Abstract
Background
Genetic factors play important role in the development of type 2 diabetes and diabetic nephropathy. Endothelial nitric oxide synthase (eNOS) gene is responsible for the bioavailability of nitric oxide and endothelial function.
Aim
To assess the association of the endothelial nitric oxide synthase (eNOS) (T786C and G894T) single nucleotide polymorphisms with Egyptian type 2 diabetes mellitus and diabetic nephropathy.
Patients and methods
A total of 200 type 2 diabetic patients and 100 apparently healthy volunteers as controls were included in the study. They were subjected to clinical examination and laboratory tests: fasting blood glucose, HBA1C, lipid profile, serum creatinine, blood urea and albumin creatinine ratio (ACR). Assessment of the T786C and G894T polymorphisms in the eNOS gene was done using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
Results
There was no significant difference in distribution of eNOS T-786C polymorphism between patients and controls; TT genotype of eNOS G894T was more frequent in diabetic patients with and without albuminuria compared to controls. Patients were divided into 3 groups according to ACR. Normoalbuminuria: 37 patients with ACR ≤ 30 mg/g, microalbuminuria: 96 patients with ACR > 30 mg/g and ≤ 300 mg/g, and macroalbuminuria: 67 patients with ACR > 300 mg/g. There was no significant difference in genotype distribution of eNOS T-786C between the 3 groups of diabetic patients. The prevalence of TT genotype of eNOS G894T was higher in microalbuminuria patients compared to other groups.
Conclusion
eNOS G894T variant may increase risk of type 2 diabetes with lack of association between eNOS T786C, eNOS G894T and DN in Egyptians.
Keyword: eNOS-DN
1. Introduction
Diabetes mellitus (DM) is a metabolic disorder characterized by hyperglycemia, oxidative stress and inflammation and it is associated with micro and macrovascular complications[1]. Endothelial dysfunction is a common finding in diabetic patients and is responsible for its vascular complication. Endothelial cells synthesize nitric oxide (NO) from l-arginine by endothelial NO synthase (NOS). Nitric oxide is a vasodilator substance, responsible for regulation of endothelial function and maintaining the balance between vascular tone, coagulation, and inflammation [2].
Diabetic nephropathy (DN) is one of the most serious complications of diabetes and is the leading cause of chronic kidney failure [3]. Diabetic nephropathy is characterized by hemodynamic (hyperfiltration and hyperperfusion) as well as structural abnormalities (glomerulosclerosis, alterations in tubulointerstitium including interstitial fibrosis) and metabolic changes [4]. There is great variability between diabetic patients in the development of DN even among comparable glycemic control. Familial clustering and interethnic variation in development of DN suggest a role for genetic factor increasing risk of disease development [5].
Several genes have been reported to be associated with DN, among which eNOS gene has drawn considerable attention. The eNOS gene is located on chromosome7q35–36 and comprises 26 exons that span 21 kb and is expressed mainly in the endothelium [6].
Evidence suggests that variants of eNOS gene may cause defective NO synthesis and decreased NO levels, enhancing the susceptibility to glomerular disease and deteriorating the renal function [7].
Of the studied eNOS polymorphisms are the T786C single-nucleotide polymorphism in the promotor region that reduces eNOS transcription, and the G894T (Glu298Asp) missense mutation in exon 7 that may be associated with a decrease in eNOS activity [8], [9].
There is a discrepancy in the results of the study assessing the potential association of the eNOS gene variants with DN depending on race and ethnic background of the population studied [10].
The aim of the present study was to assess the association of the endothelial nitric oxide synthase (eNOS) (T786C and G894T) single nucleotide polymorphisms with Egyptian type 2 diabetes mellitus and diabetic nephropathy.
2. Patients and methods
The study is a cross-sectional case-control one.
2.1. Study population
The study included 200 patients with type 2 diabetes mellitus and 100 healthy volunteers as controls, with age and sex matched to the patients. Patients were recruited from the outpatient clinic of Medical Services Unit of National Research Centre and National Institute of Diabetes and Endocrinology. We excluded patients with type 1 diabetes, autoimmune diseases, liver failure or cardiac failure. Control subjects were recruited from the general community without any family history of diabetes. Patients and controls were subjected to detailed history taking, anthropometric measurements, and thorough clinical examination and laboratory tests.
