Abstract
This study was aimed to assess cytochrome b conservation in six breeds of camels reared in Egypt and to compare its sequence with those of other livestock species. The 208-bp fragments from camel mtDNA cyto b were amplified using PCR for 54 camels belonging to 6 camel breeds reared in Egypt. The alignment of camel cyto b sequences showed the presence of two polymorphic sites resulting in four haplotypes and their nucleotide sequences were submitted to GenBank under the accession numbers: KX909894–KX909897.
The genetic distances between tested camel breeds were zero between Baladi, Fallahi and Maghrabi breeds whereas they were at low value between the other three breeds: Mowaled, Sodany and Somali. Neighbor-joining showed 4 branches; one of them include most of the tested animals and another one contains 2 Somali animals which is considered a specific haplotype for this breed. The other two branches are mixed between Sodani and Mowaled breeds.
Neighbor-joining tree was constructed between cyto b sequences of our tested camels and their sequences from livestock species include Camelus dromedaries, Camelus bactrianus, Ovis aries, Capra hircus, Bubalus bubalis, Bos Taurus and Sus scrofa. The result confirmed that our camel breeds belong to Camelus dromedaries and are clearly separated from other species.
It is concluded that cyto b sequence is highly conserved among all camel breeds reared in Egypt which belong to Camelus dromedaries in addition to the advantage of cyto b in differentiation between different livestock sources which enables it to widely use for the adulteration detection in mixed meat.
Keywords: Genetic conservation, Phylogeny relationships, Cytochrome b, Camelus dromedaries, Camel breeds in Egypt
1. Introduction
Camels have many morphological and physiological characteristics which enable them to tolerate harsh conditions in dry regions and deserts especially in Asia and Africa [5], so they play an essential role in the pastoral and agricultural system in these regions. Millions of human being depend on camels and their products in their lives because camels are considered the main source for meat and milk in dry and semidry regions in addition to their usage in transportation for large sectors of pastoral societies [26].
In Egypt, the number of camels was estimated to be 120.000 heads and this number represents 1.1%, 0.9% and 0.7% of the total camel's number reared in Arabian countries, Africa and all over the world, respectively [2]. About half of the camels in Egypt are present in the Shalateen area [17]. The production of Egyptian camels from milk, meat, hides and fibers is about 20.8, 2.3, 0.62 and 0.09 thousand tons, respectively [32]. Camel’ meat and milk possess some characteristics making them favorable where its meat produces low cholesterol and fat comparing with other meats and also its milk is more suitable for people who have allergic to bovine milk [3].
Threats to the biodiversity are increasing due to the loss of genetic diversity within the species utilized in agriculture. Mitochondrial DNA (mtDNA) plays an important role in the identification of genetic biodiversity between different breeds of domestic animals including cattle, pig, sheep, horse and goat [30], [7], [11], [31], [13]. Cytochrome b (cyto b) is a mtDNA gene which is used for phylogenetic relationship determination between different species, due to its sequence variability. Comparative studies depending on cyto b lead to the assignment of newly species and breeds as well as helping in understanding of evolutionary relationships [4].
In spite of camel's considerable contribution to food security, little is known about the genetic characterization of camels reared in Egypt. So, this work aimed to identify the genetic conservation between six camel's breeds reared in Egypt using mtDNA cyto b and also to examine the phylogenic relationships between our tested camels and other livestock species.
2. Materials and methods
2.1. Blood samples and genomic DNA extraction
Blood samples were collected from 54 camels belonging to six breeds reared in Egypt: 13 animals (Baladi), 12 (Sodany), 10 (Somali), 9 (Maghrabi), 6 (Mowaled) and 4 (Fallahi). Genomic DNA was extracted from the whole blood according to the method described by [18] with minor modifications. Briefly, Blood samples were mixed with cold 2× sucrose-triton and centrifuged at 5000 rpm for 15 min at 4 °C. The nuclear pellet was suspended in lysis buffer, sodium dodecyl sulfate and proteinase K incubated overnight in a shaking water bath at 37 °C. Nucleic acids were extracted with saturated NaCl solution. The DNA was picked up and washed in 70% ethanol. The DNA was dissolved in 1×TE buffer. The DNA concentration was determined, using Nano Drop1000 thermo scientific spectrophotometer and then diluted to the working concentration of 50 ng/μl.
