Abstract
Among others, isolate PSK1 was selected and identified by 16 S rDNA sequencing as Bacillus aryabhattai. Growth optimization of PSK1 and physicochemical parameters affected bioflocculant production was carried out by Plackett-Burman design and resulted in increasing in the activity by 4.5%. Bioflocculant production by entrapped and adsorbed immobilized microbial cells was performed using different techniques and revealed enhancement in the activity in particular with pumice adsorption. HPLC analysis of sugars and amino acids composition, FTIR and the effect of different factors on the purified PSK1 biopolymer such as presence of cations, thermal stability, pH range and clay concentration was carried out. Scanning electron microscopy (SEM) of free, immobilized cells, PSK1 bioflocculant and formed flocs were performed. The results revealed that bioflocculant PSK1 is mainly glycoprotein consists of glucose and rhamnose with a large number of amino acids in which arginine and phenylalanine were the major. SEM analysis demonstrated that PSK1 have a clear crystalline rod shaped structure. FTIR spectrum reported the presence of hydroxyl and amino groups which are preferred in flocculation process. PSK1 was soluble in water and insoluble in all other tested organic solvents, while it was thermally stable from 40 to 80 °C. Among examined cations, CaCl2 was the best coagulant. The maximum flocculation activity of the PSK1 recorded at 50 °C (92.8%), pH 2.0 (94.56%) with clay concentration range 5–9 g/l. To obtain a large amount of PSK1 bioflocculant with high flocculating activity, batch fermentation was employed. The results recorded ∼6 g/l yield after 24 h of fermentation.
Keywords: Bioflocculant, Plackett-Burman design, Entrapment, Adsorption, Bacillus aryabhattai, Flocculating activity
1. Introduction
Bioflocculation is a process in which separation of solid liquid mixture is aided by the whole microorganisms or their byproducts so-called “bioflocculants” [17]. During the last years, there was a great interest in studying bioflocculants to replace the hazardous synthetic flocculants because of their advantages such as biodegradability and safety to human and environment [16], [21]. Recently, bioflocculants are produced by various microorganisms such as bacteria, actinomycetes, fungi and algae [12], [46]. Trials for isolation of microorganisms that can produce large amount of bioflocculants with high flocculating activity are important.
Optimization of the culture medium is considered as an important way to obtain high productivity [32]. It is ineffective to use the classical method of optimization that depends on changing one factor at a time but leaving the others at constant level. This method does not clarify the interaction occurs between the factors and their influence on fermentation process. Also, long time and great effort are required because the experiments that should be carried out are in large number [7], [8], [38]. In contrast, statistical approach allows rapid identification of factors that affect the fermentation process because of small number of the required experiments [5].
Plackett-Burman design [37] is one of the statistical approaches that can be used to screen for the most significant factors for further optimization. This design is very useful when large number of factors should be investigated.
Biopolymers production by immobilized microbial cells have many advantages compared to free cells such as that the immobilized cells can be separated easily from culture media and repeatedly used for many times also, the risk of contamination can be minimized [19], [33]. Moreover, the immobilized cells can resist the surrounding toxic chemicals and retain high metabolic activities [6], [44], [50].
Batch fermentation has been widely applied in the industrial production of many biological products such as amino acids, vitamins and enzymes. This process provides simple operation and fewer possibilities of contamination because all of materials utilized in this process are sterilized in the vessel before starting the run [31]. However, there have been few reports on the biosynthesis of flocculants on a pilot scale. Therefore, this study dealt with the batch production of bioflocculant. Moreover, the present study was to isolate, characterize and produce microbial bioflocculant with high flocculating activity by the aid of statistical optimization method, immobilization techniques and batch fermentation.
2. Materials and methods
2.1. Sampling and screening for bioflocculant-producing microorganisms
Several samples were collected from different Egyptian agriculture soils. Screening for bioflocculant-producing microorganisms was carried out on nutrient agar medium (NA) containing (g/l): beef extract, 3; peptone, 5; agar, 15. However, the pre-culture and flocculants production media were containing (g/l): glucose, 10; yeast extract, 3.5; K2HPO4, 5; KH2PO4, 2; MgSO4, 0.5; NaCl, 0.1. All flasks containing the culture media were sterilized in autoclave at 120 °C for 20 min.
To screen for bioflocculant-producing microorganisms, the soil samples were serially diluted and plated on NA agar plates, then incubated at 28 °C for 24 h. Selection of bioflocculant producers were based on the mucoid morphology of appeared and obvious microbial colonies. Subsequently, selected colonies were cultured in pre-culture broth medium at 28 °C in a rotary shaker at 150 rpm for 24 h. Then, 2% inocula of the pre-cultures (OD600 nm was adjusted at 0.6) were transferred into new flasks containing 50 ml of production medium and re-incubated as described above. Kaolin suspensions (5 g/l) were then used to evaluate the flocculating activity of bacterial culture broths (the details are described in flocculation activity measurement section), and the isolates with the highest flocculating activity, were selected for further investigation.
2.2. Identification of bacteria and time course determination
Genomic DNA of the selected isolates was extracted according to the protocol of Gene Jet purification kit (Thermo K0721), which was then used as the template for 16S rDNA amplification. Primers used in the 16S rDNA amplification were 27F and 1492R [23]. The amplification process was conducted in a PCR machine using Maxima Hot Start PCR master mix (Thermo K1051). The used PCR cycles were as follow: 95 °C for 10 min; 35 cycles of 95 °C for 30 s, 65 °C for 1 min, and 72 °C for 1.5 min; and final extension at 72 °C for 10 min. The purified PCR amplicons were sequenced by ABI 3730 DNA sequencer (Applied Biosystems). Thereafter; the obtained sequences were aligned with corresponding sequences from related organisms, which were retrieved from the GenBank database using the BLAST algorithm. To construct the phylogenetic tree, ClustalX software was used to perform sequences alignment [40], and neighbor-joining phylogenetic trees were then constructed using Treeview software program.
