Abstract
Eucalyptus trees litter plays a crucial role in structuring plant populations and regulating crop quality. To help characterize the allelopathic impact of Eucalyptus plantations and understand the interactions between tree litter and understorey plant populations, we performed two different genomic approaches to determine soybean (Glycine max) crop plant response to biotic stress induced by leaf residues of Eucalyptus globulus trees. For assessing cell death, a qualitative method of DNA fragmentation test (comet assay) was employed to detect cleavage of the genomic DNA into oligonucleosomal fragments and help to characterize the apoptotic event among the experimental samples. In addition, quantitative method of genome analysis at the transcriptional level also was conducted to investigate the expression responses of soybean genome to allelochemicals. Expression of specific genes, which are responsible for the breakdown of proteins during programmed cell death PCD (cysteine proteases and their inhibitors), was examined using semi-quantitative RT-PCR (sqPCR). Results of both conducted analyses proved significant genetic effects of Eucalyptus leaf residues on soybean crop genome, revealed by steady increase in DNA damage as well as variation in the transcript levels of cysteine proteases and inhibitors. Further detailed studies using more sensitive methods are necessary for a comprehensive understanding of the allelopathic effects of Eucalyptus plantations on crops.
Abbreviations: PCD, programmed cell death; sqRT-PCR, semiquantitative reverse transcription polymerase chain reaction; CPE, papain–like cysteine proteases; VPE, vacuolar processing enzyme (legumain-like cysteine proteases); CC, cystatins; EUGL, Eucalyptus ground leaves
Keywords: Eucalyptus globulus, Glycine max, Comet assay, Cysteine proteases
1. Introduction
Allelopathy has been recognized as an important ecological mechanism that influences the type of existing vegetation in an ecosystem, plant biodiversity, the dominance and succession of plants, as well as crop management and productivity [1]. Recent reports have proved allelopathic effects revealed by forest trees on vegetation suppression and soil sickness [2], [3]. The forest tree, Eucalyptus globulus is one of the most widely cultivated trees, owing to its fast growth, wider adaptability and high productivity. Nevertheless, it spread into areas of natural vegetation and has been listed among the exotic pest plants. Regarding the ecological impact of Eucalyptus, it has been demonstrated to reduce the diversity of associated species and the productivity of understorey crops [4]. Allelochemicals are naturally released from intact living or dead Eucalyptus tissues and accumulated in soil rhizosphere at high concentrations, generating allelopathic impacts. Eucalyptus species have been evaluated for their allelopathic effects on different plant species [4], [5], [3]. Secondary metabolites including certain phenolic acids and volatile oils released from the leaves, bark and roots of certain Eucalyptus spp. have been identified as harmful biological exudates to other plant species. The potential mechanisms underlying Eucalyptus allelopathic effects on the growth of neighboring crops have been explored in many species, including weeds and crops [6], [7].
Screening bioassays are crucial tools in identifying allelopathic potential of plant species. In addition to the traditional bioassays, methods based on molecular tools have been employed to explore the allelopathic potential of a particular plant as well as the mechanisms of allelochemicals action in cells and genomes. Recently, this approach associating molecular DNA markers with classical bioassays have been used for better exploring and understanding allelopathy. Nevertheless, cytogenetic and molecular analyses have been reported as consistent data, suggesting their complementary use. Although allelopathy is an environmentally friendly method for weed control, the inducible genetic variation and the molecular mechanism for allelopathy on the plant species need to be elucidated. In this context, test plants in allelopathic research, should be sensitive and have an effective response in a short time, even when low concentrations of allelochemicals are used. Soybean [Glycine max (L.) Merr.] has been cited in literature as good candidate in allelopathy investigations [8], [3]. Meanwhile, it is one of the most important agricultural crops for oil and protein. Several genetic studies on soybean germplasm also have provided in-depth insights into functional genes and genetic mechanisms related to plant responses to biotic and abiotic stresses [8], [3].
Genotoxic damage can have long-term effects in natural ecosystems, however, there are few reports on the potential genotoxicity of Eucalyptus. For DNA damage assessment, the single cell gel electrophoresis assays (Comet assays) have been used to evaluate the genotoxicity of environmental agents in animals and terrestrial plants [9], [10], [11]. The comet assay on plants has become a valuable method for assessment of the environmental and experimental genotoxic impact. As the assay is specific and non-invasive, it has been reported as ideal to complement other test systems for DNA damage detection. Comet assay is a very sensitive and simple technique for measuring primary DNA damage events, such as single-strand and double-strand breaks, the generation of alkali-labile sites and excision repair sites and changes in chromosomal structure [11].
