fungi go ethno-botanical food use in North Eastern Tarai region of Uttar Pradesh. Proceeding of the National Symposium on Mushroom, NRCM-Solan; 1994, 3.
- 2.Muthukumaran Jayachandran, Xiao Jianbo, Baojun Xu. A critical review on health promoting benefits of edible mushrooms through gut. Microbiota Int J Mol Sci. 2017;18(9):1934. doi: 10.3390/ijms18091934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren L., Visitev A.V., Grekhov A.N., Tertov V.V., Tutelyan V.A. Anti-atherosclerotic properties of macrofungi. Voprosy Pictaniya. 1989;1:16–19. [Google Scholar]
- 4.Gareth J.E.B. Edible Mushrooms in Singapore and other South East Asian countries. The Mycologist. 1990;4:119–124. [Google Scholar]
- 5.Pegler D.N. Royal Botanic Gardens; Kew: 1983. The Genus Lentinus: A World Monograph. Kew Bulletin Additional Series X; p. 281. [Google Scholar]
- 6.Kumar Singdevsachan Sameer, Patra Jayanta Kumar, Thatoi Hrudayanath. Nutritional and Bioactive Potential of Two Wild Edible Mushrooms (Lentinus sajor-caju and Lentinus torulosus) from Similipal Biosphere Reserve, India. Food Sci Biotechnol. 2013;22(1):137–145. [Google Scholar]
- 7.Galor S.W., Yuen J., Buswell J.A., Benzie I.F.F. 2nd ed. CRC Press/Taylor and Francis; Florida: 2011. Ganoderma lucidum (Lingzhi or Reishi), a medicinal mushroom in herbal medicine: biomolecular and clinical aspects. [PubMed] [Google Scholar]
- 8.Arroyo I.J., Acevedo R.R., Perez A.A. Anti-tumor effects of Ganoderma ludicum (Reishi) in inflammatory breast cancer in in vivo and in vitro models. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0057431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao H., Zhang Q., Zhao L. Spore powder of Ganoderma lucidum improves cancer-related fatigue in breast cancer patients undergoing endocrine therapy: a pilot clinical trial. Evidence-Based Complement Altern Med. 2012 doi: 10.1155/2012/809614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai L., Sun W. Ganoderma lucidum inhibits proliferation of human ovarian cancer cells by suppressing VEGF expression and up-regulating the expression of connexin 43. BMC Complement Altern Med. 2014;14:434. doi: 10.1186/1472-6882-14-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zambino P.J., Szabo L.J. Mycologia. 1993;85:401–441. [Google Scholar]
- 12.Khush R.S., Becker E., Wach M. Appl Environ Microb. 1992;57:2971–2977. doi: 10.1128/aem.58.9.2971-2977.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh S.K., Doshi A., Yadav M.C., Kamal S. Curr Sci. 2006;9(9–10):1225–1229. [Google Scholar]
- 14.Moore D, Chiu SW. Fungal Diversity Press, Hong Kong; 2001.
- 15.Moore A.J., Challen M.P., Warner P.J., Elliott T.J. Appl Environ Microb. 2001;55:742–749. doi: 10.1007/s002530000588. [DOI] [PubMed] [Google Scholar]
- 16.Yadav M.K., Chandra Ram, Singh H.B., Yadav S.K., Yadav S.K., Naik Sushreeta, Dhakad P.K. Genetic Diversity Characterization of Pleurotus strains by Random Amplified Polymorphic DNA Fingerprinting. Int J Curr Microbiol App Sci. 2017;6(5):1260–1267. [Google Scholar]
- 17.Ben H.B.A., Garrett K. Reliable protocols for DNA extraction from freeze-dried macrofungal samples used in molecular macrofungal systematics studies. Curr Res Environ Appl Mycol. 2016;6(1):45–50. [Google Scholar]
- 18.Singer R. In: The Agaricales in Modern Taxonomy. Spoerke Vadauz, Rumack B.H., editors. CRC Press; London: 1975. pp. 65–95. (J. Cramer, 3rd eds.) [Google Scholar]
- 19.Singer R. 4th ed. Sven Koeiltz Scientific Books; Germany: 1986. The Agaricales in Modern Taxonomy; p. 981. [Google Scholar]
- 20.Natarajan K. Kavaka. 1995;33:61–125. [Google Scholar]
- 21.Jordan M. In: The Encyclopedia of Fungi of Britain and Europe. David Charles., editor. John Taylor Book Venture Ltd., Brunel House; Newton Abbot, Devon: 1995. [Google Scholar]
- 22.Saini S.S., Atri N.S. Indian J Mycol Plant Pathol. 1993;23(3):250–254. [Google Scholar]
- 23.Lamaison J.L., Polese J.M. Konemann Publication; 1999. The great encyclopedia of mushrooms; pp. 1–240. [Google Scholar]
- 24.Aessoe T., Lincoff J. Dorling Kindersley handbooks. DK adult publisher; 1998. Eye Witness Handbooks Mushroom; pp. 1–304. [Google Scholar]
- 25.Upadhyay MK. Genetic and biochemical characterization of mushrooms of Central India with special reference to Ganoderma lucidum, Ph.D Thesis, R.D. University, Jabalpur (MP), India; 2005, p. 120–145.