2.2. Polymerase chain reaction (PCR) amplification of mtDNA cyto b gene
The PCR primers used to amplify cyto b gene were synthesized according to [6]. The PCR amplifications were conducted in a 50 μL volume containing 5 μl of 10× reaction buffer, 1.5 mM MgCl2, 0.2 mMdNTPs, 0.2 μM each primer, 1.5U Taq DNA polymerase and approximately 100 ng camel genomic DNA. The amplification conditions were as follows: initial denaturation at 94 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 54 °C for 1 min, and extension at 72 °C for 1.5 min, and then the final extension at 72 °C for 10 min. The PCR products were electrophoresed on 2% agarose gel stained with ethidium bromide to test the amplification success. The amplified products were purified with a DNA purification kit (ExoSap-IT, USB Corporation) according to the manufacturer’s instructions to remove residual primers and dNTPs. Sequencing was performed in Macrogen Incorporation (Seoul, South Korea).
Forward primer: 5′-AGC CTT CTC TTC AGT CGC ACA C-3′.
Reverse primer: 5′-GCC CAT GAA AGC TGT TGC T-3′.
2.3. Data analysis
Cytochrome b sequences of tested camels were aligned using the BioEdit software [8] in order to identify and trace individual haplotype mutations. Haplotype structure, sequence variation, average number of nucleotide differences (D) and average number of nucleotide substitutions (Dxy) per site between breeds were calculated using DnaSP 5.00 software [16]. Neighbor-joining (NJ) tree for tested camel breed sequences and the phylogenetic tree between our camels and other livestock species were constructed using Mega version 5.0 software [29].
3. Results and discussion
Molecular characterization of diverse camel breeds was identified by means of microsatellite markers [23], [27], nuclear and mitochondrial markers [5]. Mitochondrial DNA (mtDNA) is characterized by its maternal inheritance, plain constitution, infrequent recombination, small molecular weight and elevated mutation rate in contrast to various nuclear markers. Therefore, mtDNA sequences have been very significant for studying of genetic analysis, investigation of quantitative trait loci, molecular evolution, disease identification, aging and apoptosis [25], [10], [33], [5]. It is considered one of the most effective tools in estimating of genetic biodiversity and phylogenetic relationships in different livestock breeds including camels [3].
The genetic characterization and biodiversity between different camel breeds reared in Egypt are an essential prerequisite to facilitate the conservation and utilization program in an effective and meaningful way. However, the Egyptian camel ecotypes are not well classified or defined, with very limited information available. Therefore, the information on genetic variability among camel breeds can facilitate the development of breeding programs and is a requirement for conservation of genetic resources and germplasm [28].
The cytochrome b (Cyt b) gene is an important part in the mtDNA genome for studying of species classification and detection of phylogenetic relations among diverse mammalian species where its nucleotide sequence is highly conserved [15], [9], [34]. In this work, 208-bp fragments from camel mtDNA cytochrome b were used for the identification of genetic diversity and conservation between 54 camels belonging to 6 camel breeds reared in Egypt named Baladi (Bal), Fallahi (Fal), Maghrabi (Mag), Mowaled (Mow), Sodany (Sod) and Somali (Som). BioEdit software was used for alignment of these 54 samples whereas the identification of variation (polymorphic) sites in these fragments was done using DnaSP 5.00 software.
The alignment declared the presence of two polymorphic sites at nucleotide No.: 37 and 187 resulting in four haplotypes. The nucleotide sequences of these four haplotypes were submitted to GenBank under the accession numbers: KX909894–KX909897. The most common haplotype was present in 47 animals whereas the other three haplotypes were present in three animals (2 from Sodany and 1 from Mowaled), two animals (1 from Sodany and 1 from Mowaled) and two animals from Somali breeds.
The haplotype diversity was 0.000 in three populations: Baladi, Fallahi and Maghrabi whereas it ranged from 0.356 in Somali, 0.439 in Sodany and 0.600 in Mowaled breeds. Within all tested breeds, the haplotype diversity and average number of pairwise differences were 0.241 and 0.31097, respectively. The result showed that nucleotide diversity ranged from 0.00172 (Somali), 0.00277 (Sodany) and 0.00417 (Mowaled) whereas it was 0.0000 in other three breeds. The total nucleotide diversity was 0.00150 for all 6 tested breeds (Table 1).