2.2.1. Determination of flocculation activity
To determine the flocculating activity, a mixture of synthetic clay suspension (5 g Sigma clay/L distilled water) with a known volume of bacteria strain in the presence of CaCl2 was stirred with rapid mixing at 230 rpm for 2 min, followed by slow mixing at 80 rpm for 3 min using Laboratory Flocculator (Flocumatic 6PLAZAS/sample, Spain) and left standing for 3 min. A sample for optical densities (OD) measurement was withdrawn using automatic pipette from a height of 3 cm below the surface of clay suspension. Relying on the upper phase OD for clay suspension that was measured at 540 nm with a spectrophotometer (7230G, Shanghai, China) the flocculation activity of the different isolated strains was screened. The flocculating efficiency was calculated according to the following equation:
where a and b are the supernatant optical densities (OD) of the control (clay suspension without any bioflocculant addition) and sample respectively, at 540 nm.
Time course experiments were performed according to that described previously by Elkady et al. [14].
The measurements were measured over 5 days of incubation. The flasks containing 125 ml production media were inoculated with 2% pre-culture and incubated at 28 °C and 150 rpm. Samples were withdrawn at different time intervals to measure flocculating activity and cell growth.
2.3. Optimization of bioflocculant production and statistical analysis by Plackett-Burman design
The influences of many factors include; carbon and nitrogen sources, pH (3–11) and inoculum size (0.5–3.5% v/v) on bioflocculant production were studied. The initial pH of the media was adjusted with diluted HCl and NaOH. The Plackett-Burman design is a fraction of two levels factorial design that permits investigation of n-1 factors using at least n experiments [37]. The main effect of each factor was determined based on the difference between the mean of measurements recorded at high level setting and the mean of measurements obtained at low level setting [2]. In this study, as stated in Table 1, seven variables included medium components and physical factors were screened in eight trials in which low and high levels of each factor were examined based on the matrix of Plackett-Burman design. The equation that used to calculate each variable main effect was:
Table 1.
Statistical analysis of Plackett-Burman experiment.
| Variable | FA (%) |
Type of media | FA (%) | |
|---|---|---|---|---|
| Main effect | t-value | |||
| Glucose | −1.2735 | −3.57557 | Optimized | 86 |
| Yeast extract | 0.3395 | 0.953205 | Control | 81.5 |
| K2HPO4 | −0.3155 | −0.88582 | Anti-optimized | 74 |
| KH2PO4 | −1.0035 | −2.8175 | ||
| MgSO4 | −1.5955 | −4.47964 | ||
| NaCl | −0.3505 | −0.98409 | ||
| Culture volume | 0.3165 | 0.888629 | ||
In which Exi was the main effect of the variable, Mi− and Mi+ were the flocculating activity in trials where the independent factor xi was present in low and high concentrations, respectively, and N was the trials number divided by two. A positive sign of the factor main effect elucidated that using this factor at high concentration was near to the optimum and the negative sign elucidated that using this factor at low concentration was near to the optimum. Microsoft Excel was used to calculate the t-values for equal unpaired samples to determine the variable significance.
2.4. Production of bioflocculant by immobilized bacterial cells
2.4.1. Immobilization by entrapment in agar
The gel was prepared by dissolving 2 g agar in 90 ml distilled water by heating at 100 °C then sterilized by autoclaving at 120 °C for 20 min. After cooling to 50 °C, 10 ml bacterial suspension was added and mixed well [9], [10], [45]. 15 ml of this mixture were aseptically poured into a petri-dish. After solidification, the gel was cut with a sterile cutter into small cubes of about 0.5 cm in length. The small gel cubes were transferred to 65 ml sterile bioflocculant production medium in 100 ml flasks. The flasks were incubated at 28 °C and 150 rpm for 18 h.
2.4.2. Immobilization by adsorption
This was carried out by preparation of several 100 ml flasks containing 65 ml bioflocculant production medium with sponge cubes, clay particles, pumice particles or luffa fibers which regarded as solid support materials for microbial cells adsorption. The flasks were then sterilized in autoclave at 120 °C for 20 min. After that, each flask was inoculated with 2% inoculum size (OD600 nm was adjusted at 0.6) from the pre-culture and then incubated at 28 °C and 150 rpm for 18 h.
2.5. Characterization, optimization and batch fermentation of bioflocculant PSK1
Purification of the extracellular bioflocculant was performed according to the method described by Abu-Elreesh et al. [3]. Detection of functional groups of the purified bioflocculant was done by FTIR spectrometer (Bruker TENSOR37). Sugars and amino acids contents of the biopolymer was determined by Agilent technologies 1200 series HPLC system after acid hydrolysis. Morphology of the free cells, immobilized cells, purified bioflocculant, clay particles and formed flocs were detected using Jeol JSM-5300 scanning electron microscope [41]. Solubility of the purified bioflocculant in distilled water and many organic solvents such as ethanol, acetone, isopropanol, dimethyl sulfoxide (DMSO), chloroform and hexane was carried out [49]. In order to achieve the maximum flocculating activity, the influence of different factors that affected bioflocculant efficiency was studied. These factors include the effect of different cations (monovalent, divalent and trivalent), clay suspension concentration (1–13 g/l), temperature (5–80 °C) and pH (2–10). Also, the stability of the bioflocculant toward different temperature ranging from 40 to 80 °C was studied.
Production of the bioflocculant by batch fermentation was carried out in 3 L fermentor filled with 1.5 L bioflocculant production medium. The fermentor (Cleverscientific, model FS-02-B03) with the medium inside it was sterilized in autoclave at 120 °C for 20 min. After that, the fermentor was inoculated with 150 ml from the pre-culture (10% inoculum size, OD600 nm was adjusted at 0.6). The temperature was fixed at 28 °C by the aid of heating jacket covering the outside part of the bioreactor. The medium was adjusted at pH 7 using diluted HCl and NaOH. In addition the agitation and aeration were adjusted at 200 rpm and 1.5 vvm, respectively. After operating the fermentor, the fermentation process was carried out for 24 h and every 2 h samples were withdrawn to measure cell growth and flocculating activity.
