Abstract
Phosphate solubilizing bacteria (PSB) has ability to convert insoluble form of phosphorous to an available form. Applications of PSB as inoculants increase the phosphorus uptake by plant in the agriculture field. In this study, isolation and identification of PSB were carried out in Indian agriculture field (Nainital region, Uttarakhand). A total of 8 phosphate solubilizing bacterial colonies were isolated on the Pikovskaya’s (PKV) agar medium, containing insoluble tricalcium phosphate (TCP). The colonies showed clear halo zones around the bacterial growth were considered as phosphate solubilizers. Out of 8 bacterial isolates, 3 isolates showed high phosphate solubilization index (PSI) ranged from 4.88 ± 0.69 to 4.48 ± 0.30, lower pH ranging 3.08 ± 0.08 to 3.82 ± 0.12 and high phosphate solubilization varied from 305.49 ± 10 μg/ml to 277.72 ± 1.45 μg/ml, were selected for further characterization. Based on the 16 S rRNA gene sequence analysis A4 isolate and H6 isolate were closely related to Alcaligenes aquatilis (99%), and C1 isolate was closely related to Burkholderia cepacia (99%). In addition, pot examination also showed the greatest efficiency in promotion of maize growth compared to uninoculated plant. Isolated PSB were able to produce different organic acids (such as gluconic acids, formic acid, and citric acid) in the culture supernatant and may consider as the principle mechanism for phosphate solubilization. This study clearly indicates that A4, C1 and H6 isolates may use as a biofertilizers in ecological agricultural systems instead of synthetic chemicals and may help to sustain environmental health and soil productivity.
Keywords: Phosphorus, Phosphate solubilizing bacteria (PSB), Pikovskaya’s, Phosphate solubilization index, Tricalcium phosphate
1. Introduction
Phosphorus (P) is one of the major essential macronutrients of plants which regulates protein synthesis and plays an important role in biological development. Along with these essential functions, P is also associated with complex signal transduction, macromolecular biosynthesis, energy transformations, respiration and nitrogen fixation in legumes in the plant [1]. Most of the P (95–99%) present in the soil is part of insoluble compound and hence cannot be utilized by plants [2]. Since P is a stable element in soils, it does not form a gas (such as ammonia), therefore cannot move far from where it is applied. The reason for the stability of phosphate compounds in soils is that they are highly reactive and reacts rapidly with other compounds (such as Al3+, Ca2+ and Fe3+), which become increasingly insoluble in the soil. Therefore, the release and mobilization of insoluble and fixed forms of phosphorus is an important aspect of increasing soil phosphorus availability. To overcome this problem, most of the farmers regularly use chemical phosphate fertilizers which get incorporated into the soil. This applied phosphorus easily transforms into an insoluble and stable form with limited availability to plants and only 5% or less of the total amount of P in soil is available for plant nutrition [3], [4]. Due to the negative environmental impacts of chemical fertilizers and their increasing costs, the use of PSB is advantageous in the sustainable agricultural practices. The use of microbial inoculants possessing P-solubilizing activities in soils is considered as an environmental-friendly alternative to further applications of chemical based P fertilizers [5]. Microbial intervention of PSB seems to be an effective way to enhance the phosphorus availability in soil.
The main mechanism of phosphate solubilization is the production of some organic acids. Among the organic acids produced, gluconic, formic acid, 2-ketogluconic, citric, oxalic, lactic, isovaleric, succinic, glycolic and acetic acids produced from P- solubilizing bacteria. Production of these organic acids results in the lowering of pH in the surroundings. The lowering in pH of the medium suggests the release of organic acids by the P-solubilizing microorganisms via the direct oxidation pathway that occurs on the outer face of the cytoplasmic membrane [6], [7]. These acids are the product of the microbial metabolism [8]. Many reports suggest a positive correlation between lowering of pH and soluble P concentration in the medium. Some of the alternate mechanisms suggested are production of chelating compounds, inorganic acids sulfuric, nitric and carbonic acids. There are several reports of phytase producing organism. Richardson AE (1997) reported insoluble P solubilization by secreting phytase enzyme in Pseudomonas sp. [9]. Several species of microorganisms are able to secrete phytohormones such as auxins, gibberellins, cytokinins, and nitric oxide which directly involved in plant growth and productivity [10], [11]. Microorganisms such as Alcaligenes sp., Aerobactor aerogenes, Achromobacter sp., Actinomadura oligospora, Burkholderia sp., Pseudomonas sp., Bacillus sp. and Rhizobium sp. are the most important phosphate solubilizers in soils. Apart from bacterial sp. some fungal stains are also reported as phosphate solubilizers such as Aspergillus sp. (A. spergillus awamori, A. niger, A. tereus, A. flavus, A. nidulans, A. foetidus, A. wentii.) and Penicillium sp. (Penicillium digitatum, P lilacinium, P balaji, and P. funicolosum). Since PSM are more suitable for high-volume production of crop, the objective of this study was to isolate the phosphate-solubilizing bacteria from rhizosphere of maize plants, to detect the phosphate-solubilizing ability, to examine the production of organic acids and to characterize the microorganisms at the phenotypic and genotypic level. In addition, this study was to examine the effect of phosphate-solubilizing bacteria as inoculants on plant growth.
2. Materials and methods
2.1. Sample site and collection
Soil samples were collected from eight different areas of Nainital region (29°23′N 79°27′E/29.38°N 79.45°E/29.38; 79.45.) of Uttarakhand, India. The plants were dug out, the excess bulk soil was removed by gently shaking, and the soil adhering the root was considered rhizosphere soil [12] and collected in sterilized plastic bags.
2.2. Isolation of phosphate solubilizing bacteria
One gram (1 g) of each soil sample was dispersed in 9 ml of autoclaved distilled water, filtered by 125 mm Whatman® qualitative filter paper, Grade 1. One ml of the above filtered solution was again transferred to 9 ml of sterile distilled water to form 10−2 dilution. Similarly 10−3, 10−4, 10−5, 10−6, 10−7 and 10−8 serials were made for each soil sample [13]. The serially diluted (10−4, 10−5 and 10−6) soil samples were plated (0.1 ml) on the Pikovskaya’s agar containing tricalcium phosphate (TCP) as the phosphate source [14]. The Pikovskaya’s medium consisted of yeast extract 0.50 (g/l), dextrose 10.00 (g/l), calcium phosphate 5.00 (g/l), ammonium sulfate 0.50 (g/l), potassium chloride 0.20 (g/l), magnesium sulfate 0.10 (g/l), manganese sulfate 0.0001 (g/l), ferrous sulfate 0.0001 (g/l), agar 15 (g/l) and dissolved in 1000 mL distilled water. The pH of the media was adjusted to 7.0 before autoclaving at 15 lbs pressure (121 °C) for 15 min. Mix well and pour into sterile Petri plates (25 ml/plate) under laminar flow hood and allowed to solidify. Plates were incubated in inverted position in incubator for up to 7 days at 27–30 °C and colonies with a clear halo were marked positive for phosphates solubilization and were considered as PSB. These selected colonies were again subcultured (2–3 times) by striking method till the pure cultures were obtained on the same PVK media when grown at 30 °C. Isolated bacteria were kept on PVK agar slant at 4 °C for further study.
2.3. Morphological characterization
The bacterial species form characteristic colonies on Pikovskaya's Agar Media. Morphology of the isolates was studied by Gram staining using kit (K001-1KT, Hi Media) by the standard procedure [15]. The stained cells were observed under compound microscope. The Gram reaction and cell morphology for efficient PSB strains were recorded. Motility of each bacterial strain was checked using HiMedia MIU medium (SL042). Bacterial strain with positive growth away from stabline causing turbidity was considered as motile.
2.4. Phenotypic identification of bacteria
Isolates were also tested for Indole test, Urease test, Catalase test, Voges-Proskauer (VP) test, Methyl Red (MR) test and Citrate test [16]. The phenotypic properties of isolates were determined as described in Bergey’s manual of systematic bacteriology [17].
2.5. Analysis of phosphate solubilizing activity
The qualitative as well as quantitative analysis of phosphate solubilizing activity of the selected isolates was conducted by plate screening method and broth culture method, respectively.
