Abstract
Sugarcane is susceptible to red rot disease caused by phytopathogenic fungus Colletotrichum falcatum Went which ultimately affect the economy of farmers as well as sugar based industry. One of the various ways to control this devastating disease is to develop disease resistance sugarcane cultivar and this requires the complete understanding of genetic makeup of pathogen. Although South Gujarat is well known sugarcane cultivating area, less published data can be found about PCR-based genetic diversity in prevalent C. falcatum accessions. So, present investigation aims at finding molecular variation among the ten accessions of C. falcatum using RAPD and ISSR molecular markers. A total of 35 RAPD and 39 ISSR primers were screened across 10 C. falcatum accessions, of which 15 RAPD and 21 ISSR primers have showed consistent amplification. Statistics related to genetic variation were estimated using NTSYS-PC by means of Dice’s coefficient. The results revealed 80.6% and 68.07% polymorphism and similarity coefficient ranged from 0.43 to 0.91 and 0.73 to 0.93 in RPAD and ISSR analysis respectively. The dendrogram generated using RAPD, ISSR and combined RAPD-ISSR grouped accessions into different clusters which reveal considerable level molecular variation among the C. falcatum accessions. It is also evident from PCA plots that accessions are rather dispersed with tested marker systems indicating good genetic base. So, in nut shell, we found considerable genetic variation and relatedness within C. falcatum accessions collected from different areas of south Gujarat, India using RAPD and ISSR markers.
Keywords: Colletotrichum falcatum, ISSR, Polymorphism, RAPD, Red rot, Sugarcane
1. Introduction
Sugarcane (Saccharum officinarum L.) belonging to family Poaceae is one of the most important agro-industrial crops under cultivation in tropical and subtropical regions in the world. In India sugarcane is grown in 21 states and it’s a second highest crop after cotton under cultivation. In Gujarat, total 4.5 Lakhs families grow sugarcane and that provides total of 5.5 Lakhs employments. Particularly South Gujarat is entire sugarcane growing belt with eleven operational sugar factories, which is highest compared to other region of state and Bardoli sugar factory is one of the Asia’s biggest sugar mill. As an industrial crop, sugarcane is used for production of sugar, bioethanol, jaiggery, molasses, cattle feed and syrups. Under field condition sugarcane is susceptible to many bacterial, viral and fungal diseases but among them fungal infections is most critical as it can affect all parts of plant. There are two main fungal diseases in sugarcane, wilt and red rot, among them red rot is oldest and severe disease of sugarcane caused by fungus Colletotrichum falcatum Went (telomorph, Glomerella tucumanensis). In India, first time this disease was reported by Barber (1901) in Andhra Pradesh [1].
Red rot pathogen infects various parts of the cane plant but it is typically considered as a stalk and a seed borne disease [2]. Infection on leaves may not affect the overall yield, while stem infection with fungus is very crucial as it directly reduces the sugar content. At early stage of the cane crop it is difficult to recognize the presence of disease in the field whereas at the later stage of sugar formation characteristic symptoms of the disease develops on the stem as reddening of the internodal tissues with intermittent red and white patches and leads to 29% reduction in sugar cane weight and 31% loss in sugar recovery [3]. Though the plant pathogen interactions studies provided an overall understanding of the genetic analysis of pathogenicity, yet the exact mechanism for development of disease is not clear. Further cultural and morphological characterization of C. falcatum isolates and use of differential host reaction to detect pathogenic variability is time consuming [4], [5]. Since, the pathogenicity is variable with environmental conditions sometimes leading to disease misinterpretation. As the genetic make-up of pathogen C. falcatum changes continuously, prompt characterization and its biological control is crucial to control the disease. Continuous evolution of newer races is also one of the key factors that make several elite varieties to become susceptible [6]. High variability and adaptability of the C. falcatum endangers the farming of different elite sugarcane cultivars. Obviously use of disease resistant variety is the important approach to prevent the disease, but due to frequent genetic mutations the pathogenicity often overcomes the resistance introduced and cultivars surrender to the pathogen soon [7], [8]. The natural selection of new, more virulent accession and collapse of host resistance is majorly due to gradual diminishing of resistance sources. [9]. Understanding of genetic variation in and diversity pattern of C. falcatum prevalent in specific area is important mean to device disease management strategy if any epidemics arise. So variety of molecular approaches have been developed and used as morphological criteria are not accurate to discriminate between Colletotrichum species. Recently, various molecular markers have been used in categorization of different Colletotrichum Spps [4], [10], [11]. Random Amplified Polymorphic DNA (RAPD) marker amplifies random DNA fragment with decamer primers using polymerase chain reaction (PCR) [12]. RAPD technique was the most widely used molecular method owing to its inexpensiveness, technical simplicity and it does not require prior sequence information and can efficiently distinguish taxa below the species level [13]. Inter-simple sequence repeats (ISSR) is also PCR-based molecular tool that amplifies genomic regions between SSR sequences repeat. ISSR markers are robust, reliable, quick, efficient and reproducible. This method was first reported as a technique for the study of molecular variation in plants and animals and later on used to find DNA markers in fungi [14], [15]. The RAPD and ISSR markers have became the markers of choice to study the genetic variations due to their technical simplicity, inexpensiveness and repeatable amplification of DNA sequence using single primer. There is a lack of information on the genetic diversity of the C. falcatum in South Gujarat. So, the present research work was carried out using RAPD and ISSR markers to find molecular variation among C. falcatum accessions collected from red rot infected sugarcane cultivars from south Gujarat.