3. Results and discussion
3.1. Screening, identification of bacteria and time course measurement
Based on the mucoid assay of colonies appeared on the screening agar plates, nine isolates PSK1, PSK4, PSK5, PSM7, WSK2, WSD6, CSK3, GSM8 and CSM9 were recorded positives. However, isolate PSK1collected from agriculture soil from potato field, Kafr ElSheikh, Egypt, formed the largest flocs and showed the lowest residual turbidity with FA >80%. Therefore, this strain was selected for further studies. Based on basic local alignment search tool (BLAST) analysis, the 16S rDNA nucleotide sequence of PSK1 had >99% similarity to Bacillus aryabhattai (its GenBank accession no. is KY681248). SEM image showed that the cells were rod shaped and arranged in chains. This result was in the same direction with many previous studies which reported that several bacterial strains closely related to the genus Bacillus able to produce bioflocculants [14], [49].
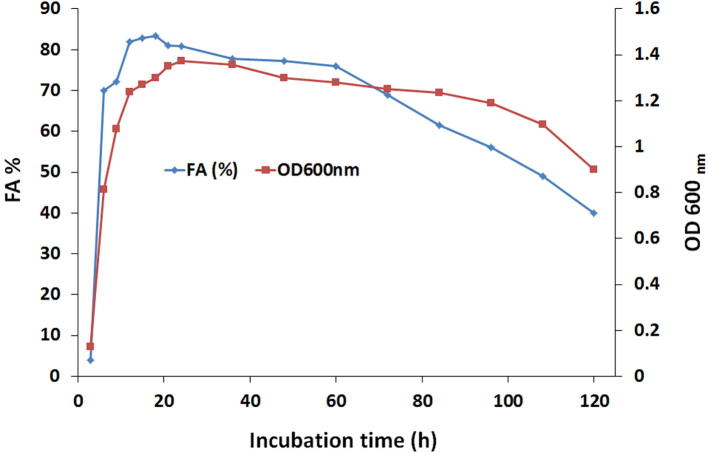
The flocculating activity and cell growth of the strain PSK1 were measured over 5 day incubation (Fig. 1). Production of bioflocculant was in parallel with cell growth until it reached the maximum value at the end of exponential phase (18 h). This phenomenon could be explained that bacteria produced the bioflocculant by biosynthesis during their growth but not by cell autolysis [29]. Also, the flocculating activity appeared to drop when bacterial cells entered the stationary phase (60 h) as the deflocculating enzymes may be secreted [20]. In contrast, some reported microorganisms needed more incubation time for production of bioflocculant with maximum flocculating efficiency. For example Nwodo et al. [36] found that 96 h incubation of Brachybacterium led to obtain bioflocculant with the highest flocculating activity.
Fig. 1.
Time course of bioflocculant production by Bacillius aryabhattai strain PSK1.
3.2. Optimization of bioflocculant production and statistical analysis by Plackett-Burman design
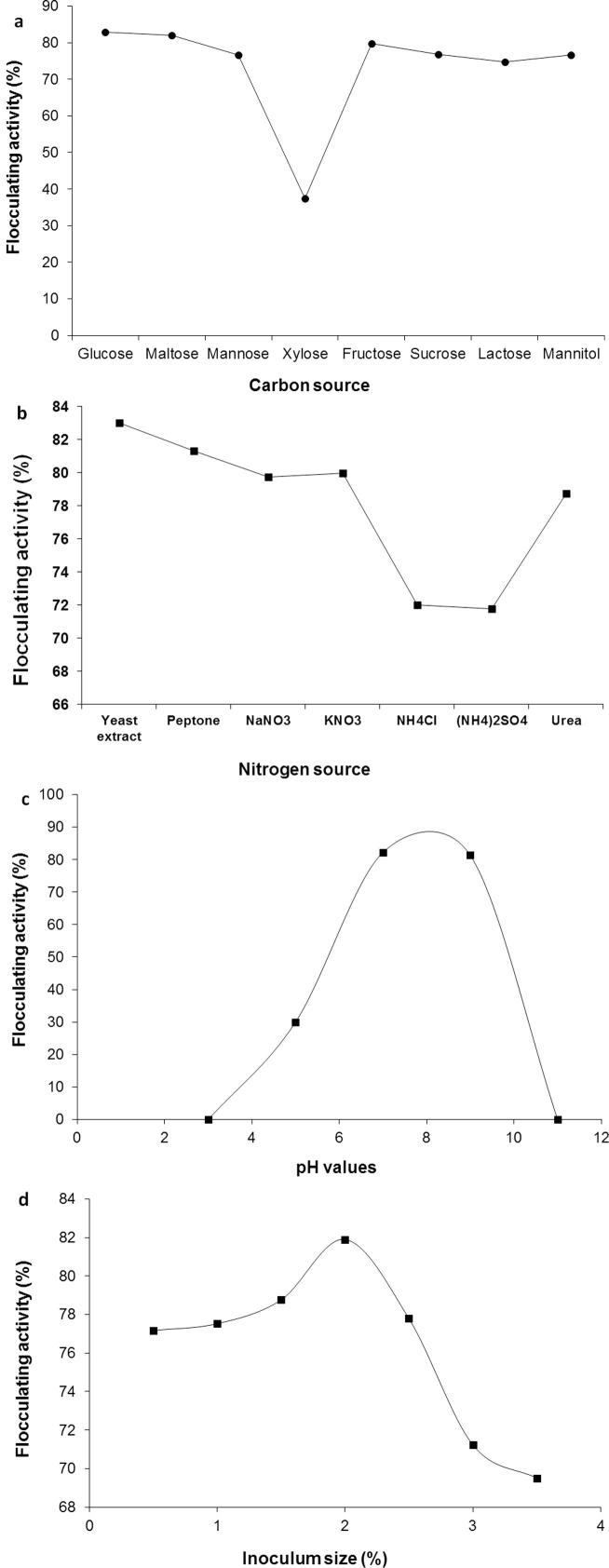
As shown in Fig. 2a, different carbon sources were noticed to influence bioflocculant production by strain PSK1. The most favorable carbon sources that enhanced the bioflocculant production were glucose and maltose. Xylose was found to have an adverse effect on bioflocculant production as flocculating activity was dropped when it was used as a sole carbon source. The same finding was reported by Ugbenyen et al. [43]. However, among examined nitrogen sources, the organic in particular yeast extract were more suitable than inorganic nitrogen sources for bioflocculant production by strain PSK1 (Fig. 2b). Previously, Abdel-Aziz et al. [1] reported that the most effective nitrogen source for bioflocculant production by Bacillus alvei NRC-14 was yeast extract. As shown in Fig. 2c, the pH values among 7–9 was the optimum, while the acidic and/or alkalinic pH caused reduction in flocculating activity. It is known that, pH dependence for bioflocculant production differed with various microorganisms because the nutrient uptake and enzymatic activity of microorganisms could be influenced by initial pH of the medium that affected the electric charge of microbial cells and also the oxido-reduction potential [46]. Moreover, 2% inoculum size was confirmed the best for bioflocculant production by strain PSK1. Small or large inoculum sizes were showed to adversely affect bioflocculant production (Fig. 2d). Small inoculum size would extend the stagnant phase, while large inoculum would cause the niche of bacteria to interfere extremely and prevented the synthesis of bioflocculant [46].