Genome analysis at the transcriptional level might be employed to provide evidence about the allelochemicals mode of action, and the mechanisms of defense against them as well. In this research, soybean (Glycine max) provides example of the expression responses of plant genome to environmental stresses. In this regard, specific genes or groups of genes that can be linked to a molecular target site could be tested. Among these, cysteine proteases are involved in a variety of processes in response to both biotic and abiotic stress [12] and responsible for the breakdown of proteins during cell death. Most of plant cysteine proteases are belonging to the papain (C1) or legumain (caspases) (C13) families, which involved in programmed cell death PCD [13], [14], [15]. Legumains are widely existed in plants and located in the vacuoles or cell wall [16]. They are known as vacuolar processing enzymes (VPE), and reveal caspase-like activity [14]. On the other hand, inhibitors of cysteine proteases (Cystatins), have crucial role in regulation of normal physiological processes, and involved in defense mechanisms against biotic and abiotic stress [17], [18], [19]. Rapid identification of soybean cysteine proteases and their inhibitors has been facilitated by the soybean genome database. This information has provided a more comprehensive analysis of the changes in transcripts encoding the cysteine protease–cystatin system proteins in soybean plants during development.
The present work aimed to evaluate the allelopathic interactions between Eucalyptus leaves residue and understorey plant populations, particularly crop plants. Comet assay was used to detect the DNA damage and apoptotic effect on soybean cells. Additionally, genome analysis of 12 proteases genes and their specific inhibitors were carried out at the transcriptional level. Therefore, it will be easily to verify that soybean crop is more or less affected by allelopathic interaction with Eucalyptus. Such information should be beneficial when planning for sowing these important crops near or beneath of eucalypt trees.
2. Materials and methods
2.1. Plant material and experimental design
Fresh mature leaves of Eucalyptus globulus trees were collected from Eucalyptus plantations, Qarwa district, Taif province, Saudi arabia. The leaves were washed, air dried, and ground to fine powder. Soybean (Glycine max) seeds were obtained from the Agricultural seed store. Pot experiment was conducted under natural conditions in plastic pots, containing mixture of clay-sandy (2:1, w/w) soil. Soybean seeds were planted in pots containing mixture of soil and Eucalyptus ground leaves (EUGL) in a percentage of 0 (control), 10, 20, 30, 40, 50, (w/w, residue/soil). Pots maintained in a growth chamber under controlled temperature (20 °C ± 2) and photoperiod of 10–14 h (light/Dark). The pots were divided into six groups including the control and the five different concentrations of Eucalyptus leaf residue. Each treatment was replicated 3 times in a completely randomized experimental design. Each pot was planted with 5 seeds of soybean at 3 cm depth. They were irrigated with water, and harvested after 3 weeks for further analyses.
2.2. DNA fragmentation test (comet assay)
The comet assay was carried out following the protocol described by Juchimiuk et al [20]. Individual soybean leaves were placed in 200 µl of cold 400 mMTris-HCl buffer, pH 7.5. To obtain low frequency of DNA damage in control cells, the leaf was gently sliced to release nuclei into the buffer under yellow light. Each slide previously coated with dried normal melting point (NMP) 1% agarose; was covered with a mixture of equal volumes of nuclear suspension and low melting point agarose (LMP) at 40 °C. The slide was coverslipped and placed on ice for at least 5 min, after then coverslip was removed. LMP agarose (0.5%) was placed on the slide; coverslip was mounted again and then removed after 5 min on ice. Slides were placed in a horizontal gel electrophoresis tank containing freshly prepared cold electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH > 13) and incubated for 15 min. Electrophoresis was performed at 16 V, 300 mA for 30 min at 4 °C. Subsequently, slides were submerged in neutralization buffer (400 mM Tris-HCl, pH 7.5) and stained with ethidium bromide (20 µg/ml) for 5 min. They were dipped in ice-cold distilled water, covered with coverslip and viewed under a fluorescence microscope with computerized image analysis system (Komet Version 3.1. Kinetic Imaging, Liverpool, UK). Images of 250 randomly selected cells (50 cells from five replicate slides) were analyzed for each treatment. The integrated intensity profiles for each cell were computed, and the comet cell components were estimated to evaluate the range of derived parameters. To quantify the DNA damage tail length (TL) and tail moment (TM) were evaluated. Tail length (length of DNA migration) is related directly to the DNA fragment size and presented in micrometers. It was calculated from the centre of the cell. Tail moment was calculated as the product of the tail length and the fraction of DNA in the comet tail.