2.5.1. Qualitative measurement of phosphate solubilization
Bacterial isolates were screened for their TCP solubilizing activity on PKV plates. Isolates were spot inoculated on the center of agar plate aseptically. All the plates were incubated at 28 °C ± 2 °C for 7 days. A clear zone around a growing colony indicated phosphate solubilization and was measured as phosphate solubilization index (SI). SI was calculated as the ratio of the total diameter (colony + halo zone) to the colony diameter [18].
All the observations were recorded in triplicate. Strains developing clear zones around their colonies could easily be identified as PSBs.
2.5.2. Quantitative measurement of phosphate solubilization
Quantitative estimation of solubilized P by bacterial isolate was done by the vanadomolybdophosphoric yellow color method [19] in pikovskaya’s broth containing 5000 µg/ml tricalcium phosphate.
The phosphate solubilizing ability of each isolate was tested using insoluble tricalcium phosphate [Ca3(PO4)2] as sole P source in Pikovskaya’s medium. 10 ml of pikovskaya’s broth containing 5000 µg/ml P in the form of tricalcium phosphate was inoculated with 0.1 ml of bacterial culture (inoculum adjusted to ∼2 × 108 CFU/ml) at 28 °C ± 2.0 °C up to 8 days. After incubation, 1 ml of the supernatant was taken out on 2nd, 4th, 6th and 8th day.
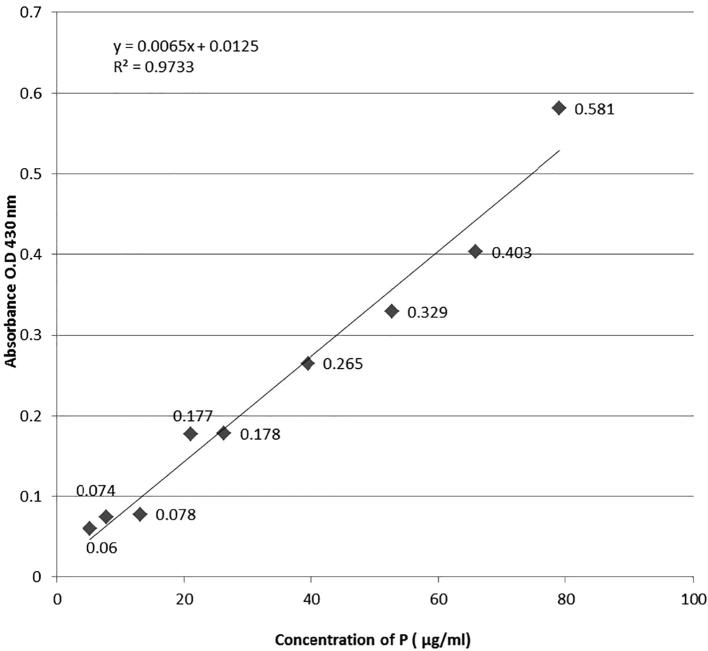
The supernatant was obtained by centrifugation at 10,000 rpm for 20 min and was passed through a 0.45 µM millipore filter and then 0.1 ml of the supernatant (filtered) was mixed with 0.25 ml of Barton’s reagent and volume was made up to 5 ml with double distilled water (ddw). After 10 min, the intensity of yellow color was read on spectrophotometer (UV–VIS Spectrophotometer-SL-159, Elico, India) at 430 nm and the amount of P-solubilized was extrapolated from the standard curve. The experiments were conducted in triplicates and values were expressed as their mean.
A standard curve was prepared by dissolving 0.02195 gm of potassium dihydrogen orthophosphate/Monopotassium phosphate (dried at 60 °C for 1 h and then cooled in desiccators) in 100 ml of double distilled water (ddw) and labeled as stock P solution. A further dilution of 15 ml of stock solution was taken and volume was made up to 25 ml with ddw and labeled as working solution. Aliquots of 0.2, 0.3, 0.5, 0.8, 1.0, 1.5, 2.0, 2.5, 3.0 ml of working solution and 0.25 ml of Barton’s reagent were added to each flask and the volume was made up to 5 ml with distilled water. After 10 min, the intensity of yellow color developed was read at 430 nm spectrophotometrically. Standard curve was prepared by plotting absorbance at 430 nm vs concentration of P.
2.5.3. Quantitative measurement of pH of the media
Initial pH and change in pH were also recorded on same interval (2nd, 4th, 6th and 8th day) by digital pH meter (OAKTON, pH 700). The experiments were conducted in triplicates and values were expressed as their mean.
2.6. Pot experiment
The experiment was carried out in a greenhouse during May–June 2016 with four replications. A pot experiment consisted of one control (non-inoculated with PSB) and 3 PSB species. Test was conducted in pots (5 cm in diameter) containing 500 gm of soil. A 15 cm depth of soil was used from the agriculture field. Unsterile soil (silt loam, pH 7.5–8.0 mg/100 g; total organic C, 0.10%; available N, 36.8 mg/100 g; available P, 5 mg/100 g; available K, 12 mg/100 g; total Ca, 39.816 mg/100 g; total Na 22 mg/100 g, total Mg, 40.1 mg/100 g) was thoroughly mixed, passed through a 2 mm sieve and dried in sunlight for 7 days, and then sterilized three times by dry heat treatment for 2 h. at 121 °C. Tricalcium phosphate (TCP) was supplied as soil P fertilizer at the rate of 160 mg kg−1 based on the nutrient requirements of maize.
For testing the efficiency of isolated phosphate solubilizing bacteria, its effect was observed on growth of maize plant in vitro. Seeds were surface sterilized with sodium hypochlorite (2%) and alcohol (70%) for 1 min. Then seeds were washed with autoclaved distilled H2O to remove small amount of chemicals. The broth culture of each 3 isolates (single colonies) was made in Pikovskaya’s medium. For this purpose, PSB suspensions were prepared by culturing on Pikovskaya’s media and incubating on an orbital shaker at 160 rpm, 30 °C, for 48 h. Then bacterial cells were harvested by centrifugation for 5 min at 13,081g (10,000 rpm), 21 °C, in RAMI centrifuge, and washed with sterile distilled water. The cell pellet was resuspended with sterile distilled water, and then cells were further adjusted to 2.0 × 108 colony forming units (CFU)/ml (at O.D 430). After making the broth culture, the equal numbers of seeds of maize were soaked in each broth culture separately for 12 hrs at 28 °C. The equal number (4 seeds in each pot) of seed was potted in the sterile soil. Previously prepared 2 mL of broth culture was uniformly applied on seeds and then again covered with uniform 3 cm layer of soil. One pot was kept as control in which the seeds were potted in sterile soil without soaking in any culture of phosphate solubilizing bacteria. Pots were watered daily to maintain soil field capacity during the study period. Agronomic variables (shoot and root length and dry weigh) were evaluated 20 days after inoculation. At the end of the study, plant growth parameters – such as% of Seed germination, shoot length, root length, shoot dry matter, root dry matter and number of leaves were calculated.
2.7. Analysis of organic acids
HPLC reverse-phase chromatography was used for the analysis of organic acids produced by PSB strains in broth medium. On 8th day, supernatant was taken from bacterial cultures that had been centrifuged at 13,000 rpm for 15 min. Samples were filtered through a 0.45 µM Millipore filter. 20 μl of filtrates was then injected into an HPLC column (Symmetry, C-18, 250 mm × 4.6 mm, 5 μm) using a glass syringe. The operating conditions consisted of 20 µM KH2PO4 and methanol (60:40v/v) were used as mobile phase at a constant flow rate of 1 ml/min and the column was operated at 30 °C. Retention time (RT) of each signal was recorded at a wavelength of 256 nm. The software used for HPLC analyses was the chemstation. HPLC profiles of the culture filtrates were analyzed by comparison with the elution profiles of pure organic acids (Bio-Rad Standard containing citric acid, gluconic acid, formic acid, pyruvic acid and oxalic acid) and the peak areas of their standards.