Fig. 2.
Influence of various carbon sources (a), nitrogen sources (b), initial pH values (c) and inoculum sizes (d) on the bioflocculant production by Bacillius aryabhattai strain PSK1.
To screen for the most important factors affecting bioflocculant production by strain PSK1, the Plackett-Burman design was employed. Seven variables include; glucose, yeast extract, K2HPO4, KH2PO4, MgSO4, NaCl and culture volume were tested using low and high level for each variable. The results in Table 1 represented the calculated main effect of each variable on flocculating activity and t-value of each factor. It was obvious that high levels of yeast extract and culture volume, and low levels of glucose, K2HPO4, KH2PO4, MgSO4 and NaCl would lead to the best bioflocculant production. Depending on these results, the predicted optimized medium for bioflocculant production was composed of (g/l): Glucose, 7; yeast extract 4.5; K2HPO4, 4.5; KH2PO4, 1.5; MgSO4, 0.25; NaCl, 0.05 and culture volume of 65 ml in 100 ml flask. So, the flocculating activity detected in optimized medium was 4.5% more than that of start production medium. In contrast, the activity resulted from anti-optimized medium showed 12% decrease in activity compared to optimized medium. Accordingly, the results obtained from the Plackett-Burman design were verified and could be applied to enhance bioflocculant production.
3.3. Production of bioflocculant by immobilized cells of strain PSK1
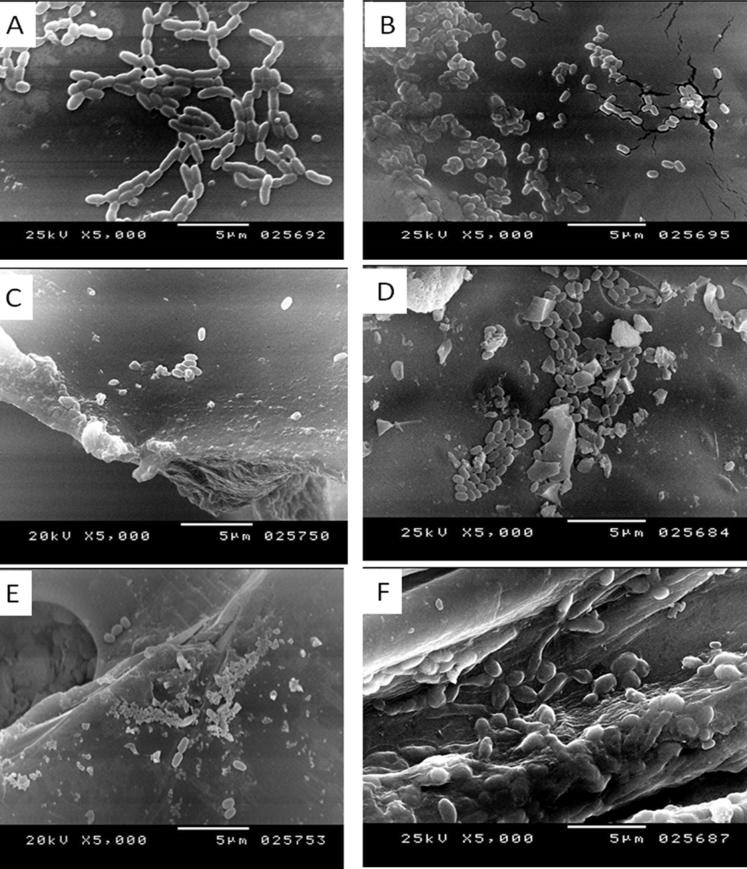
The production of bioflocculant by immobilized PSK1 cells was applied by mean of cells entrapment in agar and adsorption on different solid support materials include; pumice, sponge, clay and luffa. The results recorded in Table 2 elucidated that immobilization led to increase in flocculating activity compared to free cells. When different inoculum sizes of immobilized cells were tried, the highest FA% (91.13) was achieved by using 2% inoculum size. In case of immobilization by adsorption, the cells adsorbed on pumice particles produced bioflocculant with the best FA 91%. As shown in Fig. 3, the SEM images showed good immobilizing and growing efficiencies with all examined matrices except of sponge cubes and clay particles.
Table 2.
Production of bioflocculant by immobilized cells of Bacillus aryabhattai PSK1.
| Immobilization method | |||
|---|---|---|---|
| Entrapment in agar |
Adsorption |
||
| Inoculum size (%) of immobilized cells | FA (%) | Support material | FA (%) |
| Control (2% free cells) | 89.77 | Free cells | 89.77 |
| 1 | 88.54 | Pumice | 91 |
| 1.5 | 89.16 | Sponge | 89.96 |
| 2 | 91.13 | Clay | 90.77 |
| 2.5 | 90 | Luffa | 90.88 |
Fig. 3.
Scanning electron micrographs showing (a) free cells, (b) entrapped cells in agar, (c) adsorbed cells on sponge cubes, (d) adsorbed cells on Pumice particles, (e) adsorbed cells on Clay particles, and (f) adsorbed cells Luffa fibers of Bacillius aryabhattai strain PSK1.