2.3. RNA isolation and RT-PCR assay
Total RNA was extracted from soybean leaf tissues according to MacRae [21]. To generate c-DNA of cysteine proteases and specific inhibitors genes, specific primers were supplied by Macrogen Inc. (Korea) according to Du Plessis [22]. Five genes of papain like cysteine proteases (CP 1-5), 3 genes of legumain–like proteases (VPE1-3) and four genes of cystatins (CC1-4), were selected to generate the gene expression profiling. Total RNA was reverse transcribed using the Access RT-PCR System (Promega) and a PXE 0.5 thermocycler (Thermo Scientific) following the manufacturer’s instructions. sq RT-PCR products were visualized by conventional agarose gel electrophoresis. Quantification of generated bands was performed with GelPro32 (version 4.03).
2.4. Statistical analysis
A complete randomized design with 3 replications was used in all experiments. The analysis of variance and means were compared using the Duncan Multiple Range Test. Different letters indicate statistical difference at ρ < 0.05. The statistical analysis was done using the IBM/SPSS version 22 software. Figures were plotted by Excel software.
3. Results
3.1. DNA fragmentation test (comet assay)
The DNA migrations (comet assay) in leaf samples are shown in Fig. 1A. Because of the incompatibility about the most useful comet assay parameters for evaluating DNA damage. Tail length (TL), tail moment (TM) and % tails DNA (TD %) were measured in this study. Statistical analysis revealed a steady increase in the frequency of DNA damage in soybean nuclei proportional to concentration of EUGL used, regardless of whether the DNA damage was expressed as tail moment, tail DNA or tail length. In all treated plants, the values of all parameters increased significantly in a dose-dependent manner and were comparable to the control plants (Fig. 1B).
Fig. 1.
Damage in nuclei isolated from leaves of Glycine max Exposed in the comet Assay to various Concentrations of EUGL 1: 0 (Control); 2: 10%; 3: 20%; 4: 30%, 5: 40%; 6: 50%. A: Photomicrographs of EtBr-stained DNA from protoplasts of Glycine max cultivated with EUGL. B: analysis of DNA damage using different parameters; TL: Tail length; T. DNA: % tailed DNA; TM: Tail moment, Mean values with the same letter are not significantly different at P < 0.05 level by one-way ANOVA.
3.2. Reverse transcription-polymerase chain reaction (RT-PCR) analysis
Semi-quantitative RT-PCR was used to determine if the allelopathic effects of EUGL correlate with changes in cysteine protease transcription and which particular member of the cysteine protease gene family is induced or repressed. Also cystatins transcription under biotic stress of EUGL was also investigated.
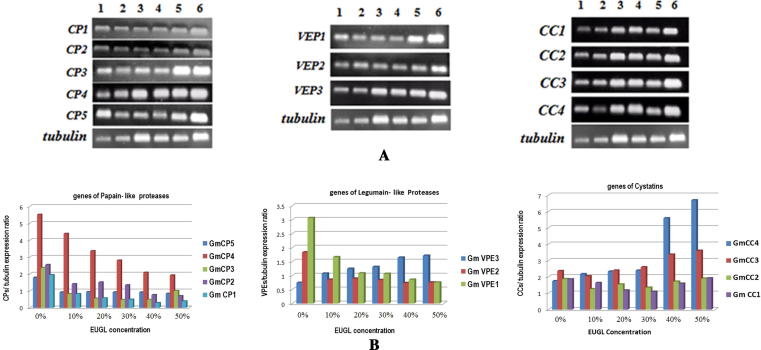
Changes in transcript amounts were detected in all members of genes including four genes of cystatins (CC1-4), five genes of papain like cysteine proteases (CP 1-5) and 3 genes of Legumain–like proteases (VPE1-3). Expression patterns and transcript amounts are presented in Fig. 2 and Table 1.
Fig. 2.
Expression pattern of 5 genes of papain–like cysteine proteases (CP1-5) 3 genes of legumain-like proteases (VEP1-3) and 4 genes of cystatins (CC1-4) in leaves of Glycine max in response to EUGL (1: control; 2: 10%; 3: 20%; 4: 30%; 5: 40% and 6: 50%). A: Transcripts accumulation; B: cysteine proteases genes and their inhibitors / tubulin expression ratio.
Table 1.