2.8. Genomic DNA isolation, PCR amplification and sequencing of 16S rDNA gene
Total genomic DNA was extracted by CTAB method. After DNA extraction, PCR reaction was performed in a veriti thermal cycler (Thermo Fisher Scientific). The universal primers (27F Forward primer 5′-AGAGTTTGATCCTGGCTCAG-3′ and 939r reverse primer 5′-CTTGTGCGGGCCCCCGTCAATTC-3′) were used for the amplification of the 16S rDNA gene fragment. PCR amplifications were carried out in 50 μl PCR reaction mixture consisted of 3 μl template DNA (100 ng – 1.0 μg), 25 μl 2X KAPA Taq ready mix [containing dNTPs (0.2 mM of each dNTP at 1X), MgCl2 (1.5 mM at 1X)], and 1 μl of both forward and reverse primers. The amplification cycle consisted of an initial denaturation step of 3 min at 93 °C, followed by 35 cycles of 30 sec at 95 °C (denaturation), 30 sec at 60 °C (annealing) and 30 sec at 72 °C (extension), with a final extension step for 3 min at 72 °C. PCR products were electrophoresed using 1.5% agarose stained with ethidium bromide (0.5 μg/ml), and visualized using a Gel luminax 312. PCR products of isolates were purified using PCR purification kit (Qiagen, Germany) according to the manufacturer's instructions and the amplified product was sequenced on Sanger sequencing platform at Pragati Biomedical, New Delhi for sequencing. All the bacterial isolates were classified by BLAST analysis of their respective 16S rRNA gene partial sequences. Various databases including eztaxon, NCBI and Ribosomal Database Classifier (RDC) were used to determine the exact taxonomical classification of individual organisms [20]. For the determination of closest type strains NCBI Blast was used [21].
2.9. Phylogenetic analysis
Sequence data were compared visually and sequences were aligned using the Clustal W software and distances were calculated according to Kimura's two-parameter method [22]. Phylogenetic trees were produced using the neighbor-joining method. Bootstrap analysis was based on 1000 resamplings. The MEGA (Molecular Evolutionary Genetics analysis) 6.05 package was used for all phylogenetic analysis [23]. The final sequence was submitted to GenBank [24].
2.10. Statistical analysis
Three/four replicates were used for each experiment. The data were analyzed by analysis of variance (ANOVA) and the means were compared with Tukey’s test at P < 0.05. In the field experiment the plots with different treatments were arranged in a randomized complete block design with three replicates per treatment.
3. Results
3.1. Isolation and identification of phosphate solubilizing bacteria (PSB)
In the present study, the collected soil samples were evaluated in vitro for P solubilizing bacteria in Pikovskaya’s (PKV) plates supplemented with 1.5% (w/v) agar. A total of 8 phosphate solubilizing bacterial colonies were isolated on PVK agar medium, containing tricalcium phosphate (TCP). Out of 8 bacterial isolates, 3 isolates (A4, C1 and H6) were found to be potent phosphate solubilizers showing clear halo zone around its colony. The halo zone formation around the bacterial colonies could be due to the production of organic acids or due to the production of polysaccharides or due to the activity of phosphatase enzymes of phosphate solubilizing bacterial strains [25], [26], [27], [28].
3.2. Analysis of phosphate solubilizing activity
3.2.1. Qualitative measurement of phosphate solubilization
Qualitative P-solubilization potential was anticipated by observing the large clear/halo zones around the bacterial colonies on Pikovskaya agar media and PSI was calculated. 3 isolates were found to have ≥4.0 phosphate solubilizing index (PSI). Maximum PSI was observed by C1 (4.88 ± 0.69) followed by H6 (4.64 ± 1.12) and A4 (4.48 ± 0.30) (Table 1).
Table 1.
Qualitative estimation of phosphate solubilization efficiency of A4, C1 and H6.
| Bacterial isolates | Colony diameter (cm) | Halo zone diameter (zone of solubilization in cm) | Phosphate solubilization index (PSI) |
|---|---|---|---|
| A4 | 1.53 ± 0.11 | 5.33 ± 0.25 | 4.48 ± 0.30 |
| C1 | 1.7 ± 0.26 | 6.46 ± 0.49 | 4.88 ± 0.69 |
| H6 | 1.73 ± 0.37 | 5.96 ± 0.72 | 4.64 ± 1.12 |
3.2.2. Quantitative measurement of phosphate solubilization
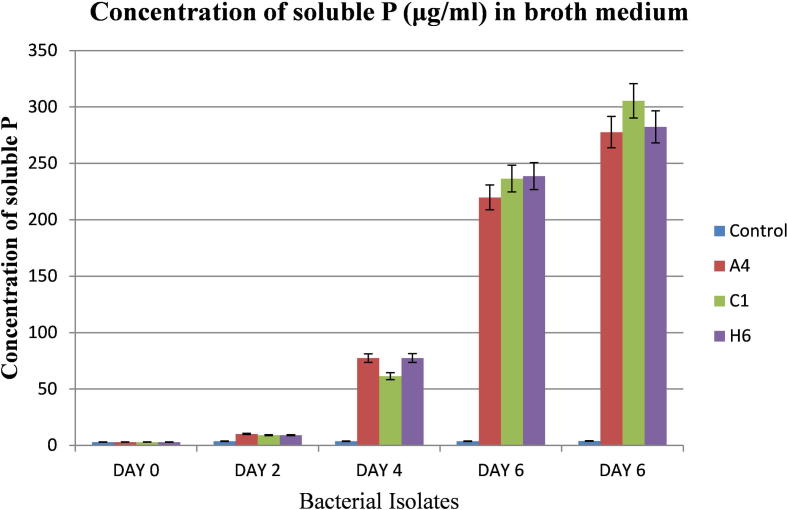
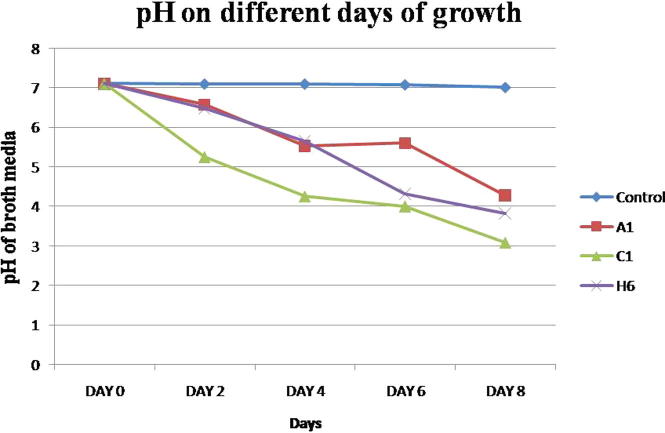
As shown in Fig. 1, Ca3(PO4)2 containing medium solubilization is slow in the first two days and then becomes fast, reaching the highest (305.49 ± 10.7281 μg/ml) in 8th day and after 8th day no further concentration of soluble P change was seen (data not presented). The amount of P-solubilized was extrapolated from the standard curve (Fig. 2). Maximum P solubilization was observed by C1 (305.49 ± 10.72 μg/ml) followed by H6 (282.38 ± 11.81 μg/ml) and A4 (277.72 ± 1.45 μg/ml). It was also found that the growth of the isolate caused a significant increase of acidity in Ca3(PO4)2 containing medium. In 8th days, pH in the culture medium declined to 3.08 ± 0.08 from an initial pH of 7.11 (Fig. 3) and after 8th day, no further pH change was seen (data not presented). Minimum pH was observed by C1 (3.08 ± 0.08) followed by A4 (3.36 ± 0.11) and H6 (3.82 ± 0.12).
Figure 1.
Phosphate solubilizing activity of A4, C1 and H6 isolates during 8 days of incubation.
Figure 2.
Standard curve.
Figure 3.
Change in pH by 3 efficient phosphate solubilizing bacterial stain in liquid broth media during 8 days of incubation.
3.3. Morphological characterization of PSB
Bacterial species were further examined for their Gram's reaction and shape. Characteristically, all the isolates were Gram negative and of rod shaped with flagellum, motile and most of were shining. The results are summarized in Table 2.
Table 2.
Morphological characterization and biochemical characterization of A4, C1 and H6 isolates.
| Bacterial isolates | Cell shape | Gram staining | Motility test | Indole test | Urease test | Catalase test | Voges-proskauer (VP) test | Methyl red (MR) test | Citrate test |
|---|---|---|---|---|---|---|---|---|---|
| A4 | Rod shaped, Larger, Single flagellum, Shining, Yellowish colored, Smooth | − | Motile | + | + | + | + | − | + |
| C1 | Rod shaped, Small, Single flagellum, Shining, Cream colored, Smooth | − | Motile | − | + | + | + | − | + |
| H6 | Rod shaped, Larger, Single flagellum, Shining, Yellowish colored, Smooth | − | Motile | + | + | + | + | − | + |
3.4. Biochemical characterization of PSB
These isolates were further characterized by a series of biochemical reactions and identified as genus Burkholderia and Alcaligenes. These bacteria were well known identified as phosphate solubilizers in many studies. [29], [30], [31], [32]. All 3 bacteria have shown positivity for urease, catalase test, Voges-Proskauer (VP) test and citrate test. Bacteria isolate C1 was negative for indole test and Methyl Red (MR) test. However, bacteria isolate A4 and H6 were negative for Methyl Red (MR) test. The results are summarized in Table 2.