2.2. Laboratory tests
Fasting blood glucose, HbA1c, complete lipid profile, blood urea nitrogen (BUN), and serum creatinine were assessed using quantitative enzymatic- colorimetric determination by Stanbio Lab. An early morning urine sample was taken for assessment of urinary albumin, urinary creatinine and calculation of albumin/creatinine ratio (ACR) (urine albumin (mg/dl)/urine creatinine (g/dl)) [11].
2.3. Detection of the T786C and G894T polymorphisms in the eNOS gene
The genomic DNA was extracted from peripheral blood leukocytes using QIAamp® DNA Mini Kit (QIAGEN, Germany) according to the manufacturer instructions. The genotyping of the two polymorphisms was carried out using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis as described by Cruz-González et al. [12]. Two sets of primers were used to amplify the locus in the promotor region which contains the −786T > C variant and exon 7 of the eNOS gene which harbor the G894T. The primers used and the size of the PCR products and restriction enzyme used are listed in [Table 1].
Table 1.
Sequence of primers, size of the PCR products and restriction enzyme used in genotyping of the -T786C, G894T.
| Sequence of primers | Size of PCR product (bp) | Restriction enzyme | |
|---|---|---|---|
| T786C | Sense: 5′ TGGAGAGTGCTGGTGTACCCCA3′ Antisense: 5′ GCCTCCACCCCACCCTGTC 3′ |
180 | MspI |
| G894T | Sense: 5′ AACCCCCTCTGGCCCACTCCC3′ Antisense:5′ TCCATCCCACCCAGTCAAT 3′ |
200 | MboI |
PCR was carried out in a 50 µl total final volume containing 200 µM dNTPs (Finzyme, Finland), 10 pmol of each primer, 2U of Taq polymerase (Finzyme, Finland) and 500 ng DNA. Thermal cycling conditions were as follows: Denaturation at 95 °C for 10 min, followed by 30 cycles of denaturation at 95 °C for 50 s, annealing at 57 °C for 50 s, and elongation at 72 °C for 50 s followed by a final elongation of 5 min. The PCR fragment containing the -786T > C polymorphism was subjected to digestion with MspI, which cuts the PCR product when the T at position -786 is replaced by a C. The fragment containing exon 7 was digested with MboI, which cuts only in the presence of T at position 894. Ten µl of the successfully amplified PCR products was digested with 5 units of the enzyme (Fermentas, Germany) and the fragments were run in 3.5% agarose gel stained with ethidium bromide, and analyzed under ultraviolet light (Figs. 1 and 2).
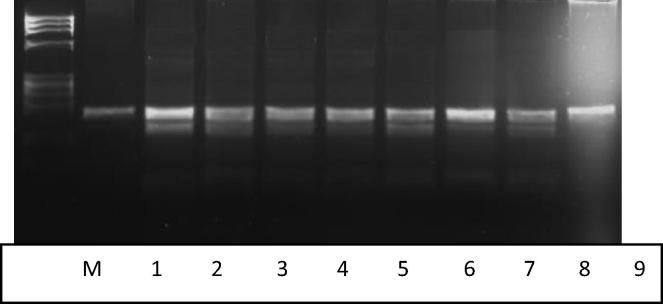
Fig. 1.
A 3.5% agarose gel illustrating digestion of the PCR products with MspI for detection of the -786T > C. Lane 1: Undigested PCR product (200 bp). Lane 2–8: 7 samples with the T/C genotype. Lane 9: one sample with T/T genotype. Lane M: Size marker (Phix 174 DNA-Hae III digest).