Transcription of Cysteine proteases and cystatins in leaves of soybean, determined by sq RT-PCR. ↑ indicates upregulation and ↓ indicates downregulation of gene expression after 3 weeks of EUGL exposure. The values refer to means ± S.E. (n = 3). For a given measurement, means followed by the same letter within a row are not significantly different at P ≤ 0.05 level by one-way ANOVA.
| Gene | EUGL concentration | |||||
|---|---|---|---|---|---|---|
| Relative gene expression | ||||||
| 0 (control) | 10% | 20% | 30% | 40% | 50% | |
| Gm CP1 | 1.94 ± 0.26f | 0.82 ± 0.17e↓ | 0.57 ± 0.026d↓ | 0.49 ± 0.026c↓ | 0.29 ± 0.026b↓ | 0.39 ± 0.028a↓ |
| Gm CP2 | 2.54 ± 0.029d | 1.4 ± 0.021b↓ | 1.5 ± 0.029c↓ | 1.34 ± 0.026b↓ | 0.76 ± 0.026a↓ | 0.70 ± 0.025a↓ |
| Gm CP3 | 2.37 ± 0.029d | 0.82 ± 0.026b↓ | 0.55 ± 0.023a↓ | 0.47 ± 0.026a↓ | 0.48 ± 0.022a↓ | 1.00 ± 0.056c↓ |
| Gm CP4 | 5.52 ± 0.023f | 4.39 ± 0.023e↓ | 3.36 ± 0.023d↓ | 2.81 ± 0.029c↓ | 2.08 ± 0.05b↓ | 1.92 ± 0.015a↓ |
| Gm CP5 | 1.80 ± 0.021c | 0.92 ± 0.018b↓ | 0.94 ± 0.012b ↓ | 0.92 ± 0.015b↓ | 0.92 ± 0.015b↓ | 0.84 ± 0.012a↓ |
| Gm VPE1 | 3.07 ± 0.032d | 1.66 ± 0.034c↓ | 1.08 ± 0.050b↓ | 1.06 ± 0.036b↓ | 0.85 ± 0.020a↓ | 0.75 ± 0.015a↓ |
| GmVPE2 | 1.83 ± 0.022d | 0.85 ± 0.017b,c↓ | 0.89 ± 0.016c↓ | 0.84 ± 0.012b↓ | 0.73 ± 0.013a↓ | 0.75 ± 0.018a↓ |
| Gm VPE3 | 0.74 ± 0.012a | 1.06 ± 0.039b↑ | 1.24 ± 0.021c↑ | 1.31 ± 0.021c↑ | 1.64 ± 0.021d↑ | 1.71 ± 0.035d↑ |
| Gm CC1 | 1.85 ± 0.029a | 1.63 ± 0.018d↓ | 1.17 ± 0.015b↓ | 1.1 ± 0.030a | 1.6 ± 0.021d↓ | 1.91 ± 0.019a |
| Gm CC2 | 1.87 ± 0.028e | 1.25 ± 0.028a↓ | 1.54 ± 0.023c↓ | 1.34 ± 0.032b↓ | 1.71 ± 0.015d↓ | 1.89 ± 0.017e |
| Gm CC3 | 2.36 ± 0.032b | 2.03 ± 0.13a↓ | 2.39 ± 0.020b | 2.60 ± 0.017c↑ | 3.36 ± 0.015d↑ | 3.60 ± 0.036e↑ |
| Gm CC4 | 1.73 ± 0.029a | 2.17 ± 0.032b↑ | 2.33 ± 0.012c↑ | 2.39 ± 0.028 c↑ | 5.59 ± 0.026d↑ | 6.69 ± 0.022e↑ |
No increase in the amount of transcripts of papain–like proteases was found in soybean leaves after 3 weeks EUGL exposure. In contrast, transcript amount of all papain like cysteine proteases, declined during plant development of exposed soybean seeds. This induced repression was most prominently in case of higher concentration of EUGL. Similarly, all legumain-like proteases showed a decrease in transcription in stressed plants but one of these proteases (VPE3) showed a dramatic increase in transcription. On the other hand, the amount of transcripts of the cystatins (CC3 and CC4), greatly increased due to treatment. Such increase was highly significant in higher concentrations, compared to control plants. However, increase in the other two cystatins (CC1, CC2) was not remarkable.