3.5. Pot examination
For determination of plant parameters, all maize plants were harvested after 20 days and the growth parameters, such as shoots and roots height, fresh and dry weight and number of leaves and leaves length were recorded. The shoots were separated from the roots at 0.5 cm above the surface of soil. The roots were washed out with tap water to remove the soil particles. The dry weight of roots and shoots was measured after drying in an oven for 48 h at 60 °C and compared with control plant. A4 isolates showed shoots height 5.75 ± 0.46 cm, shoots fresh weight 1.07 ± 0.23 gm, shoots dry weight 0.46 ± 0.01 gm, roots height 12.9 ± 1.8 cm, roots fresh weight 0.52 ± 0.17 cm and roots dry weight 0.25 ± 0.02 cm and leaves height 11.15 ± 0.94 cm. C1 isolates showed shoots height 6.5 ± 0.58 cm, shoots fresh weight 1.09 ± 0.41 cm, shoots dry weight 0.48 ± 0.01 cm, roots height 13.875 ± 1.6 cm, roots fresh weight 0.61 ± 0.04 gm and roots dry weight 0.52 ± 0.12 gm and leaves height 13.075 ± 0.62 cm. H6 isolates showed shoots height 6.15 ± 0.5 cm, shoots fresh weight 1.04 ± 0.1 gm, shoots dry weight 0.43 ± 0.17 gm, roots height 13.27 ± 1.34 cm, roots fresh weight 0.54 ± 0.05 gm and roots dry weight 0.26 ± 0.04 gm and leaves height 12.35 ± 0.51 gm. After comparing, we found that there was significant difference (p < 0.05) between treated seed and without treated seed (control). The results are summarized in Table 3.
Table 3.
Effect of phosphate solubilizing bacteria (A4, C1 and H6) on plant growth.
| Seed weight (g) | % Of seed germination | Shoot |
Root |
Number of leaves | Maximum leaves length (cm) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Height (cm) | Fresh weight (g) | Dry weight (g) | Maximum length (cm) | Fresh weight (g) | Dry weight (g) | |||||
| Control | 0.36 ± 0.02 (a) | 100% | 3.4 ± 0.18 (a) | 0.58 ± 0.02 (a) | 0.20 ± 0.07 (a) | 7.4 ± 0.14 (a) | 0.31 ± 0.01 (a) | 0.13 ± 0.01 (a) | 3 | 5.85 ± 0.53 (a) |
| A4 | 0.33 ± 0.01 (a) | 100% | 5.75 ± 0.46 (b) | 1.07 ± 0.23 (b) | 0.46 ± 0.01 (b) | 12.9 ± 1.8 (b) | 0.52 ± 0.17 (b) | 0.25 ± 0.02 (b) | 3 | 11.15 ± 0.94 (b) |
| C1 | 0.35 ± 0.02 (a) | 100% | 6.5 ± 0.58 (b) | 1.09 ± 0.41 (b) | 0.48 ± 0.01 (b) | 13.875 ± 1.6 (b) | 0.61 ± 0.04 | 0.52 ± 0.12 | 3 | 13.075 ± 0.62 (b) |
| H6 | 0.34 ± 0.03 (a) | 100% | 6.15 ± 0.5 (b) | 1.04 ± 0.1 (b) | 0.43 ± 0.17 (b) | 13.27 ± 1.34 (b) | 0.54 ± 0.05 (b) | 0.26 ± 0.04 (b) | 3 | 12.35 ± 0.51 (b) |
Values are expressed as means ± standard deviation of four independent data. Data were analyzed by ANOVA and the means were compared using Tukey’s test at P < 0.05.
Mean values (mean ± S.D) sharing the same letter do not differ significantly at P ≤ 0.05.
3.6. Production of organic acid
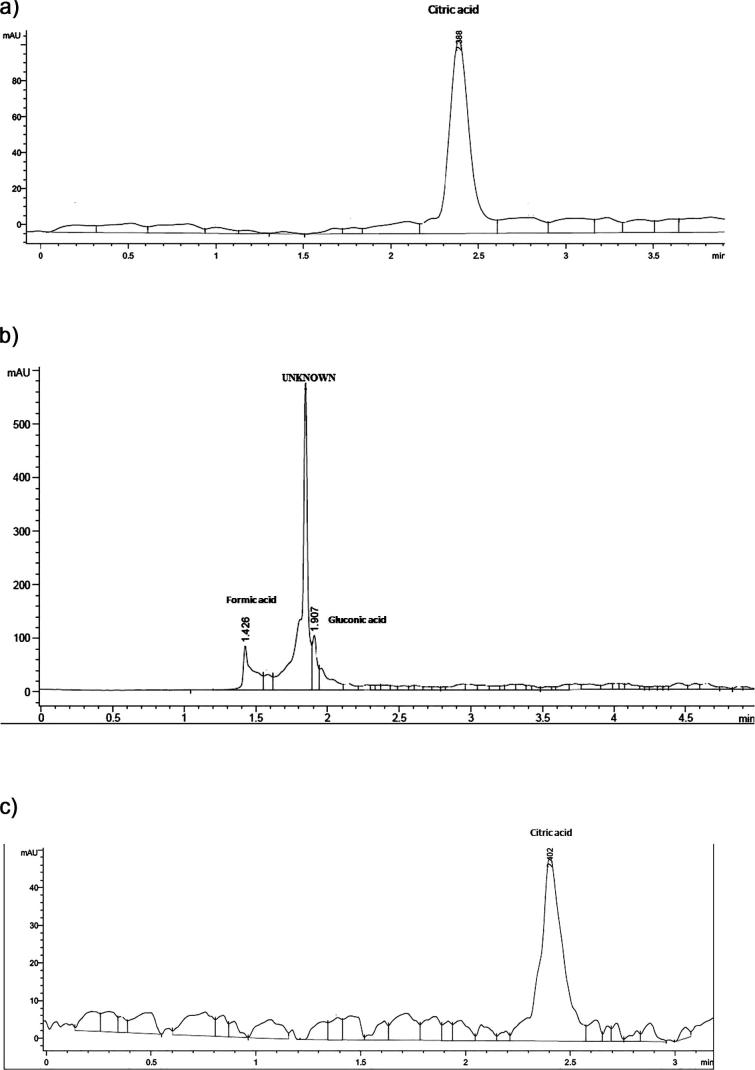
Analyses of organic acids production by HPLC are presented in the chromatogram in Fig. 4 and were compared with their standard. The results revealed that three different kinds of organic acids - gluconic acid, formic acid and citric acid along with unknown acid were produced from PSB isolates. C1 isolates showed the presence of two organic acids (gluconic acid and formic acid) and one unknown acid. Two isolates (A4 and H6) produce only one organic acid (citric acid). The results are summarized in Table 4.
Figure 4.
(a) HPLC chromatogram of organic acids produced from A4 isolates. The corresponding peak detected in culture medium was of citric acid. (b) HPLC chromatogram of organic acids produced from C1 isolate. The corresponding peaks detected in culture medium were of gluconic acid and formic acid including unknown peak. (c) HPLC chromatogram of organic acids produced from H6 isolates. The corresponding peak detected in culture medium was of citric acid.
Table 4.