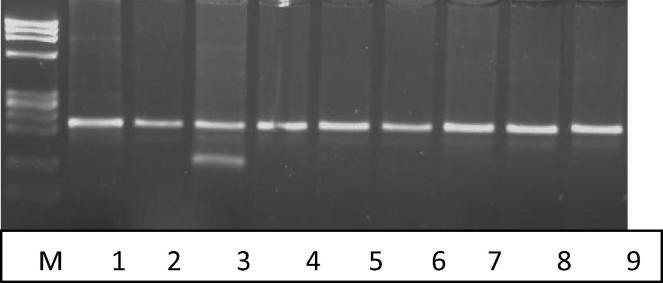
Fig. 2.
A 3.5% agarose gel illustrating digestion of the PCR products of exon 7 with MboI for detection of the G894T. Lane 1: Undigested PCR product (200 bp). Lane 2, 4–9: 7 samples with the C/C genotype. Lane 3: one sample with C/T genotype. Lane M: Size marker (Phix 174 DNA-Hae III digest).
Some samples were performed in duplicates to ensure quality, and all these duplicated samples showed same results in both runs.
The study was approved by the ethical committee of the National Research Centre and all participants signed written informed consent before enrollment in the study.
2.4. Statistical methods
Data were entered and analyzed using SPSS software computer program version 18. Quantitative data were represented as mean ± standard deviation (mean ± SD). Comparison of means was done using independent t-test and ANOVA (analysis of variance). Qualitative data were presented as frequency and percentage. Qualitative variables were compared using chi-square test to assess significant association and odds ratio was used to assess the strength of association. The alpha level of significance was set at p-value <0.05.
Conformity with the Hardy–Weinberg law of genetic equilibrium (HWE) was assured by a non-significant chi-square test comparing the observed versus the expected genotypes among studied cases and controls. A minimum level of statistical significance was considered at a p level of <0.05.
3. Results and analysis of the results
The study included 200 patients with type 2 diabetes, mean age 52.6 years, 86 males and 114 females and 100 apparently healthy volunteers as control group, mean age 51.6 years, 44 males and 56 females.
The results of the study showed that 47 (23.5%) of the patients were smokers, 145 (72.5%) of the patients had a positive family history of type 2 diabetes. Hypertension was present in 134(67%) patients, dyslipidemia in 74 (37%) patients, ischemic heart diseases in 83 (41.5%) patients and peripheral neuropathy in 139 (69.5%) patients.
According to ACR, patients were divided into 3 groups:
-
–
Normoalbuminuria: 37 patients with ACR ≤ 30 mg/g, 14 males and 23 females, mean age: 53.8 ± 7.9 years.
-
–
Microalbuminuria: 96 patients with ACR > 30 mg/g and ≤300 mg/g, 26 males and 70 females, mean age: 51.4 ± 9.2.
-
–
Macroalbuminuria: 67 patients with ACR > 300 mg/g, 46 males and 21 females, mean age: 54.6 ± 7 years.
Demographic and biochemical characteristics of the studied groups are presented in Table 2.
Table 2.
Demographic and biochemical characteristics of the studied groups (values are in mean ± SD).
| Normoalbuminuria n = 37 |
Microalbuminuria n = 96 |
Macroalbuminuria n = 67 |
Controls n = 100 |
Test | p | |
|---|---|---|---|---|---|---|
| Duration of diabetes (years) (mean ± SD) | 8.6 ± 6.2 | 9.4 ± 6.0 | 11.8 ± 6.3b | NA | 96.9 | 0.02 |
| Smokers n (%) (n = 64) | 7 (18.9) | 17 (17.7) | 23 (34.3) | 17 (17) | 2.0 | 0.03 |
| Waist circumference (cm) (mean ± SD) | 107.7 ± 12.1a | 108.6 ± 10.5a | 108.6 ± 12.3a | 97.5 ± 9.1 | 23.4 | <0.001 |
| BMI (kg/m2) (mean ± SD) | 32.1 ± 4.5a | 32.2 ± 5.1a | 32.7 ± 5.8a | 30.1 ± 2.8 | 5.9 | <0.001 |
| Fasting glucose (mg/dl) (mean ± SD) | 200.5 ± 96.1a | 210.9 ± 97.7a | 237.4 ± 100.2a,b,c | 94.5 ± 13.4 | 101.7 | <0.001 |
| HbA1C (%) (mean ± SD) | 9.1 ± 1.9a | 9.3 ± 2.0a | 9.6 ± 1.7a | 6.0 ± 0.6 | 188.0 | <0.001 |
| Serum urea (mg/dl) (mean ± SD) | 26.0 ± 6.8 | 23.8 ± 7.4 | 25.5 ± 8.3 | 24.3 ± 8.4 | 1.1 | 0.37 |
| S creatinine (mg/dl) (mean ± SD) | 0.98 ± 0.31 | 1.05 ± 0.38a | 1.1 ± 0.43a | 0.93 ± 0.27 | 4.0 | 0.01 |
BMI: body mass index, HBA1C: glycated hemoglobin.