4. Discussion
Litter composition affects plant growth by either providing beneficial nutrients or by allowing harmful allelopathic leaching. As noticeable litter fall accumulation is often observed under deciduous trees, interactions between them and understorey plant populations are worthy of study. Eucalyptus trees are evergreen, and propagated only from seeds. Previous investigations explained the poor performance of crops beneath the tree area on the basis of the allelopathic effect of intact living or dead Eucalyptus tissues [23], [24]. They released allelochemicals which accumulate in soil rhizosphere in high concentrations to produce allelopathic effects. Moreover, chopped Eucalyptus parts have been found to release allelochemicals more rapidly than intact parts [25], [3].
Many researchers demonstrated that the most principal allelochemicals in Eucalyptus are phenolic glycosides [3], [26]. They also reported the release of high levels of stable phenolic compounds from litter of Eucalyptus species rather than other plant parts. Thus, the overall effect of one plant species on another may be the product of multiple and complex interaction of these compounds that may act simultaneously. Various physiological and biochemical processes have been reviewed to elucidate the mechanism of allelochemicals action in the plants [27], [28], [7]. In this respect, this study has been designed to explore the genetic response of soybean genome to the allelopathic effects of Eucalyptus ground leaves (EUGL) using genotoxic and molecular-based approaches, which might help to understand the mode of action of allelochemicals and the mechanisms of defense against them.
One of the main features of PCD is the condensation of nuclear chromatin as a result of endonucleolytic degradation of nuclear DNA (nDNA). In our investigation, the comet assay was performed to detect the nDNA fragmentation. Plant comet assay has been employed to a variety of adverse environmental factors including allelochemicals [29], [30]. It has been reported as a sensitive method to detect internucleosomal damage which is specific for PCD. Because of the conflict about the most useful parameters of the comet assay for evaluating DNA damage, tail length, tail moment and % tails DNA were measured in this study. Results revealed a steady increase in the frequency of DNA damage in soybean nuclei proportional to concentration of EUGL used, regardless of the used parameter to evaluate DNA damage. Other researchers reported a rise in frequency of comets with increasing doses of leaf extract, in assessment of mutagenicity of plant aqueous extracts [31], [29], [30]. Therefore, as suggested by internucleosomal fragmentation of genomic DNA, an active process of cell death might be induced by EUGL, which is referred to an autolytic kind of PCD process [32]. Progressive internucleosomal fragmentation was documented in plant cells during PCD in response to different abiotic and biotic environmental stressors [33], [34]. It has been suggested that the same enzymatic apparatus might be involved in internucleosomal fragmentation during both slow programmed cell death and rapid accidental death [35]. In plant cells, DNA breakage is caused by either rapid accidental vacuole disintegration or programmed vacuolar collapse. This is congruent with the present results of comet pattern which indicate DNA cleavage and therefore, appearance of senescence and PCD in EUGL exposed plants. Breakage of the genomic DNA into discrete fragments prior to membrane disintegration is considered as a main hallmark for apoptosis. Hence, prelytic DNA fragmentation mechanisms should be assayed. Recently, one of these mechanisms involved a caspase activity (cysteine proteases), has been identified [36].
In this context, a comprehensive characterization of the cysteine protease–cystatin system in soybean leaves in response to biotic stress of EUGL was undertaken in this study. After treatment with phytotoxins, detection of the expression profiling of plant genomes is possible at the transcriptional level [37]. Although several researchers have investigated cysteine protease expression during development in response to different biotic and abiotic stress factors, none of the studies investigated specifically papain-like or Legumains–like cysteine proteases or their potential inhibitors, the cystatins. Therefore, this study focused on transcript profiles of these two classes of cysteine proteases and their potential inhibitors in response to allelopathic effects of EUGL.
Results of sqRTPCR exhibited upregulation of transcript amounts of only one member of cysteine proteases (Legumains-like-VPE3) during plant development of exposed soybean seeds. In contrast, transcripts of Legumains-like cysteine proteases (VPE1, 2) and all papain-like cysteine proteases measured in this study were strongly down regulated comparable to up-regulation of VPE3. Upregulation of cysteine proteases under biotic and abiotic stress has been reported by other studies [38], [22]. Plant caspases have remained unidentified even though there have been numerous efforts to identify proteinases that exhibit caspase activities. Vacuolar processing enzyme (VPE) has been shown to reveal a caspase activity which is essential for induced hypersensitive cell death [39]. Certain types of VPEs have been reported as the principal candidates that could be responsible for the caspase like activities observed, and are very likely involved in PCD [40]. In apoptosis, caspases are considered to be responsible for amplifying proteolytic cascade resulting in cleavage of numerous substrates, and the typical morphological features of apoptosis were appeared [39]. They cleave the supporting proteins of nuclear membrane, causing disintegration of the nucleus. Additionally, caspases cleave a protein Inhibitor of Caspase Activated DNase (ICAD of CAD-ICAD complex) that normally holds the CAD-a DNA degrading enzyme in an inactive form, allowing DNase to degrade the DNA in the cell nucleus [41]. Since VPEs are closely related to senescence and stressed-condition including induced PCD in plants [42], it can be inferred from analysis of both gene expression profiling and the comet pattern that Legumain-like cysteine protease VPE3 has a main role in DNA degradation, indicating an induced apoptotic response to allelopathic stress generated by EUGL.