Variety and quantity of organic acid produced by A4, C1 and H6 isolates.
| Sample | Name of organic acid | Retention time (Min) |
Amount of organic acid in culture filtrate (μg/ml) | pH of the liquid media | |
|---|---|---|---|---|---|
| Standard | Culture filtrate | ||||
| A4 | Citric acid | 2.39 | 2.38 | 1.68 | 3.36 |
| C1 | Gluconic acid | 1.89 | 1.90 | 0.803167052 | 3.1 |
| Formic acid | 1.49 | 1.42 | 7.48114555 | ||
| H6 | Citric acid | 2.39 | 2.40 | 0.682604544 | 3.45 |
3.7. Genomic DNA isolation, PCR amplification and sequencing of 16S rDNA gene
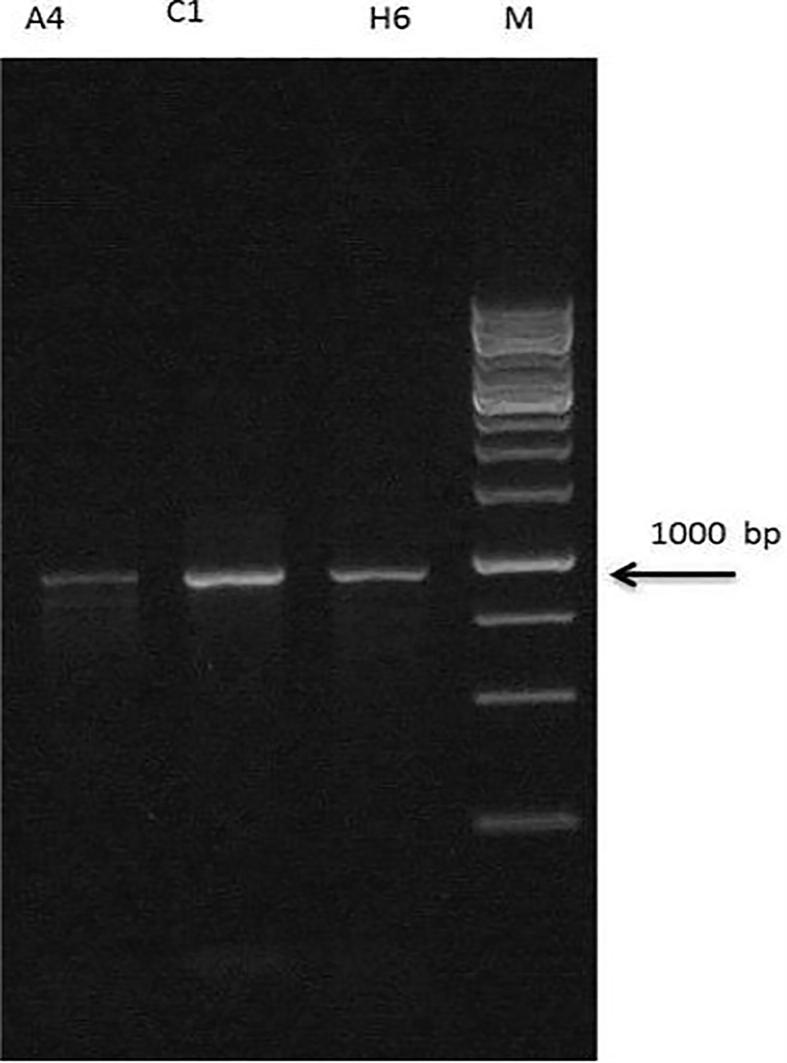
The extracted DNA was analyzed on agarose gel electrophoresis. A good quality of intact DNA having high molecular weight was obtained. The genomic DNA of 3 bacterial isolates was amplified by 16S rRNA and was run on agarose gel (1.5%). The isolates indicated the positive signal in the form of discrete and distinct 1000 bp bands on gel. Results are shown in Fig. 5.
Figure 5.
Electrophoresis amplification result of three selected isolates based on 16 S rRNA genes. M: Marker 1 KB, Well 1–3: Isolate A4, Isolate C1 and Isolate H6.
3.8. Genotypic identification and phylogenetic analysis
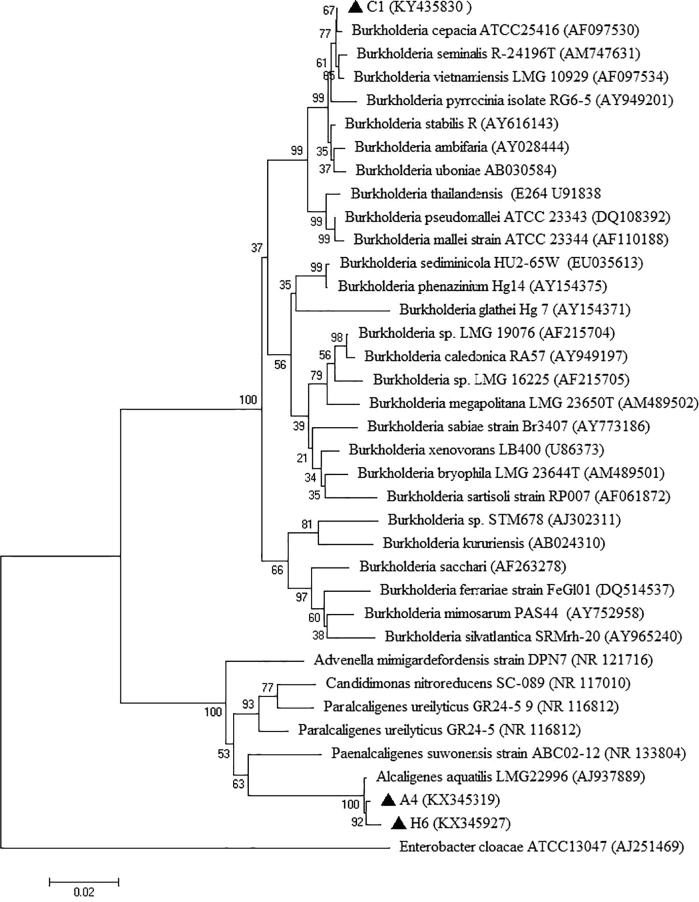
The Blast search performed against GenBank revealed a large number of similar 16S rRNA gene sequences. The blast results of most promising bacterial isolates showed >99% similarities between available GenBank entries in which C1 isolate was identified as Burkholderia cepacia and A4 and H6 were identified as Alcaligenes aquatilis. Results are shown in Table 5. These sequences were submitted to the NCBI database and the accession numbers were obtained. Fig. 6 showed a phylogenetic tree including the isolates (C1, A4 and H6) from this study and some closely related sequences obtained from NCBI. Two distant phylogenetic groups corresponding to the genera Burkholderia sp. and Alcaligenes sp. were obtained. In the phylogenetic group of genus Burkholderia, isolate C1 was closely related to Burkholderia cepacia. The isolates A4 and H6 formed a cluster together and were closely related to Alcaligenes aquatilis. The number beside the node is the statistical bootstrap value. In brackets are the GenBank accession numbers of the 16S rRNA genes. The tree was rooted using Enterobacter cloacae ATCC13047 (AJ251469) as outgroup.
Table 5.
Molecular characterization of A4, C1 and H6 isolates.
| Isolate | Most closely related organism |
16 S rRNA accession no. | Strain | ||
|---|---|---|---|---|---|
| Species | % Similarity | Sequence query coverage (%) | |||
| A4 | Alcaligenes aquatilis | 99% | 97% | KX345319 | LMG22996 |
| C1 | Burkholderia cepacia | 99% | 97% | KY435830 | ATCC 25416 |
| H6 | Alcaligenes aquatilis | 99% | 96% | KX345927 | LMG 22996 |
Figure 6.
Phylogenetic tree based on 16S rDNA gene sequences showing the position of Burkholderia cepacia (C1), Alcaligenes aquatilis (A4) and Alcaligenes aquatilis (H6) strains with regard to related species, which was generated based on pairwise nucleotide distance of the Kimura 2-parameter using the neighbor-joining method included in the MEGA 6.05 software package. The scale bar indicates 0.02 substitutions per nucleotide position. The number beside the node is the statistical bootstrap value. In brackets are the GenBank accession numbers of the 16S rRNA genes.
4. Discussion
PSB is a phosphate-solubilizing microorganism which can be routinely screened by a plate assay method using Pikovskaya medium. The bacteria will grow on this medium and form a clear zone around the colony [33], [34]. These bacteria can convert tricalcium phosphate in the medium from insoluble to soluble forms [35]. Previous reports described some Burkholderia strains as being efficient phosphate solubilizers [36], [37]. Phosphate solubilization potential has been attributed to the strains ability to reduce pH of the surroundings, either by releasing organic acids or protons [38]. Organic acids, such as gluconic acid, formic acid, oxalic acid, and citric acid, secreted by PSB can directly solubilize mineral phosphate as a result of anion exchange or indirectly chelate both Fe and Al ions associated with phosphate. This leads to increased P availability, which ultimately increases plant P uptake.