Significant difference with controls (p < 0.05 in post hoc test).
Significant difference with microalbuminuria (p < 0.05 in post hoc test).
Significant difference with normoalbuminuria (p < 0.05 in post hoc test).
3.1. Genotype distribution of eNOS (T-786C) and eNOS (G894T) in type 2 diabetes and controls
The genotype and allele distribution of eNOS (T-786C) polymorphism were not in agreement with the Hardy–Weinberg equilibrium (p = 0) and this could be due to the absence of C/C genotype in the studied patients. On the other hand the genotype and allele distribution of eNOS (G894T) were in Hardy–Weinberg equilibrium (p = 0.2).
There was no significant difference in distribution of eNOS T-786C polymorphism between patients and controls (Table 3). None of the patients nor controls was carrying C/C genotype of eNOS (T-786C) and we did not observe association between eNOS (T-786C) and DM (OR: 1.2 95%CI: 0.9–1.6 p = 0.2). As regards eNOS G894T polymorphism, TT genotype was more frequent in diabetic patients with and without albuminuria compared to controls (Table 4). Patients who were carrying T/T genotype had an increased likelihood of having DM compared to patients carrying G/T genotype (OR: 5.7 95%CI: 1.1–29.1 p = 0.04).
Table 3.
Illustration of the three different genotypes of the eNOS (T-786C) polymorphism in diabetic patients and controls.
| Patients (n = 200) | Controls (100) | X2 | p | |
|---|---|---|---|---|
| T/T n (%) | 48 (24.0) | 33 (33.0) | 2.7 | 0.09 |
| T/C n (%) | 152 (76.0) | 67 (67.0) | ||
| C/C n (%) | 0 (0.0) | 0 (0.0) | ||
| C allele carrier n (%) | 152 (76.0) | 67 (67.0) | 2.2 | 0.12 |
Table 4.
Illustration of the three different genotypes of the eNOS (G894T) polymorphism in diabetic patients and controls.
| Patients (n = 200) | Controls (100) | X2 | p | |
|---|---|---|---|---|
| G/G n (%) | 122 (61.0) | 46 (46.0) | 13.3 | 0.001** |
| G/T n (%) | 64 (32.0) | 52 (52.0) | ||
| T/T n (%) | 14 (7) | 2 (2) | ||
| T allele carrier n (%) | 78 (39.0) | 54 (54) | 5.9 | 0.01* |
p highly significant.
p highly significant.
3.2. Distribution of eNOS (T-786C) and eNOS (G894T) in type 2 diabetes with and without nephropathy and controls
We assessed if there is any relation between eNOS (T-786C), eNOS (G894T) and DN, and we found higher prevalence of TT genotype of eNOS (G894T) in the 3 groups of diabetic patients than in controls and the highest prevalence was in patients with microalbuminuria, however there was no significant relation between eNOS (T-786C) and DN (Table 5).
Table 5.