No previous research has focused on investigating cystatins expression in allelopathic plant response. Some cystatins (CC1 and CC2) investigated in this study revealed notable down-regulation, but not affected at high concentration. In contrast, transcripts of the cystatins CC3, CC4 were strongly up-regulated in response to high concentration of EUGL. The cystatin plays a crucial role in regulation of cysteine protease activity throughout plant development and senescence. The other actively transcribed cystatins were only capable of inhibiting specific types of cysteine proteases activity (papain-like family or C1-family) which required for PCD involved in the plant stress hypersensitive response [22]. In our study, transcription of the CC3 and CC4 strongly increased coinciding with the decline in papain-like cysteine proteases (CPs) transcripts.
In conclusion, allelopathic interactions of Eucalyptus globulus allelochemicals include variable genetic effects on soybean plant. The current study revealed that the high accumulation of Eucalyptus leaves on the soil surface may be responsible for retardation of growth of understory plants and consequently reduces the plant yield. Future studies with DNA microarrays with the altered genomic structure identified in this research will be helpful to elucidate the global response of plant genomes after treatment with allelochemicals. For example, generation of transcriptome profiles library for allelochemicals with different molecular target sites would be useful in the determination of the molecular targets. Moreover, identification of allelochemicals that could be responsible for the observed effects in soybean would be of interest and help to clarify allelochemicals mode of actions or mechanism of defense response induced by plant genomes against them.
Acknowledgement
This work was supported by a financial grant from Scientific Research Deanship at Taif University. Project No. 1/436/4298.
Acknowledgments
Conflict of interest
The authors declare that they have no conflicts of interests.
Acknowledgments
Authors’ contributions
Authors participated in both the laboratory and pot experiments.1st author carried out the design of the study and coordination, and draft the manuscript. All authors read and approved the final manuscript.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Chou C.H. Roles of allelopathy in plant biodiversity and sustainable agriculture. Crit Rev Plant Sci. 1999;18:609–636. [Google Scholar]
- 2.Baltzinger M., Archaux F.D., Dumas Y. Tree litter and forest understorey vegetation: a conceptual framework to understand the effects of tree litter on a perennial geophyte, Anemone nemorosa. Ann Bot. 2012:1–10. doi: 10.1093/aob/mcs047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hegab M.M., Gabr M.A., Al-Wakeel S.A., Hamed B.A. Allelopathic potential of Eucalyptus rostrata leaf residue on some metabolic activities of Zea mays L. Univ J Plant Sci. 2016;4(2):11–21. [Google Scholar]
- 4.Sasikumar K., Vijayalakshmi C., Parthiban K.T. Allelopathic effects of Eucalyptus on blackgram (Phaseolus mungo L.) Allelopathy J. 2002;9:205–214. [Google Scholar]
- 5.Zhang C., Fu S. Allelopathic effects of leaf litter and live roots exudates of Eucalyptus species on crops. Allelopathy J. 2010;26(1):91–100. [Google Scholar]
- 6.Raj A., Jhariya M.K., Bargali S.S. Bund based agroforestry using eucalyptus species: a review. Curr Agric Res. 2016;4(2):2118. [Google Scholar]
- 7.Ashraf R., Sultana B., Yaqoob S., Iqbal M. Allelochemicals and crop management: a review. Curr Sci Perspect. 2017;3(1):1–13. [Google Scholar]
- 8.Wu X.L., He C.Y., Wang Y.J., Zhang Z.Y., Dongfang Y., Zhang J.S. Construction and analysis of a genetic linkage map of soybean. Acta Genet Sin. 2001;28:1051–1061. [PubMed] [Google Scholar]
- 9.Gichner T. Differential genotoxicity of ethyl methanesulphonate, N-ethyl-N nitrosourea and maleic hydrazide in tobacco seedlings based on data of the Comet assay and two recombination assays. Mutat Res. 2003;538:171–179. doi: 10.1016/s1383-5718(03)00117-7. [DOI] [PubMed] [Google Scholar]
- 10.Sturchio E, Boccia P, Zanellato M, Mancinelli R, Campiglia E, Cvalieri A. Evaluation of genotoxic effect of Lavendar (Lavandula spp.) essential oil. In: Proceedings of the 10th international conference on environmental science and technology KOS Island, Greece; 2007.