In the present study, maize rhizosphere was selected for isolation of phosphate solubilizing bacteria. This habitat was chosen due to greater possibility of occurrence of phosphate solubilizing bacteria. Panhwar et al. found considerably higher number of PSB population in the rhizosphere in comparison with non-rhizospheric or bulk soil [39]. Barea et al. also screened several rhizospheric bacteria for phosphate solubilizing potential [40]. In this study, 8 bacterial isolates produced a halo zone around colonies, of these the 3 best isolates were selected based on the basis of phosphate solubilizing index (>4), lower pH (<4) and phosphate solubilizing ability (>250 μg/ml). We have found the soluble P concentration was inversely correlated with pH in culture medium during the growth period of bacteria. This was reported earlier by Chen et al. and Xiang et al. [41], [42]. The data showed negative correlation (r = −0.68) between pH and soluble P content of the medium and the positive correlation (r = 0.96) between soluble P content and PSI, suggested that acidification of the medium can facilitate phosphate solubilization. However, our results contrasted with the report of Tao et al., which documented that there was no correlation between the pH of culture medium and the P mineralization by the organic phosphorus mineralizing bacteria, suggesting that mineralization of organic phosphorus may have different mechanisms of phosphorus solubilizing [43].
In addition, biochemical tests performed for the PSB isolates led to their probable identification up to genus level. Results for some of the common tests performed are listed in Table 2. As shown in Table 2, Isolate C1, A4 and H6 were gram negative, rod-shaped and motile. Isolate C1 was negative for indole test and methyl red (MR) test, and positive for urease test, catalase test, Voges-Proskauer (VP) test and citrate test. Isolates A4 and H6 were negative for methyl red (MR) test and positive for other tests.
Increased growth and P uptake of several crop plants due to PSB inoculation have been reported in a number of studies conducted under both growth chamber and greenhouse conditions [44], [45], [46], [38], [47], [48].
The effects of PSB on the growth of maize in a pot trial found that all 3 isolates showed significant stimulating effect on maize growth (P ≤ 0.05) in root and shoot height, and root and shoot fresh and dry weight and leaves length with respect to control (Table 3). Generally, available phosphorus in soil can be increased by low-molecular-mass organic acids produced from PSB [49]. The results of this study demonstrated that three types of organic acids were produced along with one unknown acid by PSB (on day 8). This was a significant characteristic of these bacteria, including a drop in pH and soluble phosphorus which indicates that organic acid production may have a central role in the solubilization of an insoluble phosphate source. Fig. 4b demonstrates that C1 isolates produced two organic acids such as gluconic acid (0.80 μg/ml) and formic acid (7.48 μg/ml) and one unknown acid, while A4 isolate (Fig. 4a) and H6 isolate (Fig. 4c) produced only citric acid along with two different concentrations 1.68 μg/ml and 0.68 μg/ml, respectively and the results are summarized in Table 4.
In this study, multiple organic acid isolate of C1 showed phosphate solubilizing (305.49 ± 10 μg/ml) activity and a pH (3.08 ± 0.08) drop greater than single organic acid isolates of A4 (277.72 ± 1.45 μg/ml, pH 3.36 ± 0.11) and H6 (282.38 ± 11.81 μg/ml, pH 3.82 ± 0.12). This result similar with the findings of Chen et al. [41], [50], who reported that a combination of multiple organic acids could accomplish mineral solubilization and pH drop better than a single organic acid. However, the phosphate solubilization ability may also depend on other factors such as substrate, medium, temperature, time, and other mechanisms in addition to organic acid production.
Valverde et al. [51] reported that Burkholderia sp. produced gluconic acid, acetic acid and citric acid, and showed that these bacterial genera had high efficiency in solubilizing insoluble phosphate. Other organic acids could be produced by them as well, such as butyric acid, lactic acid, succinic acid, malic acid, glycolic acid, fumaric acid [52], propionic acid, formic acid [53] and unknown organic acids [54]. Furthermore, C1 was the best isolate for significantly enhancing all plant growth parameters (higher than uninoculated plants); including shoot height (6.5 ± 0.58 cm), shoot fresh weight (1.09 ± 0.41 gm), and shoot dry weight (0.48 ± 0.01 gm), root length (13.875 ± 1.6 cm), root fresh weight (0.61 ± 0.04 gm) root dry weight (0.52 ± 0.12 gm) and leaves height (13.075 ± 0.62 cm). Regarding the ability of mineral phosphate solubilization and types of organic acids of C1, we found that this isolate had high ability for solubilization of tricalcium phosphate, and produced gluconic acid and formic acid. This indicated that gluconic acid and formic acid may be related to the mechanism of PSB in promoting maize growth. Our findings correspond to results reported by Afzal and Bano [55] who reported that wheat (Triticum aestivum) inoculated with PSB (Pseudomonas sp. strain 54RB) significantly increased root and shoot weight, plant height, spike length, grain yield and P uptake higher than the control. Similar results were also found in cowpea (Vigna unguiculata) which revealed the enhancement of nodulation, root and shoot biomass, straw and grain yield and P and N uptake of plants inoculated with Gluconacetobacter sp. and Burkholderia sp. [56].
Moreover, the cultures were also identified using the modern 16S rRNA technique. According to the sequence of the 16S rRNA gene, 1 isolates belong to Burkholderia sp. and other two belongs to the Alcaligenes sp. Sequences from 3 isolates were almost 99% similar to other 16S rRNA sequences from the NCBI database. As shown in Table 5, bacterial isolates of C1 were identified as Burkholderia cepacia (99% sequence homology) and were belong to B. cepacia complex (Bcc) and bacterial isolates of A4 and H6 were identified as Alcaligenes aquatilis (99% sequence homology). In the rhizosphere of maize, Burkholderia cepacia represents probably one of the predominant bacterial species [57]. Some studies also revealed that B. cepacia is present in large numbers associated with the roots and the rhizosphere of maize, [58], [59]. Furthermore, B. cepacia has been reported to compete, survive, and colonize roots of various maize cultivars [60], to enhance the productivity of several crop plants [61], and to antagonize and suppress all the major soilborne fungal pathogens of maize, such as those belonging to the genus Fusarium [62], [63]. In addition to phosphate solubilization, Burkholderia sp., especially Burkholderia cepacia has potential for antimicrobial activity and promoting plant growth of maize [64].
Phylogenetic tree based on 16S rRNA sequences of C1, A4 and H6 was constructed with other closely related species of Burkholderia and Alcaligenes, obtained from NCBI (Fig. 6). This phylogenetic analysis revealed that C1 isolates closely matched with Burkholderia cepacia, B. seminalis and B. vietnamiensis and both A4 and H6 isolates closely matched with Alcaligenes aquatilis which are completely separated from Enterobacter sp. by 100 bootstrap values. However, 16S rRNA based identification has limited utility in the genus Burkholderia, especially within the B. cepacia complex, where it cannot be used as a mean to accurately distinguish this to the species level [65], [66]. In order to identify C1 to the species level, molecular analysis of phylogenetic marker, recA, should be used for further study. The recA gene has been widely applied in bacterial systematics [67] and has proven very useful for the identification of B. cepacia complex species, with phylogenetic analysis of sequence variation within the gene enabling discrimination of all nine current species within the B. cepacia complex [65].
Moreover, decreasing pH in media and production of organic acid (Citric acid) also conform the phosphate solubilizing activity of A4 and H6 isolates and may consider first time as novel phosphate solubilizing bacteria. In summary, the rhizosphere soil of maize in uttarakhand region was a good source of Burkholderia sp. with phosphate solubilizing potential. Previous studies have also reported that some Burkholderia species are efficient phosphate solubilizers [68].
Based on the results found in present study, we concluded that Alcaligenes aquatilis may consider as novel phosphate solubilizer. In addition, Burkholderia cepacia strain had great potential for use as soil inoculants when compared to others isolates. These PSB isolates hold good prospects in future for sustainable agricultural practice with minimal chemical inputs and enhances organic farming. Production and utilization of biofertilizer formulation using these rhizobacterial isolates in agricultural fields can increase soil fertility and can increase the crop yield. However, before using these bacterial isolates as a biofertilizer, it should be investigated for crop productivity.