- 26.Williams J.G.K., Kubelik A.R., Lilvak K.J., Rafalski J.A., Tingey S.V. Nucleic Acids. 1990 doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammer O., Harper D.A.T., Ryan P.D. Palaeontol Electron. 2001;4(1):1–9. [Google Scholar]
- 28.Nei’s M. Columbia University Press, New York; 1988.
- 29.Tamura K., Peterson D., Peterson N., Stecher G., Nei’s M., Kumar S. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nei’s M, Li W. Proceeding of National Academy of Sciences 1979;76:5269–73. [DOI] [PMC free article] [PubMed]
- 31.Excoffier L., Laval G., Schneider S. Evaluat Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 32.Peakall R., Smouse P. Bioinformatics (Oxford, England). 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore D., Chiu S.W. Fungal Diversity Press; Hong Kong: 2001. Filamentous fungi as food. Exploitation of Filamentous Fungi; pp. 223–251. [Google Scholar]
- 34.Chandra S., Ghosh K., Acharya K. Nat Sci. 2010;8(7):90–95. [Google Scholar]
- 35.Hsew R.S., Wang H.H., Wang H.F., Moncalvo J.M. Appl Environ Microb. 1996;62:1354–1363. doi: 10.1128/aem.62.4.1354-1363.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasim G., Ali M., Mehmood N. Pak J Bot. 2010;42(5):3307–3315. [Google Scholar]
- 37.Chiu S.W., Ma A., Lin F., Moore D. Mycol Res. 1996;100:1393–1399. [Google Scholar]
- 38.Zhao M.W., Chen M.J., Wang N. J Nanjing Agri Univ. 2003;26:60–63. [Google Scholar]
- 39.Zheng L., Jia D., Fei X., Luo X., Yang Z. Microbiol Res. 2009;164:312–321. doi: 10.1016/j.micres.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Dobzhansky T. Columbia University Press; New York: 1970. Genetics of Evolutionary Process; pp. 1–505. [Google Scholar]
- 41.Ayala F.J., Kiger J.A. Modern Genet. 1984;3:1–923. [Google Scholar]
- 42.Nelson B.C., Dias E.S., Marcos A.G., Ferreira A., Eira D. Brazilian J Microbiol. 2002;33(2):131–133. [Google Scholar]
- 43.Bostein D., White R.L., Skolnick M., Davis R.W. Am J Human Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 44.Weber A., Karst J., Gilbert B., Kimmins J.P. Oecologia. 2005;143:148–156. doi: 10.1007/s00442-004-1777-y. [DOI] [PubMed] [Google Scholar]
- 45.Bhattacharya S., Bandopadhyay T.K., Ghosh P.D. Emirates J. Food Agri. 2010;22(1):13–24. [Google Scholar]
- 46.Najaphy A., Ashrafiparchin R., Farshadfar E. Biotechnol Biotechnol Equip. 2011;10:2634–2638. [Google Scholar]
- 47.Wijarat P., Keeratinijakal V., Toojinda T., Vanavichit A., Tragoonrung S. Thai J Genet. 2011;4(2):115–125. [Google Scholar]
- 48.Tripathi N., Chouhan D.S., Saini N., Tiwari S. Biotechnology. 2012;2:327–336. [Google Scholar]
- 49.Sadeghi A., Cheghamirza K. Ann Biolog Res. 2012;3(7):3267–3273. [Google Scholar]
- 50.Lal S., Mistry K.N., Chaturvedi S.P. Int J Biol Pharma Allied Sci. 2013;2(2):373–385. [Google Scholar]
- 51.Yang K.J., Zhang X.P., Zhang Z.X. J Plant Res Environ. 2007;16:79–80. [Google Scholar]
- 52.Muthusamy S., Kanagarajan S. Electron J Biotechnol. 2008;11(3):1–10. [Google Scholar]
- 53.Sarwat M., Das S., Srivastava P.S. Plant Cell Rep. 2008;27:519–528. doi: 10.1007/s00299-007-0478-5. [DOI] [PubMed] [Google Scholar]
- 54.Praveen C.V., Chakrabarty D., Jena S.N., Mishra D.K., Singh P.K., Sawant S.V., Tuli R. Indian Crops Product. 2009;29:581–589. [Google Scholar]
- 55.Cruse-Sanders J.M., Hamrick J.L. Am J Bot. 2004;91(4):540–548. doi: 10.3732/ajb.91.4.540. [DOI] [PubMed] [Google Scholar]
- 56.Hu Y., Wang L., Xie X., Yang J., Li Y., Zhang H. Genetic diversity of wild populations of Rheum tanguticum endemic to China as revealed by ISSR analysis. Biochem Syst Ecol. 2010;38:264–274. [Google Scholar]
- 57.Gregory A, Burke T, Ferris R, Robson J, Smithers R, Whitlock R. The conservation of genetic diversity: Science and policy needs in a changing world Gregory. JNCC report No. 2006, p. 1–38.