Distribution of T-786C and G894T polymorphisms of eNOS gene in the 3 groups of the diabetic patients and controls.
| Genotypes n (%) | Normoalbuminuria n = 37 |
Microalbuminuria n = 96 |
Macroalbuminuria n = 67 |
Controls n = 100 |
X2 | p |
|---|---|---|---|---|---|---|
| T-786C | ||||||
| TT | 11 (29.7%) | 21 (21.9%) | 16 (23.9%) | 33 (33%) | 6.5 | 0.3 |
| TC | 26 (70.3%) | 75 (78.1%) | 51 (76.1%) | 67 (67%) | ||
| CC | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||
| C allele carrier | 26 (70.3%) | 75 (78.1%) | 51 (76.1%) | 67 (67%) | ||
| G894T | ||||||
| GG | 16 (45.9%) | 57 (59.4%) | 48 (71.6%) | 46 (46%) | 20.3 | 0.002* |
| GT | 18 (48.6%) | 31 (32.3%) | 15 (22.4%) | 52 (52%) | ||
| TT | 2 (5.4%) | 8 (8.3%) | 4 (6%) | 2 (2%) | ||
| T allele carrier | 20 (54.1%) | 39 (40.6%) | 19 (28.4%) | 54 (54%) | 12.6 | 0.005* |
4. Discussion
The prevalence of DN among type 2 diabetic patients varies between different ethnic populations which could be attributed to genetic background. The eNOS gene has been reported to be associated with DN in some ethnic populations [6].
In the present study, we found association of TT genotype of G894T with type 2 DM and no significant association with DN. No association was found between eNOS T786C and DM or DN. The potential association of eNOS genes and diabetes revealed contradictory results. Thameem and colleagues reported association of eNOS T-786C gene with type 2 diabetes in Mexican American patients [13]. Among Japanese patients, Ohtoshi and colleagues [14] reported no differences in allele frequencies of eNOS T786C gene between type 2 diabetic patients and nondiabetics. However, eNOS G894T polymorphism was found to increase risk of type 2 DM among Chinese individuals with impaired glucose tolerance [15]. On the other hand, several studies reported association of eNOS T786C and G894T variant with DN [7], [16], [17], [18].
Moreover in Iranian study, GT genotype of eNOS G894T was associated with increased risk of microalbuminuria and macroalbuminuria [19].
Most meta analysis studies revealed association of G894T polymorphisms in the eNOS gene with susceptibility to DN in Asian populations, but not in Caucasian populations [6], [20], [21].
However in one of these meta analysis, the control group consisted of healthy nondiabetic subjects and notdiabetic patients without nephropathy. Therefore, the association of the polymorphisms may be with DM itself not DN [20].
The CC genotype of T786C variant was not recorded in our studied population which is different from other populations studied. Tanus-Santos and colleagues [22] examined the distribution of T786C variant of eNOS gene in a sample of different ethnic individuals and they found that Caucasians had higher prevalence (42.0%) than Africans Americans (18%) than Asians (14%).
The absence of association between T786C and G894T polymorphisms with DN in the present study might be due to the presence of other non studied functional mutations in the eNOS gene that had not been assessed in the present study such as 4b/a polymorphism. It has been reported that the interaction of these variants within haplotypes may be the major determinant of disease susceptibility instead of single polymorphism [23].
Moreover other risk factors may affect the development of DN such as duration of diabetes, control of diabetes, smoking and BMI [24]. The mechanism responsible for the potential association between eNOS polymorphisms and risk of DN is not yet fully elucidated. Variants of eNOS gene may lead to decrease nitric oxide levels due to defective synthesis so increasing the susceptibility to glomerular disease and deteriorating the renal function [17]. However, changes in eNOS expression do not always correlate with actual NO synthesis because NO synthesis by eNOS depends on the availability of adequate substrate and cofactors [25]. Moreover, decreased synthesis of eNO may be due to accumulation of inhibitors of eNOS such as asymmetric dimethylarginine (ADMA) [26].
Endothelial NOS polymorphism has been implicated in the risk of type 2 diabetes. In the present study, we found association of TT genotype of eNOS G894T variant with type 2 diabetes, however T allele frequency did not reveal significant difference between patients and controls which may assume a recessive mode of inheritance. A study conducted among southern Brazilian population found association between G894T eNOS gene polymorphism and features of the metabolic syndrome and assuming a recessive mode of inheritance [27]. Additionally, a positive association between G894T polymorphism and metabolic syndrome has been demonstrated in Chinese and Japanese populations [28], [29]. Another study conducted among Indian population demonstrated higher prevalence of the mutant genotypes (eNOS-894 GT/TT) in type 2 diabetic patients compared to healthy controls [2]. Similarly, Monti and colleagues [30] reported association of eNOS gene polymorphisms with type 2 diabetes.