3.4. Purification and characterization of bioflocculant PSK1
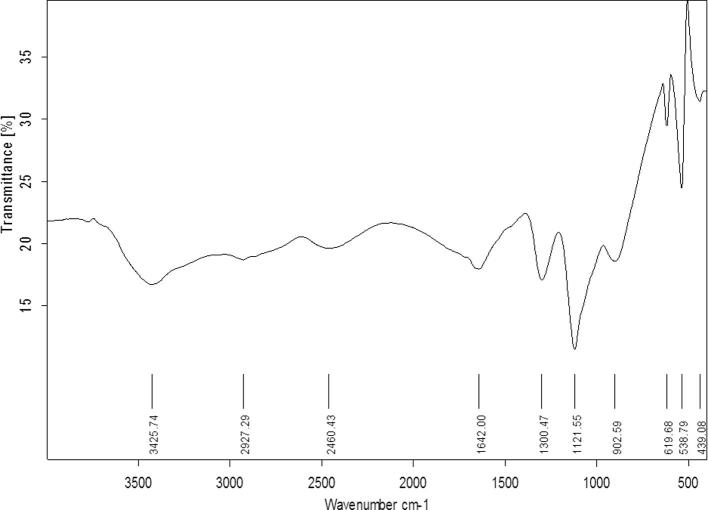
Purification of the bioflocculant from the free-culture supernatant of strain PSK1 was carried out using cold absolute ethanol. So, FA >90% and 2 g/l biopolymer yield were achieved using 4 volumes of ethanol for precipitation. The obtained yield was more than that recorded in many previous reports [13], [25]. As shown in Fig. 4, FTIR spectroscopy of purified PSK1 biopolymer indicted a broad stretching intense peak at 3425 cm−1demonstrated the presence of hydroxyl and amino groups. The weak stretching band detected at 2927 cm−1 revealed the presence of aliphatic C—H stretching. The broad peak at 2460 cm−1 was characteristic of N—H group. The observed asymmetrical stretching peak around 1642 cm−1 was an indication of C O stretching vibration in amide group. The strong stretching band at 1300 cm−1 and 1121 cm−1 confirmed the presence of C—O group. The peak displayed at 902 cm−1 was characteristic of C—H group. The strong absorption peak at 538 cm−1 was confirmed to be characteristic of sugar derivatives. Many previous researches reported the presence of hydroxyl and amino groups in the microbial bioflocculants such as that produced by Bacillus mojavensis 32A [14] and Bacillus velezensis 40B [48].
Fig. 4.
FTIR spectrum of the purified PSK1 bioflocculant.
HPLC analysis indicated that PSK1 bioflocculant is glycoprotein composed of two sugars, glucose (76.67% w/v) and rhamnose (23.24% w/v), with a large number of amino acids in which arginine, phenylalanine, threonine and tyrosine were the major (Table 3). The glycoprotein bioflocculants were reported previously to be produced by many microorganisms [4], [30], [35], [36]. However, due to its contents of sugars and proteins, PSK1 biopolymer was soluble in water and insoluble in organic solvents. The same behavior was noticed with bioflocculants produced by Pseudomonas sp strain 38A [15], Bacillus velezensis 40B [48] and Bacillus mojavensis strain 32A [14].
Table 3.
Amino acids composition of the purified PSK1 bioflocculant as indicated by HPLC analysis.
| Amino acid | % (w/w) |
|---|---|
| Aspartic acid | 7.877 |
| Asparagine | 2.389 |
| dl-Serine | 7.475 |
| Glutamine | 2.4 |
| Glycine | 0.621 |
| dl-Threonine | 14.217 |
| l-Alanine | 0.04 |
| l-Arginine | 17.937 |
| Tyrosine | 10.778 |
| dl-2-Amino-N-Butyric acid | 2.978 |
| l-Lysine | 3.904 |
| Tryptophan | 4.051 |
| Phenyl alanine | 16.698 |
| Isoleucine | 2.967 |
| l-Leucin | 5.667 |
As shown in Fig. 5, SEM images were taken to show the morphology of purified bioflocculant, clay particles and flocs formed due to contribution of the purified bioflocculant in flocculation of clay particles. It was obvious that the purified bioflocculant appeared to have a crystalline rod shaped structure (Fig. 5a) and the clay particles were distributed separately in water (Fig. 5b) then aggregated in large flocs by the aid of the purified bioflocculant (Fig. 5c).
Fig. 5.
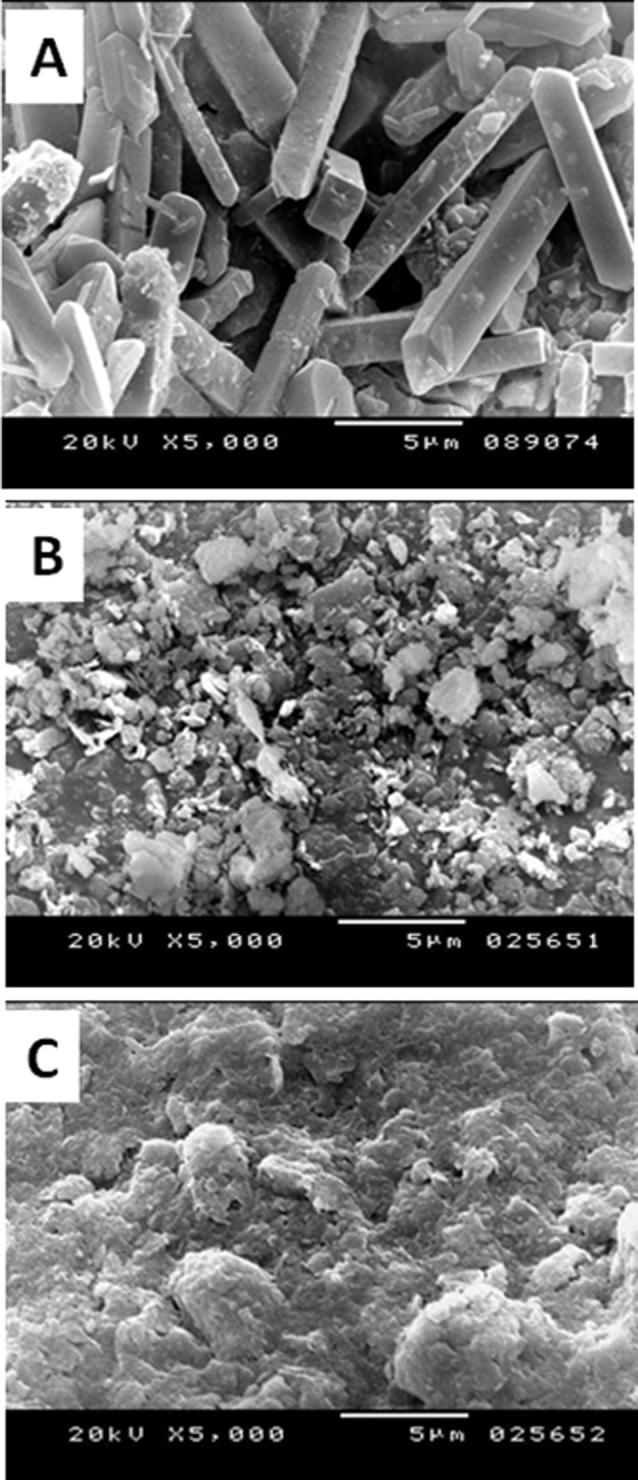
Scanning electron micrographs of (a) the purified PSK1 bioflocculant, (b) clay particles and (c) the flocs formed by flocculation of clay particles by the purified bioflocculant.