- 11.Araujo S.D., Benko-Iseppon A.M., Brasileiro-Vidal A.C. Genotoxicity and mutagenicity assays for selection of chemical compounds with therapeutic potential: a short commentary. Biochem Anal Biochem. 2015;4:208. [Google Scholar]
- 12.Roberts I.N., Veliz C.G., Criado M.V., Signorini A., Simonetti E., Caputo C. Identification and expression analysis of 11 subtilase genes during natural and induced senescence of barley plants. J Plant Physiol. 2017;211:70–80. doi: 10.1016/j.jplph.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Hara-Nishimura I., Hatsugai N., Nakaune S., Kuroyanagi M., Nishimura M. Vacuolar processing enzyme: an executor of plant cell death. Curr Opin Plant Biol. 2005;8(4):404–408. doi: 10.1016/j.pbi.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Martinez M., Diaz-Mendoza M., Carrillo L., Diaz I. Carboxy terminal extended phytocystatins are bifunctional inhibitors of papain and Legumain cysteine proteinases. FEBS Lett. 2007;581(16):2914–2918. doi: 10.1016/j.febslet.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 15.Chichkova N.V., Shaw J., Galiullina R.A. Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J. 2010;29:1149–1161. doi: 10.1038/emboj.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müntz K., Shutov A.D. Legumains and their functions in plants. Trends Plant Sci. 2002;7:340–344. doi: 10.1016/s1360-1385(02)02298-7. [DOI] [PubMed] [Google Scholar]
- 17.Diop N.N., Kidric M., Repellin A., Gareil M., d’Arcy-Lameta A., PhamThi A.T. A multicystatin is induced by drought-stress in cowpea (Vigna unguiculata (L.) Walp) leaves. FEBS Lett. 2004;577:545–550. doi: 10.1016/j.febslet.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Massonneau A., Condamine P., Wisniewski J.P., Zivy M., Rogowsky P.M. Maize cystatins respond to developmental cues, cold stress and drought. Biochem Biophys Acta. 2005;1729:186–199. doi: 10.1016/j.bbaexp.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Jangpromma N., Saito A., Araki T., Jaisil P., Songsri P., Daduang S. Molecular cloning and characterization in eukaryotic expression systems of a sugarcane cysteine protease inhibitor gene involved in drought tolerance. Turk J Bot. 2014;38:724–736. [Google Scholar]
- 20.Juchimiuk J., Gnys A., Maluszynska J. DNA damages induced by mutagens in plant and human cell nuclei in acellular comet assay. Folia Histochem Cytobiol. 2006;44:127–131. [PubMed] [Google Scholar]
- 21.MacRae E. Extraction of plant RNA, from: methods in molecular biology. In: Hilario E., Mackay J., editors. vol. 353. Springer; 2007. p. 15. (Protocols for nucleic acid analysis using nonradioactive probes). [DOI] [PubMed] [Google Scholar]
- 22.Du Plessis M. Cysteine proteases activity and gene expression studies in soybean nodules during development and drought stress. MSc. thesis. Faculty of Natural and Agricultural sciences. University of Pretoria; 2013.