Table 1.
The genetic diversity data of tested camel breeds.
| Breeds | No. of sequences (N) | No. of polymorphic sites (S) | No. of haplotypes (H) | Haplotype diversity (HD) | Average number of pairwise differences (K) | Nucleotide diversity (π) |
|---|---|---|---|---|---|---|
| Baladi | 13 | 0 | 1 | 0.000 | 0.00000 | 0.00000 |
| Fallahi | 4 | 0 | 1 | 0.000 | 0.00000 | 0.00000 |
| Maghrabi | 9 | 0 | 1 | 0.000 | 0.00000 | 0.00000 |
| Mowaled | 6 | 2 | 3 | 0.600 | 0.86667 | 0.00417 |
| Sodany | 12 | 2 | 3 | 0.439 | 0.57576 | 0.00277 |
| Somali | 10 | 1 | 2 | 0.356 | 0.35556 | 0.00172 |
| Total | 54 | 2 | 4 | 0.241 | 0.31097 | 0.00150 |
The genetic distances were expressed by average number of nucleotide difference (D) and the average number of pairwise differences (Dxy) between breeds (Table 2). The distance was zero between Baladi, Fallahi and Maghrabi. The highest distance was between Mowaled and Sodany breeds (D: 0.639 and Dxy: 0.00307) followed by distance between Mowaled and Somali (D: 0.633 and Dxy: 0.00306). The genetic distances were between Mowaled and each of Baladi, Fallahi and Maghrabi as well as between Sodany and Somali (D: 0.500 and Dxy: 0.00242). The lowest genetic distances were recorded between Somali and each of Baladi, Fallahi and Maghrabi breeds (D: 0.200 and Dxy: 0.00097) followed the distance between Sodany and each of Baladi, Fallahi and Maghrabi breeds (0.333 and Dxy: 0.00161).
Table 2.
Average pairwise differences between populations.
| Baladi | Fallahi | Maghrabi | Mowaled | Sodany | Somali | |
|---|---|---|---|---|---|---|
| Baladi | – | 0.000 | 0.000 | 0.00242 | 0.00161 | 0.00097 |
| Fallahi | 0.000 | – | 0.000 | 0.00242 | 0.00161 | 0.00097 |
| Maghrabi | 0.000 | 0.000 | – | 0.00242 | 0.00161 | 0.00097 |
| Mowaled | 0.500 | 0.500 | 0.500 | – | 0.00307 | 0.00306 |
| Sodany | 0.333 | 0.333 | 0.333 | 0.639 | – | 0.00242 |
| Somali | 0.200 | 0.200 | 0.200 | 0.633 | 0.500 | – |
Average number of nucleotide differences between breeds D (below). Average number of nucleotide substitutions per site between breeds Dxy (above).