2. Materials and methods
2.1. Fungal accessions
Ten accessions of C. falcatum were isolated from red rot infected stems of nine sugarcane cultivars used in present study [16]. Accessions one to nine were collected different region of South Gujarat and Cf8436 was obtained from Sugarcane Breeding Institute, Coimbatore (Table 1). Pure cultures of each accession were made by single spore isolation and were maintained on oatmeal agar slants at 4 ± 1 °C until used for DNA isolation.
Table 1.
Details of Colletotrichum falcatum accessions collected from different regions of South Gujarat for genetic diversity analysis.
| Sr No | Isolate | Place of collection | Host Cultivar | Latitude (°N), Longitude (°E) |
|---|---|---|---|---|
| 1 | cfNAV | Navsari, Gujarat | Co 671 | 20.9467° N, 72.9520° E |
| 2 | cfVES | Vesma, Gujarat | CoS 707 | 21.0314° N, 72.9752° E |
| 3 | cfPAR | Pardi, Gujarat | Co 671 | 20.5230° N, 72.9594° E |
| 4 | cfTIM | Timbarva, Gujarat | Co 86002 | 21.1864° N, 73.1364° E |
| 5 | cfMAR | Maroli, Gujarat | CoN 10071 | 21.0243° N, 72.8899° E |
| 6 | cfGAN | Gandevi, Gujarat | Co 86032 | 20.8077° N, 72.9992° E |
| 7 | cfKAM | Kamrej, Gujarat | Co 94004 | 21.2676° N, 72.9609° E |
| 8 | cfCHA | Chalthan, Gujarat | Co 94008 | 21.1544° N, 72.9623° E |
| 9 | cfMAD | Madhi, Gujarat | Co 86249 | 21.1170° N, 73.1076° E |
| 10 | Cf8436 | Coimbatore, Tamil Nadu | Co 8436 | 11.0045° N, 76.9616° E |
2.2. Genomic DNA extraction
Total fungal DNA was isolated from the mycelia grown for 5–7 days at room temperature on complete media broth (CMB) (yeast extract at 6 g/l, Casien Acid Hydrolysate at 6 g/l and Sucrose at 10 g/l) using modified method of Raeder and Broda [17]. The mycelium harvested using filter paper (Whatman no. 1) was dried overnight in a centrifugal evaporator. Using liquid nitrogen the mycelium was grounded into a fine powder in a pestle and mortar. Dried powder of each isolate (70 mg) was suspended in 750 μl of DNA extraction buffer (700 mM NaCl, 50 mM Tris (pH 8.0), 10 mM EDTA (pH 8.0), 2% cetyl trimethyl ammonium bromide, 1% Polyvinyl pyrolidone (PEP) and 1% β-mercaptoethanol) and incubated for 45 min with intermittent shaking at 62 °C. After incubation, suspension was subjected to centrifugation at 10,000 rpm for 15 min and supernatant was collected into a fresh tube. Later, the supernatant was extracted twice with chloroform: isoamyl alcohol (24:1) and later two volumes of absolute ethanol were added to the aqueous extract and the mixture was placed on ice for 35 min. The contents were centrifuged at 12,000 rpm for 10 min to collect the precipitated DNA. The DNA pellet was washed with 70% ethanol, briefly air-dried and resuspended suspended in 200 μl of 1X Tris-EDTA (TE). The RNA contamination from the genomic DNA was removed by adding 5 μl of RNase (10 μg/ml) to each tube, DNA was redisolved by taping the pellet and incubated for 1 h at 37 °C. Further, genomic DNA was purified by phenol: chloroform: isoamyl alcohol (25:24:1) treatment and precipitation with 3 M ammonium acetate and absolute alcohol and finally dissolved in TE buffer. The quantity and quality of isolated DNA was analysed on spectrophotometer and agarose gel (0.8%) electrophoresis, respectively. Purified DNA samples were stored at 4 °C for further RAPD and ISSR analysis.