3.5. Factors influencing the flocculating activity of the bioflocculant PSK1
As illustrated in Table 4, the cations Ca2+, Mg2+, K+ and Na+ increased flocculating activity of the bioflocculant, while the presence of Fe3+ caused inhibition of the activity. However, K+ and Na+ enhanced the flocculating efficiency but with less effect than Ca2+ which was the best stimulating cation and led to the highest flocculating activity >89%. It is known that Ca2+ could neutralize and stabilize negative charges of the functional groups of bioflocculant which resulted in forming bridges between suspended particles [47]. So, the biopolymer and the suspended particles could form solid complexes by the aid of Ca2+ ions [26]. However, Fe3 may be changed the charges of suspended particles' surfaces and coverage the bioflocculant adsorbing sites. The competition between positively charged suspended particles and less adsorbing sites decreased the flocculating activity [18].
Table 4.
Factors influencing the flocculating activity of PSK1 bioflocculant.
| Cation | FA (%) | Clay concentration (g/l) | FA (%) | Clay suspension temperature (°C) | FA (%) | Clay suspension pH | FA (%) |
|---|---|---|---|---|---|---|---|
| – | 71.76 | 1 | 54.73 | 5 | 87 | 2 | 94.56 |
| CaCl2 | 90 | 3 | 84.21 | 25 | 90 | 3 | 93.28 |
| MgSO4 | 86.59 | 5 | 90 | 30 | 90.5 | 4 | 92.52 |
| CuSO4 | 60.49 | 7 | 90.4 | 40 | 91.5 | 5 | 90.8 |
| FeSO4 | 66.13 | 9 | 91 | 50 | 92.8 | 6 | 90.3 |
| FeCl3 | 0 | 11 | 85.29 | 60 | 92 | 7 | 89.8 |
| NaCl | 83.21 | 13 | 68.23 | 70 | 91.6 | 8 | 89 |
| KCl | 86.9 | 80 | 91 | 9 | 82.88 | ||
| 10 | 78.66 |
Estimation of the optimum clay concentration revealed that there was a parallel increase in flocculating activity with increasing clay concentration up to 9 g/l. However, lower or higher concentrations adversely influenced the flocculating efficiency. Previously, Elkady et al. [14] reported that flocculating activity of the bioflocculant produced by Bacillus mojavensis declined when clay concentration exceed 5 g/l. This observation may be due to increasing in the mutual repulsive force between clay particles as the clay density in the suspension increased [24]. As a result of this repulsive force, the clay particles were kept in constant motion and the bridging effect of the bioflocculant less frequently occurred because more bioflocculant was adsorbed on single clay particle [39].
The PSK1 flocculating activity was improved gradually and reached the maximum value (92.8%) at 50 °C. Lower or higher temperature was not preferred for the flocculating activity. Some previous reports indicated that clay suspension temperature influenced the flocculating activity. For example, the bioflocculant produced by Bacillus circulans achieved high activity at 20 °C then the activity dropped by increasing temperature above 20 °C [25]. Farag et al. [15] found that the bioflocculant produced by Pseudomonas sp. strain 38A recorded the highest activity when clay suspension temperature was 75 °C. Chemical kinetics could explain these results. Both the rate of suspended particles collision and bioflocculant diffusion were enhanced when temperature increased which led to increase in the reaction rate [39]. However, at too high temperature, the protein fraction of the bioflocculant was denatured and the hot movement of clay particles was increased [28].
The effect of pH on the purified bioflocculant PSK1 was studied. High flocculating activity was observed at acidic pH and the maximum activity was achieved at pH 2 (94.56%). Alkaline pH was unfavorable for the bioflocculant performance as the flocculating activity decreased and reached the lowest value at pH 10. This phenomenon may be due to adsorption of H+ ions which reduced the distance between clay particles and enhanced the bridging effect of the bioflocculant [11].
Our results elucidated that the bioflocculant PSK1 was thermally stable when heated at the temperature range of 40–80 °C of various time periods (Table 5). The flocculating activity showed an increase of 1% more than that of the control after heating the bioflocculant at 70 °C for 30 min. This result may be due to the release of poly-substances when the biopolymer was heated at higher temperature [18]. On the other hand, the flocculating activity was slightly decreased by 2.9% after heating the bioflocculant at 80 °C for 45 min. This decline in activity may be due to that the polysaccharide chain of the bioflocculant could be degraded at high temperature which reduced the ability of bioflocculant to form bridges with suspended particles [27]. Also, high temperature could denature the protein fraction of the biopolymer which reduced the flocculating efficiency [42].
Table 5.