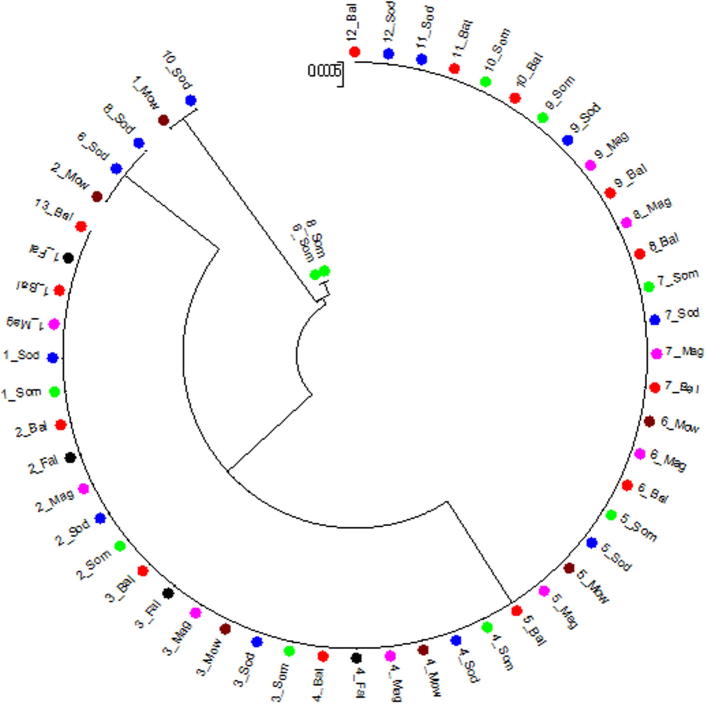
Neighbor-joining tree was constructed using Mega 5.0 software for the tested 54 camels (Fig. 1). The tree showed 4 branches: one of them includes most of the tested animals (47 animals) and another one contains 2 Somali animals which is considered a specific haplotype for this breed. The other two branches are mixed between Sodany and Mowaled breeds. This tree declared the highly genetic conservation between the tested camel breeds due to the absence of genetic distance between three of tested breeds or present at low values between the other three breeds as shown in Table 2. Similarly, the molecular phylogeny based on mtDNA cyto b gene sequences was used for the detection of genetic conservation and relationships among different camel breeds including Camelus dromedaries in Pakistan [3] and Camelus bactrianus in China [20] and among other livestock species and breeds [12], [19], [14].
Fig. 1.
Neighbor-joining (NJ) tree of the tested camel breed sequences. Baladi in  , Sodany in
, Sodany in  , Somali in
, Somali in  , Maghrabi in
, Maghrabi in  and Mowaled in
and Mowaled in  and Fallahi in black.
and Fallahi in black.
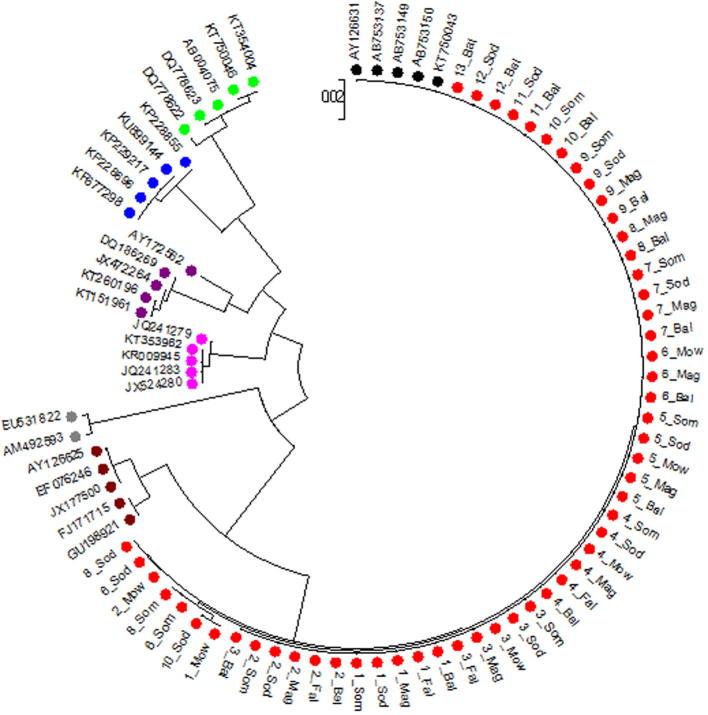
The Neighbor-joining tree of tested camels in this work and other livestock species was conducted (Fig. 2). The sequences of our camel cytochrome b were aligned with reference sequences for Camelus dromedaries: KT750043, AB753150, AB753149, AY126631 and AB753137; Camelus bactrianus: JX177500, U198921, FJ171715, EF076246 and AY126625; Ovis aries: KP228855, KU899144, KP229217, KF677298 and KP228696; Capra hircus: DQ778623, KT750046, DQ778622, AB004075 and KT354004; Bubalus bubalis: JQ241279, KT353962, KR009945, JX524280 and JQ241283; Bos taurus: AY172562, JX472264, DQ186269, KT151961 and KT260196 and Sus scrofa: AM492593 and EU531822.
Fig. 2.