5. Conclusion
In conclusion, 3 bacterial strains from maize rhizosphere soil in Nainital region were isolated, purified, characterized and identified by 16S rRNA gene sequencing. These bacterial strains were identified as belonging to the genera Burkholderia and Alcaligenes. From this study, we have isolated efficient phosphate solubilizing bacteria, which released high amounts of P in broth media and released 3 different kinds of organic acid, were detected from the culture medium by HPLC analysis. The results of our isolates highlight their importance as phosphate solubilizers and recommend these strains as biofertilizers. Further studies could be performed to evaluate their effect on plant growth promoting properties under greenhouse conditions as well as the field ones.
Acknowledgments
The authors gratefully acknowledge Ms. Supriya Pandey and Mr. Avinash Negi for their support during this work.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Khan M.S., Zaidi A., Ahemad M., Oves M., Wani P.A. Plant growth promotion by phosphate solubilizing fungi current perspective. Arch. Agron. Soil Sci. 2010;56:73–98. [Google Scholar]
- 2.Corona M.E.P., Klundert I.V.D., Verhoeven J.T.A. Availability of organic and inorganic phosphorus compounds as phosphorus sources for carex species. New Phytol. 1996;133:225–231. doi: 10.1111/j.1469-8137.1996.tb01889.x. [DOI] [PubMed] [Google Scholar]
- 3.Pundarikakshudu R. Studies of the phosphate dynamics in a vertisol in relation to the yield and nutrient uptake of rainfed cotton. Exp. Agric. 1989;25:39–45. [Google Scholar]
- 4.Boronin А.M. Rhizosphere bacteria of the genus Pseudomonas enabling plant growth and development. Sorovsky Educ. Mag. 1998;10:25–31. [Google Scholar]
- 5.Zaidi A., Khan M.S., Ahemad M., Oves M., Wani P.A. Recent advances in plant growth promotion by phosphate-solubilizing microbes. In: Khan M.S., editor. Microbial Strategies for Crop Improvement. Springer-Verlag; Berlin Heidelberg: 2009. pp. 23–50. [Google Scholar]
- 6.Maliha R., Samina K., Najma A., Sadia A., Farooq L. Organic acids production and phosphate solubilization by phosphate solubilizing microorganisms under in vitro conditions. Pak. J. Biol. Sci. 2004;7:187–196. [Google Scholar]
- 7.Babu-Khan S., Yeo T.C., Martin W.L., Duron M.R., Rogers R.D., Goldstein A.H. Appl. Environ. Microbiol. 1995;61:972–978. doi: 10.1128/aem.61.3.972-978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trolove S.N., Hedley M.J., Kirk G.J.D., Bolan N.S., Loganathan P. Progress in selected areas of rhizosphere research on P acquisition. Aust. J. Soil Res. 2003;41:471–499. [Google Scholar]
- 9.Richardson A.E., Hadobas P.A. Soil isolates of Pseudomonas spp. that utilize inositol phosphates. Can. J. Microbiol. 1997;43:509–516. doi: 10.1139/m97-073. [DOI] [PubMed] [Google Scholar]
- 10.Fibach-Paldi S., Burdman S., Okon Y. Key physiological properties contributing to rhizosphere adaptation and plant growth promotion abilities of Azospirillum brasilense. FEMS Microbiol. Lett. 2012;326:99–108. doi: 10.1111/j.1574-6968.2011.02407.x. [DOI] [PubMed] [Google Scholar]
- 11.Rojas A., Holguin G., Glick B.R., Bashan Y. Synergism between Phyllobacterium sp. (N2-fixer) and Bacillus licheniformis (P-solubilizer), both from a semiarid mangrove rhizosphere. FEMS Microbiol. Ecol. 2001;35:181–187. doi: 10.1111/j.1574-6941.2001.tb00802.x. [DOI] [PubMed] [Google Scholar]
- 12.Von der Weid I., Paiva E., Nobrega A., van Elsas J.D., Seldin L. Diversity of Paenibacillus polymyxa strains isolated from the rhizosphere of maize planted in Cerrado soil. Res. Microbiol. 2000;151:369–381. doi: 10.1016/s0923-2508(00)00160-1. [DOI] [PubMed] [Google Scholar]
- 13.Hou Y.H., Wang Q.F., Shen J.H., Miao J.L., Li G.Y. Molecular identification of a halotolerant bacterium NJ82 from Antarctic Sea ice and preliminary study on its salt tolerance. Microbiology. 2008;35:486–490. [Google Scholar]
- 14.Pikovskaya R.I. Mobilization of phosphorous in soil in the connection with vital activity of some microbial species. Mikorobiologiya. 1948;17:362–370. [Google Scholar]
- 15.Isenberg H.D. American Society for Microbiology; Washington D.C.: 1995. Essential Procedures for Clinical Microbiology. [Google Scholar]
- 16.Mc Fadden. Williams and Wilkins; Baltimore, USA: 1980. Biochemical Tests for Identification of Medical Bacteria; pp. 51–54. [Google Scholar]
- 17.Krieg N.R., Holt G. 9th ed. Williams and Wilkins; Baltimore: 1994. Bergey’s Manual of Determinative Bacteriology. [Google Scholar]
- 18.Edi-Premono, Moawad M.A., Vleck L.G. Effect of phosphate solubilizing Pseudomonas putida on the growth of maize and its survival in the rhizosphere. Indones. J. Crop Sci. 1996;11:13–23. [Google Scholar]
- 19.Subba Rao N.S. Phosphate solubilization by soil microorganisms. Oxford & IBH Publishing Co; 1982. pp. 1–149. (Advances in Agricultural Microbiology). [Google Scholar]
- 20.Hamid A.A.A., Hamdan S., Ariffin S.H.Z., Huyop F. Molecular prediction of dehalogenase producing microorganism using 16S rDNA analysis of 2,2-dichloropropionate (dalapon) degrading bacterium isolated from volcanic soil. J. Biol. Sci. 2010;10:190–199. [Google Scholar]
- 21.Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 22.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halder A.K., Chakrabartty P.K. Solubilization of inorganic phosphate by Rhizobium. Folia Microbiol. 1993;38:325–330. [Google Scholar]
- 26.Goenadi D.H., Sisweto I., Sugiarto Y. Bioactivation of poorly soluble phosphate rocks with a phosphorus-solubilizing fungus. Soil Sci. Soc. Am. J. 2000;64:927–932. [Google Scholar]
- 27.Paul D., Sinha S.N. Isolation of phosphate solubilizing bacteria and total heterotrophic bacteria from river water and study of phosphatase activity of phosphate solubilizing bacteria. Adv. Appl. Sci. Res. 2013;4:409–412. [Google Scholar]
- 28.Paul D., Sinha S.N. Phosphate solubilization potential and phosphatase activity of some bacterial strains isolated from thermal power plant effluent exposed water of river Ganga CIBTech. J. Microbiol. 2013;2:1–7. [Google Scholar]
- 29.Peix A., Mateos P.F., Rodriguez-Barrueco C., Martinez-Molina E. Velazquez and E. Growth promotion of common bean (Phaseolus vulgaris L.) by a strain of Burkholderia cepacia under growth chamber conditions. Soil Biol. Biochem. 2001;33:1927–1935. [Google Scholar]
- 30.Collavino M.M., Sansberro P.A., Mroginski L.A., Aguilar O.M. Comparison of in vitro solubilization activity of diverse phosphate-solubilizing bacteria native to acid soil and their ability to promote Phaseolus vulgaris growth. Biol. Fertil. Soils. 2010;46:727–738. [Google Scholar]
- 31.Kothamasi D., Kothamasi S., Bhattacharyya A., Kuhad R.C., Babu C.R. Arbuscular mycorrhizae and phosphate solubilising bacteria of the rhizosphere of the mangrove ecosystem of Great Nicobar island, India. Biol. Fertil. Soils. 2006;42:358–361. [Google Scholar]
- 32.Kumar V., Behl R.K., Narula N. Establishment of phosphate-solubilizing strains of Azotobacter sp. chroococcum in the rhizosphere and their effect on wheat cultivars under greenhouse conditions. Microbiol. Res. 2001;156:87–93. doi: 10.1078/0944-5013-00081. [DOI] [PubMed] [Google Scholar]
- 33.Pikovskaya R.I. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Microbiologia. 1948;17:362–370. [Google Scholar]
- 34.Nilsson W.B., Paranjype R.N., DePaola A., Strom M.S. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 2003;41:442–446. doi: 10.1128/JCM.41.1.442-446.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pal S.S. Interactions of an acid tolerant strain of phosphate solubilizing bacteria with a few acid tolerant crops. Plant Soil. 1998;198:167–177. [Google Scholar]