Thermal stability of the purified PSK1 bioflocculant.
| Temperature (°C) | Time of exposure (min) | FA (%) |
|---|---|---|
| Control | – | 92.89 |
| 40 | 15 | 92.5 |
| 30 | 93 | |
| 45 | 92 | |
| 50 | 15 | 93 |
| 30 | 93.4 | |
| 45 | 92.2 | |
| 60 | 15 | 89 |
| 30 | 93.2 | |
| 45 | 93.8 | |
| 70 | 15 | 93.2 |
| 30 | 93.8 | |
| 45 | 91.9 | |
| 80 | 15 | 90.9 |
| 30 | 92.8 | |
| 45 | 90 | |
3.6. Bioflocculant production by batch fermentation
As shown in Fig. 6, the flocculating activity and flocculant yield of PSK1 was increased gradually with increasing cell growth and reached its maximum value (FA = 91.5%) after 18 h of fermentation. Then, FA% started to decrease while the bioflocculant yield continued to increase. The reasons for this observation were that when the bioflocculant was used in excess amount, most of binding sites of clay particles was oversaturated which resulted in a decrease in other particles attractive force. Also, the presence of excess bioflocculant with incomplete dispersion of it made only clay particles around the bioflocculant accomplished the flocculation in a very short time and the most of other particles did not share in the flocculation so flocculating activity dropped [22]. Previously, Ning et al. [34] found that batch fermentation for production of REA-11 bioflocculant by Corynebacterium glutamicum resulted in obtaining the highest flocculating activity after 30 h of fermentation which was a long time compared to that required for the bioflocculant PSK1 (18 h).
Fig. 6.
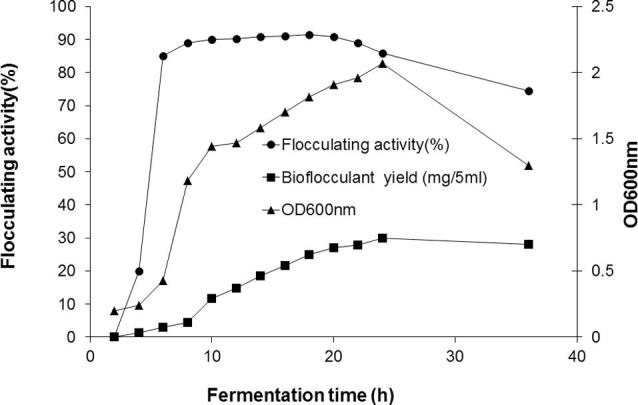
Production of bioflocculant by Bacillus aryabhattai strain PSK1 using batch fermentation approach.
4. Conclusion
A bioflocculant producing strain, Bacillus aryabhattai PSK1, was isolated from soil samples and showed high flocculating activity for turbidity removal from clay suspension after optimization of physicochemical factors affecting the bioflocculant production. Also, statistical optimization by Plackett-Burman design and production of bioflocculant by immobilized cells of strain PSK1 and by batch fermentation were carried out and resulted in increasing in bioflocculant efficiency. Characterization of the purified bioflocculant PSK1 revealed that it was a glycoprotein bioflocculant which had hydroxyl and amino groups that were preferred for flocculation. Using the suitable cation for flocculation, and optimum clay concentration, temperature and pH enhanced the flocculating activity up to 94.56%. Also, PSK1 bioflocculant was found to have very high thermal stability over wide range of temperatures.
Acknowledgment
Sincere thanks to Department of Environmental biotechnology, Genetic Engineering and Biotechnology Research Institute, City of Scientific Research and Technological Applications, Egypt, and Faculty of Science, Alexandria University, Egypt, for providing all facilities required to accomplish this study.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Abdel-Aziz S.M., Hamed H.A., Mouafi F.E., Abdelwahed N.A.M. Life Sci. J. 2011;8(4):883–890. [Google Scholar]
- 2.Abdel-Fattah Y.R., Soliman N.A., Gaballa A.A., Sabry S.A., EI-Diwany A.I. Acta Microbiol. Pol. 2002;51:353–366. [PubMed] [Google Scholar]
- 3.Abu-Elreesh G., Zaki S., Farag S., Elkady M.F., Abd-El-Haleem D. Afr. J. Biotechnol. 2011;10(34):6558–6563. [Google Scholar]
- 4.Aljuboori A.H.R., Idris A., Abdullah N., Mohamad R. Bioresour. Technol. 2013;172:489–493. doi: 10.1016/j.biortech.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Aravindan R., Viruthagiri T. J. Chem. Technol. Biotechnol. 2007;82:460–470. [Google Scholar]
- 6.Cassidy M.B., Lee H., Trevors J.T. J. Ind. Microbiol. 1996;16:79–101. [Google Scholar]
- 7.Chakravarthi R., Sahai V. Process Biochem. 2002;38:481–486. [Google Scholar]
- 8.Chandrasekhar K., Arthur Felse P., Panda T. Bioprocess Eng. 1999;20:203–207. [Google Scholar]
- 9.Chapatwala K.D., Babu G.R.V., Wolfram J.H. J. Ind. Microbiol. 1993;11:69–72. [Google Scholar]
- 10.Chibata I., Tosa T., Sato T. In: Manual of Industrial Microbiology and Biotechnology. Demain A.L., Solomon N.A., editors. ASM; Washington, DC: 1986. pp. 217–226. [Google Scholar]
- 11.David H.F.L., Liptak B.G. Lewis Publishers; New York: 2000. Wastewater Treatment. [Google Scholar]
- 12.Deng S.B., Bai R.B., Hu X.M., Luo Q. Appl. Microbiol. Biotechnol. 2003;60:588–593. doi: 10.1007/s00253-002-1159-5. [DOI] [PubMed] [Google Scholar]
- 13.Deng S., Yu G., Ting Y.P. Coll. Surf. B Biointerfaces. 2005;44:179–186. doi: 10.1016/j.colsurfb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Elkady M.F., Farag S., Zaki S., Abu-Elreesh G., Abd-El-Haleem D. Bioresour. Technol. 2011;102:8143–8151. doi: 10.1016/j.biortech.2011.05.090. [DOI] [PubMed] [Google Scholar]
- 15.Farag S., Zaki S., Elkady M.F., Abd-El-Haleem D. J. Adv. Biol. 2014;4(1):286–295. [Google Scholar]