Neighbor-joining (NJ) tree of 54 tested camels and reference sequences. Tested camel sequences in  , Camelus dromedaries in black, Camelus bactrianus in brown Ovis aries in
, Camelus dromedaries in black, Camelus bactrianus in brown Ovis aries in  , Capra hircus in
, Capra hircus in  , Bubalus bubalis in
, Bubalus bubalis in  , Bos Taurus in
, Bos Taurus in  and Sus scrofa in
and Sus scrofa in  .
.
This cyto b phylogenetic tree gave highly informative results where it declares the genetic relationships between different species and confirmed their taxonomy. It showed 4 separated branches, the first one includes Camelus dromedaries which belongs to all tested camel breeds in this study and Camelus bactrianus, the second contains Ovis aries and Capra hircus, the third includes Bubalus bubalis and Bos taurus and the last one contains Sus scrofa. These results confirmed the previous studies which declared that mtDNA sequences might allow the identification of various mammalian species [24]. They reported that PCR-RFLP technique for mtDNA cyto b gene can be used for the differentiation between different species such as Bos taurus, Ovis aries, Capra hircus, Capra capreolus and Capra elaphus.
Cyto b polymorphism was used to differentiate between biological materials of different species including cattle, sheep, goats and deer [21]. They reported the efficiency of this technique in the differentiation of some biological materials such as skin, blood stains and meat derived from different mammalian species. One of the most serious problems faced meat consumers is the adulteration of high quality and most favorable meat with cheap and undesired one in the meat industry. The molecular genetics techniques can solve this problem because this adulteration cannot be detectable by eye or other simple methods. Panwar et al. [22] used PCR of mtDNA cyto b gene for goat and sheep meat identification. They reported the usefulness of this technique for detection of adulteration in mixed meat and suggested the usage of this technique for differentiation between different species. In this way, Abdel-Hameid et al. [1] used mtDNA cyto b for the identification of meat species including camel, donkey and rats.
It is concluded that cyto b sequence is highly conserved among all camel breeds reared in Egypt where the genetic distance between them is absent or present at low values. On the other hand, Neighbor-joining tree declared that all tested camels belong to Camelus dromedaries and the phylogenetic tree showed the usefulness of cyto b sequences in the differentiation between different mammalian species and this advantage could be used for the detection of adulteration in mixed meat from different livestock sources.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Abdel-Hameid Z.G., Hassanane M.S., El-Ashmaoui H.M., Barakat I.A.H., Abd El-Baset S., Elsalkh B. Egypt J. Anim. Prod. 2011;48(1):41–54. [Google Scholar]
- 2.A. Anon, CARDN/ACSAD/Camel/, 2000, p 69.
- 3.Babar M.E., Hussain T., Wajid A., Nawaz A., Nadeem A., Shah S.A., Shahid M.A., Ahmad N., Javed K., Abdullah M. J. Anim. Plant Sci. 2015;25(2):591–594. [Google Scholar]
- 4.Castresana J. Mol. Biol. Evol. 2001;18(4):465–471. doi: 10.1093/oxfordjournals.molbev.a003825. [DOI] [PubMed] [Google Scholar]
- 5.Chuluunbat B., Charruau P., Silbermayr K., Khorloojav T., Burger P.A. Anim. Genet. 2014;45:550–558. doi: 10.1111/age.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Morshedy A.E.D., El-Daly E.S.A., El-Atabany A.I., Tharwat A.E. J. Am. Sci. 2011;7:339–343. [Google Scholar]