- 23.Anaya A.L. Allelopathy as a tool in the management of biotic resource agroecosystems. Crit Rev Plant Sci. 1999;18(6):697–739. [Google Scholar]
- 24.Shetta N.D., Alshahrani T.S., Aref I.M., Nasser R.A. Allelopathic potential of Calotropis procera and Eucalyptus species on germination and growth of some timber trees. Allelopathy J. 2017;40(1):81–94. [Google Scholar]
- 25.Espinosa-Garcia F.J. Révision sobre la alelopatia d’EucalyptusL’Herit. Bol Soc Bot Mexico. 1996;58:55–74. [Google Scholar]
- 26.Reigosa M.J., González L., Souto X.C. Pastoriza: allelopathy in forest ecosystems. In: Narwal S.S., editor. Allelopathy in ecological agriculture and forestry. Kluwer Academic Publishers; Netherlands: 2000. pp. 183–193. [Google Scholar]
- 27.Einhellig F.A. The physiology of allelochemicals action: clues and views. In: Reigosa M.J., Bonjoch N.P., editors. Proc. 1st European allelopathy sym. on “Physiological Aspects of Allelopathy”. GAMESAL, S. A.; Vigo (Spain): 2001. pp. 3–26. [Google Scholar]
- 28.Petriccione M., Ciniglia C. Comet assay to assess the genotoxicity of Persian Walnut (Juglansregia L.) Husks with statistical evaluation. Bull Environ Contam Toxicol. 2012;89:166–171. doi: 10.1007/s00128-012-0637-4. [DOI] [PubMed] [Google Scholar]
- 29.Gichner T., Znidar I., Wagner E., Plewa M. The use of higher plants in the comet assay. In: Dhawan A., Anderson D., editors. Issues in toxicology no. 5. The comet assay in toxicology. Royal Society of Chemistry; London: 2009. pp. 98–119. [Google Scholar]
- 30.Tang Z., Zhang J., Yu J., Wang C., Zhang D. Allelopathic effects of volatile organic compounds from Eucalyptus grandis rhizosphere soil on Eisenia fetida assessed using avoidance bioassays, enzyme activity, and comet assays. Chemosphere. 2017;173:307–317. doi: 10.1016/j.chemosphere.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Dal-Souto Frescura V., Kuhn A.W., Laughinghouse H.D. Evaluation of the allelopathic, genotoxic, and antiproliferative effect of the medicinal species Psychotria brachypoda and Psychotria birotula (Rubiaceae) on the germination and cell division of Eruca sativa (Brassicaceae) Int J Cytol Cytosyst Cytogen. 2013;66(2):138–144. [Google Scholar]
- 32.Van Doorn W.G. Classes of programmed cell death in plants compared to those animals. J Exp Bot. 2011;62:4749–4761. doi: 10.1093/jxb/err196. [DOI] [PubMed] [Google Scholar]
- 33.Fojtova A., Kovarik A. Genotoxic effect of cadmium is associated with apoptotic changes in tobacco cells. Plant Cell Environ. 2000;23:531–537. [Google Scholar]
- 34.Santos C.L.V., Pourrut B., Ferreira de Oliveira J.M.P. The use of comet assay in plant toxicology: recent advances. Front Genet. 2015;6:216. doi: 10.3389/fgene.2015.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuthanova A., Opatrny Z., Fischer L. Is internucleosomal DNA fragmentation an indicator of programmed death in plant cells? J Exp Bot. 2008;59(8):2233–2240. doi: 10.1093/jxb/ern090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unsal N.P., Buyuktuncer E.D., Tufekci M.A. Programmed cell death in plants. J Cell Mol Biol. 2005;4:9–23. [Google Scholar]
- 37.Duke S.O., Baerson S.R., Pan Z., Kagan I.A., Sánchez-Moreiras A., Reigosa M.J., Pedrol N., Schulz M. Genomic approaches to understanding allelochemicals effects on plants. In: Zeng R.S., Mallik A.U., Luo S.M., editors. Allelopathy in sustainable agriculture and forestry. Springer; New York (NY): 2008. pp. 157–167. [Google Scholar]
- 38.Jones J.T., Mullet J.C. A salt and dehydration-inducible pea gene Cyp 15a, encodes a cell-wall protein with sequence similarity to cysteine proteases. Plant Mol Biol. 1995;28:1055–1065. doi: 10.1007/BF00032666. [DOI] [PubMed] [Google Scholar]
- 39.Nanda S., Mishra S., Varshney V.P., Singh R.B. A biotechnological approach to apoptosis of somatic and germ cells in living organisms. Open Nutraceuticals J. 2010;3:81–93. [Google Scholar]
- 40.Rojo E., Martın R., Carter C. VPEc exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol. 2010;14:1897–1906. doi: 10.1016/j.cub.2004.09.056. [DOI] [PubMed] [Google Scholar]
- 41.Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S. A caspase-activated DNase that degrades DNAduring apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 42.Misas-Villamil J.C., Toenges G., Kolodziejek I., Sadaghiani A.M., Kaschani F., Colby Activity profiling of vacuolar processing enzymes reveals a role for VPE during oomycete infection. Plant J. 2013;73:689–700. doi: 10.1111/tpj.12062. [DOI] [PubMed] [Google Scholar]