- 36.Torres A.R., Araújo W.L., Cursino L., Hungria M., Plotegher F., Mostasso F.L., Azevedo J.L. Diversity of endophytic enterobacteria associated with different host plants. J. Microbiol. 2008;46:373–379. doi: 10.1007/s12275-007-0165-9. [DOI] [PubMed] [Google Scholar]
- 37.Khalimi K., Suprapta D.N., Nitta Y. Effect of Pantoea agglomerans on growth promotion and yield of rice. Agric. Sci. Res. J. 2012;2:240–249. [Google Scholar]
- 38.Hariprasad P., Niranjana S.R. Isolation and characterization of phosphate solubilizing rhizobacteria to improve plant health of tomato. Plant Soil. 2009;316:13–24. [Google Scholar]
- 39.Panhwar Q.A., Othman R., Abdul Rahman Z., Meon S., Ismail M. Isolation and characterization of phosphate solubilizing bacteria from aerobic rice. Afr. J. Biotechnol. 2011;11:2711–2719. [Google Scholar]
- 40.Barea J.M., Navarro E., Montoya E. Production of plant growth regulators by rhizosphere phosphate solubilizing bacteria. J. Appl. Microbiol. 1976;40:129–134. doi: 10.1111/j.1365-2672.1976.tb04161.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y.P., Rekha P.D., Arun A.B., Shen F.T., Lai W.A., Young C.C. Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 2006;34:33–41. [Google Scholar]
- 42.Xiang W.L., Liang H.Z., Liu S., Luo F., Tang J., Li M.Y., Che Z.M. Isolation and performance evaluation of halotolerant phosphate solubilizing bacteria from the rhizospheric soils of historic Dagong Brine Well in China. World J. Microbiol. Biotechnol. 2011;27:2629–2637. [Google Scholar]
- 43.Tao G.C., Tian S.J., Cai M.Y., Xie G.H. Phosphate-solubilizing and -mineralizing abilities of bacteria isolated from soils. Pedosphere. 2008;18:515–523. [Google Scholar]
- 44.Dey R., Pal K.K., Bhatt D.M., Chauhan S.M. Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol. Res. 2004;159:371–394. doi: 10.1016/j.micres.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Fernández L.A., Zalba P., Gómez M.A., Sagardoy M.A. Phosphate-solubilization activity of bacterial strains in soil and their effect on soybean growth under greenhouse conditions. Biol. Fertil. Soils. 2007;43:805–809. [Google Scholar]
- 46.Vikram A., Hamzehzarghani H. Effect of phosphate solubilizing bacteria on nodulation and growth parameters of greengram (Vigna radiata L. Wilczek) Res. J. Microbiol. 2008;3:62–72. [Google Scholar]
- 47.Bharucha U., Patel K., Trivedi U.B. Optimization of indole acetic acid production by Pseudomonas putida UB1 and its effect as plant growth-promoting rhizobacteria on mustard (Brassica nigra) Agric. Res. 2013;2:215–221. [Google Scholar]
- 48.Sivakumar T., Shankar T., Vijayabaskar P., Ramasubramanian V. Plant growth promoting activity of nickel tolerant Bacillus cereus TS1. J. Agric. Technol. 2012;8:2101–2113. [Google Scholar]
- 49.Mia M.A.B., Shamsuddin Z.H., Mahmood M. Effects of rhizobia and plant growth promoting bacteria inoculation on germination and seedling vigor of lowland rice. Afric. J. Biotechnol. 2012;11:3758–3765. [Google Scholar]
- 50.Song O.K., Lee S.J., Lee Y.S., Lee S.C., Kim K.K., Choi Y.L. Solubilization of insoluble inorganic phosphate by Burkholderia cepacia DA23 isolated from cultivated soil. Brazilian J. Microbiol. 2008;39:151–156. doi: 10.1590/S1517-838220080001000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A. Valverde, P. Delvasto, A. Peix, E. Vel zquez, I. Santa-Regina, A. Ballester, C. Rodr guez-Barrueco, C. Garc a-Balboa, J.M. Igual, Burkholderia ferrariae sp. nov., isolated from an iron ore in Brazil, Int. J. Syst. Evol. Microbiol. 56 (2006) 2421–2425. [DOI] [PubMed]
- 52.E. Frossard, L.M. Condron, A. Oberson, S. Sinaj, J.C. Fardeau, Processes governing phosphorus availability in temperate soils, J. Environ. Qual. 29, 15–23.
- 53.Rodriguez H., Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999;17(2000):319–339. doi: 10.1016/s0734-9750(99)00014-2. [DOI] [PubMed] [Google Scholar]
- 54.E. Perez, M. Sulbaran, M.M. Ball, L.A. Yarzábal, Isolation and characterization of mineral phosphatesolubilizing bacteria naturally colonizing a limonitic crust in the south-eastern Venezuelan region, Soil Biol. Biochem. 39 (2007) 2905–2914.
- 55.Afzal A., Bano A. Rhizobium and phosphate solubilizing bacteria improve the yield and phosphorus uptake in wheat (Triticum aestivum) Int. J. Agri. Biol. 2008;10:85–88. [Google Scholar]
- 56.Linu M.S., Stephen J., Jisha M.S. Phosphate solubilizing Gluconacetobacter sp., Burkholderia sp. and their potential interaction with cowpea (Vigna unguiculata (L.) Walp.) Int. J. Agric. Res. 2009;4:79–87. [Google Scholar]
- 57.Nacamulli C., Bevivino A., Dalmastri C., Tabacchioni S., Chiarini L. Perturbation of maize rhizosphere microflora following seed bacterization with Burkholderia cepacia MCI 7. FEMS Microbiol. Ecol. 1997;23:183–193. [Google Scholar]
- 58.K.P. Hebbar, M.H. Martel, T. Heulin, Burkholderia cepacia, a plant growth promoting rhizobacterial associate of maize, in: M.H. Ryder, P.M. Stephens, G.D. Bowen (Eds.), Improving Plant Productivity with Rhizosphere Bacteria), CSIRO Division of Soils, Adelaide, 1994, pp. 201–203.
- 59.Di Cello F., Bevivino A., Chiarini L., Fani R., Paffetti D., Tabacchioni S., Dalmastri C. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 1997;63:4485–4493. doi: 10.1128/aem.63.11.4485-4493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hebbar K.P., Davey A.G., Merrin J., Dart P.J. Rhizobacteria of maize antagonistic to Fusarium moniliforme, a soil-borne fungal pathogen: colonization of rhizosphere and roots. Soil Biol. Biochem. 1992;24:989–997. [Google Scholar]
- 61.Tabacchioni S., Bevivino A., Chiarini L., Visca P., Del Gallo M. Characteristics of two rhizosphere isolates of Pseudomonas cepacia and their potential plant-growth-promoting activity. Microb. Rel. 1993;2:161–168. [Google Scholar]
- 62.Hebbar K.P., Martel M.H., Heulin T. Suppression of pre- and postemergence damping-off in corn by Burkholderia cepacia. Eur. J. Plant Pathol. 1998;104:29–36. [Google Scholar]
- 63.Klinger J.M., Stowe R.P., Obenhuber D.C., Groves T.O., Mishra S.K., Pierson D.L. Evaluation of the Biolog automated microbial identification system. Appl. Environ. Microbiol. 1992;58:2089–2092. doi: 10.1128/aem.58.6.2089-2092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhattacharyya P.N., Jha D.K. Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 2012;28:1327–1350. doi: 10.1007/s11274-011-0979-9. [DOI] [PubMed] [Google Scholar]
- 65.LiPuma J.J., Dulaney B.J., McMenamin J.D., Whitby P.W., Stull T.L., Coenye T., Vandamme P. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 1999;37:3167–3170. doi: 10.1128/jcm.37.10.3167-3170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mahenthiralingam E., Bischof J., Byrne S.K., Radomski C., Davies J.E., Av-Gay Y., Vandamme P. DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 2000;38:3165–3173. doi: 10.1128/jcm.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karlin S., Weinstock G.M., Brendel V. Bacterial classifications derived from recA protein sequence comparisons. J. Bacteriol. 1995;177:6881–6893. doi: 10.1128/jb.177.23.6881-6893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caballero-Mellado J., Onofre-Lemus J., Estrada-de Los Santos P., Martínez-Aguilar L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 2007;73:5308–5319. doi: 10.1128/AEM.00324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]