- 7.GiuVra E., Kijas J.M.H., Amarger V., Carlborg Ö., Jeon J.T., Anderson L. Genetics. 2000;154:1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall T.A. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 9.Haus T., Akom E., Agwanda B., Hofreiter M., Roos C., Zinner D. Am. J. Primatol. 2013;75(4):350–360. doi: 10.1002/ajp.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X.H., Han X.L., Ma Y.H. Acta Ecol. Anim. Domast. 2009;30:9–13. [Google Scholar]
- 11.Hiendleder S., Kaupe B., Wassmuth R., Janke A. Proc. R. Soc. Lond. B. 2002;269:893–904. doi: 10.1098/rspb.2002.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh H.M., Chiang H.L., Tsai L.C., Lee J.C. Forensic Sci. Int. 2001;122(1):7–18. doi: 10.1016/s0379-0738(01)00403-0. [DOI] [PubMed] [Google Scholar]
- 13.Joshi M.B., Rout P.K., Mandal A.K., Tyler-Smith C., Singh L., Thangaraj K. Mol. Biol. Evol. 2004;21:454–462. doi: 10.1093/molbev/msh038. [DOI] [PubMed] [Google Scholar]
- 14.Kim J.H., Byun M.J., Kim M.J., Suh S.W., Ko Y.G., Lee C.W., Jung K.S., Kim E.S., Yu D.J., Kim W.H., Choi S.B. Asian Aust. J. Anim. Sci. 2013;26(2):163–170. doi: 10.5713/ajas.2012.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li A., Zhao Q., Tang S., Zhang Z. J. Genet. 2005;84:137–142. doi: 10.1007/BF02715839. [DOI] [PubMed] [Google Scholar]
- 16.Libardo P., Rozas J. Bioinformatics. 2009;25:151–1452. [Google Scholar]
- 17.Mahran O.M. Assuit Vet. Med. J. 2004;50(102):172–184. [Google Scholar]
- 18.Miller S.A., Dykes D.D., Polesky H.F. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minarovič T., Trakovická A., Rafayová A., Lieskovská Z. Anim. Sci. Biotech. 2010;43(1):296–299. [Google Scholar]
- 20.Ming L., Yi L., Guo F.C., Siriguleng S., Jirimutu L. Genet. Mol. Res. 2016;15(3) doi: 10.4238/gmr.15038983. gmr.15038983. [DOI] [PubMed] [Google Scholar]
- 21.Natonek-Wioeniewska M., S£ota E., Kalisz B. Folia biologica (Kraków) 2010;58(1-2):47–50. doi: 10.3409/fb58_1-2.47-50. [DOI] [PubMed] [Google Scholar]
- 22.Panwar N., Gahlot G.C., Gahlot K., Ashraf M., Singh A. Indian J. Anim. Res. 2015;49(4):537–541. [Google Scholar]
- 23.Parikh R.C., Patel N.A., Padheriya Y.D., Wadhwani K.N., Rank D.N. J. Camel Prac. Res. 2012;19(2):153–157. [Google Scholar]
- 24.Pfeiffer I., Burger J., Brenig B. BMC Genet. 2004;5:30. doi: 10.1186/1471-2156-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quan J.X., Zhang Y.P., Han J.L., Men Z.M. Yi Chuan Xue Bao. 2000;27:383–390. [PubMed] [Google Scholar]
- 26.Sulieman M., Malik Y.A., Ahmed S.S., Makkawi A.A., Gornas N. Open Access Library J. 2014;1:e667. doi.org/10.4236/oalib.1100667. [Google Scholar]
- 27.Sushma P., Sharique A.A., Priyanka B., Jyoti J., Upasna S., Vijh R.K., Intl D.H.R. J. Biomed. Life Sci. 2014;5(1):286–296. [Google Scholar]
- 28.Taberlet P., Coissac E., Pansu J., Pompanon F. C. R. Biol. 2011;334(3):247–254. doi: 10.1016/j.crvi.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Troy C.S., MacHugh D.E., Bailey J.F., Magee D.A., Loftus R.T., Cunningham P., Chamberlain A.T., Sykes B.C., Bradley D.G. Nature. 2001;410:1088–1091. doi: 10.1038/35074088. [DOI] [PubMed] [Google Scholar]
- 31.Vilà C., Leonard J.A., Götherström A., Marklund S., Sandberg K., Liden K., Wayne R.K., Ellegren H. Science. 2001;291:474–477. doi: 10.1126/science.291.5503.474. [DOI] [PubMed] [Google Scholar]
- 32.Wardeh M.F. ACSAD Camel New-sletter. 1992;9:15–19. [Google Scholar]
- 33.Zhao E., Yu Q., Zhang N., Kong D., Zhao Y. Trop. Anim. Health Prod. 2013;45:1715–1722. doi: 10.1007/s11250-013-0420-5. [DOI] [PubMed] [Google Scholar]
- 34.Zhong X., Wang N., Hu D., Wang J. Korean J. Parasitol. 2014;52:205–209. doi: 10.3347/kjp.2014.52.2.205. doi.org/10.3347/kjp.2014.52.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]