Nitric oxide modulates peripheral and hepatic glucose metabolism and affects insulin secretion. Activation of eNOS especially that of skeletal muscle mitochondria increases muscle blood flow, with increased delivery of glucose, to the muscle cells. Thus, genetic defect in the eNOS might have a role in the occurrence of hyperinsulinemia and insulin resistance [10], [30].
To the best of our knowledge, this is the first study, to relate endothelial nitric oxide synthase eNOS G894T single-nucleotide polymorphism with type 2 diabetes mellitus patients among Egyptians.
5. Conclusion
The result of the study suggested that eNOS G894T variant may increase risk of type 2 diabetes with lack of association between eNOS T786C, eNOS G894T and DN in Egyptians. We need further studies with larger number of patients and assessing intron 4b/a variant of eNOS gene and correlating the polymorphisms with nitric oxide levels.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Reddy Marpadga A., Natarajan Rama. Cardiovasc. Res. 2011;90:421–429. doi: 10.1093/cvr/cvr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.T. Angeline, H.R. Krithiga, W. Isabel, A.J. Asirvatham, A. Poornima1, Oxidative Med. Cell. Longevity 2011 (2011) 4 (Article ID 462607). http://dx.doi.org/10.1155/2011/462607. [DOI] [PMC free article] [PubMed]
- 3.Gray S.P., Cooper M.E. Nat. Rev. Nephrol. 2011;7:71–73. doi: 10.1038/nrneph.2010.176. [DOI] [PubMed] [Google Scholar]
- 4.Cooper M.E. Diabetologia. 2001;44(11):1957–1972. doi: 10.1007/s001250100000. [DOI] [PubMed] [Google Scholar]
- 5.Mooyaart A.L., Valk E.J., van Es L.A., Bruijn J.A., deHeer E., Freedman B.I., ekkers O.M., Baelde H.J. Diabetologica. 2011;54:544–553. doi: 10.1007/s00125-010-1996-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hey Y., Fany Z., Zhang J., Zhang Q., Zheng M., Li Y., Zhang D., Gu1 S., Yang H. Mutagenesis. 2011;26(2):339–349. doi: 10.1093/mutage/geq100. [DOI] [PubMed] [Google Scholar]
- 7.Ahluwalia T.S., Ahuja M., Rai T.S., Sud K.K., Khullar B.M. Mol. Cell. Biochem. 2008;314:9–17. doi: 10.1007/s11010-008-9759-8. [DOI] [PubMed] [Google Scholar]
- 8.Tesauro M., Thompson W.C., Rogliani P., Qi L., Chaudhary P.P., Moss J. Proc. Natl. Acad. Sci. USA. 2000;97:2832–2835. doi: 10.1073/pnas.97.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamblin N., Cuilleret F.J., Helbecque N., Dallongeville J., Lablanche J.M., Amouyel P., Bauters C., Van Belle E. BMC Cardiovasc. Disord. 2005;5:27. doi: 10.1186/1471-2261-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahimia Z., Rahimia Z., Shahvaisi-Zadeha F., Sadegheic S., Vessalc M., Yavaria N. Dis. Markers. 2013;34:437–443. doi: 10.3233/DMA-130988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Diabetes Association, Diabetes Care 27 (Suppl. 1) (2004) S79–S83. [DOI] [PubMed]