- 16.Fujita M., Ike M., Jang J.H., Kim S.M., Hirao T. World Sci. Technol. 2001;44(10):237–243. [PubMed] [Google Scholar]
- 17.Gao J., Bao H., Xin M., Liu Y., Li Q., Zhang Y. J. Zhejiang Univ. Sci. B. 2005;7(3):186–192. doi: 10.1631/jzus.2006.B0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong W.X., Wang S.G., Sun X.F., Liu X.W., Yue Q.Y., Gao B.Y. Bioresour. Technol. 2008;99:4668–4674. doi: 10.1016/j.biortech.2007.09.077. [DOI] [PubMed] [Google Scholar]
- 19.Karel S.F., Libicki S.B., Robertson C.R. Chem. Eng. Sci. 1985;40:1321–1354. [Google Scholar]
- 20.Kurane R., Nohata Y. Agric. Biol. Chem. 1991;55:1127–1129. [Google Scholar]
- 21.Kurane R., Toeda K., Suzuki T., Takeda K. Agric. Biol. Chem. 1986;50:2309–2313. [Google Scholar]
- 22.Kwon G.S., Moon S.H., Hong S.D., Lee H.M., Kim H.S., Oh H.M., Yoon B.D. Biotechnol. Lett. 1996;18(12):1459–1464. [Google Scholar]
- 23.Lane D.G. In: Nucleic Acid Techniques in Bacterial Systematic. Stackebrandt E., Goodfellow M., editors. John Wiley and Sons Inc.; New York: 1991. pp. 115–148. [Google Scholar]
- 24.Letterman R.D. fifth ed. Mc Graw-Hill Book Company; New York: 1999. Water Quality and Treatment. [Google Scholar]
- 25.Li Z., Chen R.W., Lei H.Y., Shan Z., Bai T., Yu Q., Li H.L. World J. Microbiol. Biotechnol. 2009;25:745–752. [Google Scholar]
- 26.Li W.W., Zhou W.Z., Zhang Y.Z., Wang J., Zhu X.B. Bioresour. Technol. 2008;99:6893–6899. doi: 10.1016/j.biortech.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 27.Liu W., Wang K., Li B., Yuan H., Yang J. Bioresour. Technol. 2010;101(3):1044–1048. doi: 10.1016/j.biortech.2009.08.108. [DOI] [PubMed] [Google Scholar]
- 28.Liu W., Yuan H., Yang J., Li B. Bioresour. Technol. 2009;100(9):2629–2632. doi: 10.1016/j.biortech.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 29.Lu W.Y., Zhang T., Zhang D.Y., Li C.H., Wen J.P., Du L.X. Biochem. Eng. J. 2005;27:1–7. [Google Scholar]
- 30.Mabinya L.V., Cosa S., Nwodo U., Okoh A.I. Int. J. Mol. Sci. 2012;13:1054–1065. doi: 10.3390/ijms13011054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macauley-Patrick S., Finn B. In: Practical Fermentation Technology. McNeil B., Harvey L.M., editors. John Wiley and Sons Inc.; UK: 2008. pp. 69–95. [Google Scholar]
- 32.Montesinos J.L., Lafuente J., Gordillo M.A., Valero F., Sola C. Biotechnol. Bioeng. 1995;48:573–584. doi: 10.1002/bit.260480604. [DOI] [PubMed] [Google Scholar]
- 33.Moriwaki C., Pelissari F.M., Gonçalves R.A.C., Gonçalves J.E., Matioli G. J. Mol. Catal. B: Enzym. 2007;49:1–7. [Google Scholar]
- 34.Ning H., Xiao-Jie W., Xu D.G., Ying-hua L., Qing-biao L. Chem. Res. Chinese U. 2004;20(2):152–155. [Google Scholar]
- 35.Nwodo U.U., Agunbiade M.O., Green E., Mabinya L.V., Okoh A.I. Int. J. Mol. Sci. 2012;13:8679–8695. doi: 10.3390/ijms13078679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nwodo U.U., Agunbiade M.O., Green E., Nwamadi M., Rumbold K., Okoh A.I. Mater. J. 2013;6:1237–1254. doi: 10.3390/ma6041237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plackett R.L., Burman J.P. Biom. 1946;33:305–325. [Google Scholar]
- 38.Rodriguez L., Teixeira J., Oliveira R., van der Mei H. Process Biochem. 2006;41:1–10. [Google Scholar]
- 39.Ronald R. A Wiley-Interscience Publication; John Wiley and Sons, New York: 1987. Hand Book of Separation Process Technology. [Google Scholar]
- 40.Saitou N., Nei M. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Tahmasebi P., Javadpour F., Sahimi M. Transp. Porous Med. 2015;110:521–531. [Google Scholar]
- 42.Tang W., Song L., Li D., Qiao J., Zhao T., Zhao H. PLoS ONE J. 2014;9(12):1–19. doi: 10.1371/journal.pone.0114591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ugbenyen A., Cosa S., Mabinya L., Babalola O.O., Aghdasi F., Okoh A. Int. J. Environ. Res. Pub. Health. 2012;9:2108–2120. doi: 10.3390/ijerph9062108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Luo Q.F., Zhao J.H., Zhang X.H., Huang L.J. Biomed. Environ. Sci. 2006;19:147–152. [PubMed] [Google Scholar]
- 45.Woodward J. J. Microbiol. Methods. 1988;8:91–102. [Google Scholar]
- 46.Xia S., Zhang Z., Wang X., Yang A., Chen L., Zhao J., Leonard D., Renault N.J. Bioresour. Technol. 2008;99:6520–6527. doi: 10.1016/j.biortech.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 47.Yim J.H., Kim S.J., Ahn S.H., Lee H.K. Bioresour. Technol. 2007;98:361–367. doi: 10.1016/j.biortech.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 48.Zaki S.A., Elkady M.F., Farag S., Abd-El-Haleem D. J. Environ. Biol. 2013;34:51–58. [PubMed] [Google Scholar]
- 49.Zaki S., Farag S., Abu-Elreesh G., Elkady M., Nosier M., Abd-El-Haleem D. Int. J. Environ. Sci. Technol. 2011;8(4):831–840. [Google Scholar]
- 50.Zhou L., Guiying L., Taicheng A., Jiamo F., Guoying S. Recent Pat. Eng. 2008;2:28–35. [Google Scholar]