- 12.Cruz-González I., Corral E., Sánchez-Ledesma M., Sánchez-Rodríguez A., Martín-Luengo C., González-Sarmiento R. BMC Cardiovasc. Disord. 2009;2009(9):35. doi: 10.1186/1471-2261-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thameem F., Puppala S., Arar N.H., Stern M.P., Blangero J., Duggirala R., Abboud H.E. Diab. Vasc. Dis. Res. 2008;5(2):109–113. doi: 10.3132/dvdr.2008.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohtoshi K.1., Yamasaki Y., Gorogawa S., Hayaishi-Okano R., Node K., Matsuhisa M., Kajimoto Y., Hori M. Diabetologia. 2002;45(11):1594–1601. doi: 10.1007/s00125-002-0922-6. [DOI] [PubMed] [Google Scholar]
- 15.Tso A.W., Tan K.C., Wat N.M., Janus E.D., Lam T.H., Lam K.S. Metabolism. 2006;55(9):1155–1158. doi: 10.1016/j.metabol.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Ezzidi I., Mtiraoui N., Hadj Mohamed M.B., Mahjoub T., Kacem M., Almawi M.Y. J. Diabetes Complications. 2008;22:331–338. doi: 10.1016/j.jdiacomp.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Shin Y.S., Baek S.H., Chang K.Y. Diabetes Res. Clin. Pract. 2004;65:257–265. doi: 10.1016/j.diabres.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Shoukry A., Shalaby S.M., Abdelazim S., Abdelazim M., Ramadan A., Ismail M.I., Fouad M. Genet. Test Mol. Biomarkers. 2012;16:574–579. doi: 10.1089/gtmb.2011.0218. 18. [DOI] [PubMed] [Google Scholar]
- 19.Jafari Y., Rahimi Z., Vaisi-Raygani A., Rezaei M. Mol. Cell. Biochem. 2011;353:23–34. doi: 10.1007/s11010-011-0770-0. [DOI] [PubMed] [Google Scholar]
- 20.Zintzaras E., Papathanasiou A.A., Stefanidis I. Genet. Med. 2009;11(10):695–706. doi: 10.1097/GIM.0b013e3181b2046b. [DOI] [PubMed] [Google Scholar]
- 21.Zeng Z., Li L., Zhang Z., Li Y., Wei Z., Huang K., He L., Shi Y. Hum. Genet. 2010;127(4):373–381. doi: 10.1007/s00439-009-0783-x. [DOI] [PubMed] [Google Scholar]
- 22.Tanus-Santos J.E., Desai M., Flockhart D.A. Pharmacogenetic. 2001;11:719–725. doi: 10.1097/00008571-200111000-00011. [DOI] [PubMed] [Google Scholar]
- 23.Zintzaras E., Kitsios G., Stefanidis I. Hypertension. 2006;48:700–710. doi: 10.1161/01.HYP.0000238124.91161.02. [DOI] [PubMed] [Google Scholar]
- 24.Passaro A., Calzoni F., Volpato S. J. Intern. Med. 2003;254:264–271. doi: 10.1046/j.1365-2796.2003.01184.x. [DOI] [PubMed] [Google Scholar]
- 25.Welch W.J., Wilcox C.S. J. Clin. Invest. 1997;100(9):2235–2242. doi: 10.1172/JCI119761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fliser D. J. Nephrol. 2010;23(4):369–376. [PubMed] [Google Scholar]
- 27.Piccoli J.C., Gottlieb M.G.V., Castro L., Bodanese L.C., Manenti E.R.F., Bogo M.R., Peres A., Rocha M.I.U.M., Cruz I.B.M. Arq. Bras. Endocrinol. Metabol. 2008;52(8):1367–1373. doi: 10.1590/s0004-27302008000800027. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh M.C., Hsiao J.Y., Tien K.J., Chang S.J., Lin P.C., Hsu S.C., Liang H.T., Chen H.C., Lin S.R. Metabolism. 2008;57(8):1125–1129. doi: 10.1016/j.metabol.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Imamura A., Takahashi R., Murakami R., Kataoka H., Cheng X.W., Numaguchi Y., Murohara T., Okumura K. Eur. J. Endocrinol. 2008;158(2):189–195. doi: 10.1530/EJE-07-0632. [DOI] [PubMed] [Google Scholar]
- 30.Monti L.D., Barlassina C., Citterio L., Galluccio E., Berzuini C., Setola E., Valsecchi G., Lucoti P., Pozza G., Bernardinelli L., Casari G., Piatti P. Diabetes. 2003;52(5):1270–1275. doi: 10.2337/diabetes.52.5.1270. [DOI] [PubMed] [Google Scholar]