2.3. PCR amplification and agarose gel electrophoresis of PCR products
PCR was performed in a programmable gradient thermocycler (Eppendorf) using 200 µl PCR tubes (Himedia). In total 35 RAPD primers (Operon technologies, USA) and 39 ISSR primers were screened for genetic analysis of C. falcatum accessions. PCR for RAPD and ISSR was carried out in of 25 µl volume containing buffer MgCl2, primer, deoxynucleotides mixtures and Taq polymerase (Bangalore GeNei, India, GeNei TM), and genomic DNA. The optimal annealing temperature was found to vary according to the G + C content of the di- or tri-nucleotide repeats of containing ISSR primers. PCR products were separated on 1.5% agarose gels and visualized in Gel Doc system (BioRad). A 100-base pair ladder (Bangalore GeNei Pvt Ltd, Bangalore, India) was used as a standard DNA marker. Analysis was replicated twice to confirm reproducibility of results.
2.4. Data analysis
Amplification was scored based on bands from gel photographs. The presence of band at an amplicon level was scored as 1 and its absence as 0, and only those bands consistently reproducible amplifications were included in the final analysis. The data was analysed using standard procedure in NTSYS-PC by means of Dice’s coefficient [18]. Dendrogram was constructed using unweighted pair group method with arithmetic averages (UPGMA) algorithm based on the genetic distance of Nei and Li [19]. Polymorphism information content (PIC) was calculated using EIGEN module of NTSYSpc 2.02 to confirm primer efficiency according to the formula given by Khaleghi et al. [20].
3. Results
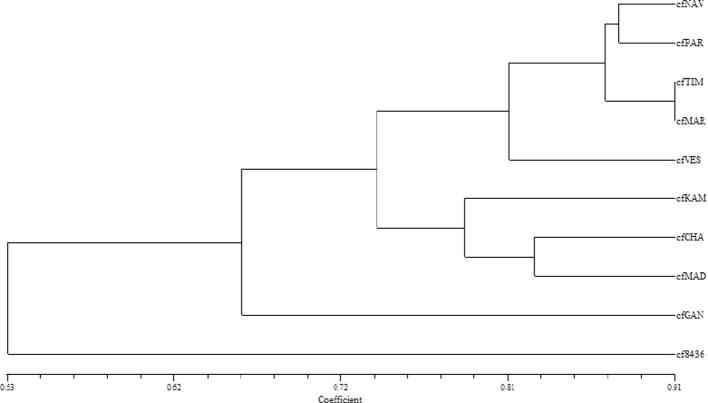
DNA based markers are almost unlimited in number and are not affected by environment as a result they are used to detect and measure variability among individual isolates. The present study was carried out with the ten different C. falcatum accessions to evaluate the molecular variation among them. Different Colletotrichum species including Colletotrichum gloeosporioides, Colletotrichum lindemuthianum and Colletotrichum accutatum and have been reported to possess a high degree of molecular variation when evaluated by RAPD analysis [21], [22], [23], [24]. A total of 35 random RAPD primers were tested among them 15 primers gave apparent and reproducible loci amplification. The distinct patterns obtained for each strain can be taken as specific fingerprinting that describe them. RAPD amplifications using 15 primers yielded total 67 scorable loci, of which13 monomorphic loci (Table 2). The bands were distinct and easy to score. The number of scorable bands for RAPD primer varied from 2 (RPI15) to 7 (RPI1 and RPI20). In present study, total of 434 bands were amplified by 15 RAPD primers. Primer RPI1 amplified the maximum number of bands (55), while primer RPI5 amplified least number of bands (12). Out of 15 primers, 8 primers (RPI5, RPI10, RPI11, RPI14, RPI15, RPI17, RPI18 and RPI19) showed the 100% percent polymorphism. This polymorphism is often due to the existence of genetic variants represented by the number of alleles at a locus and their frequency of distribution in a population. The highest genetic similarity coefficient (0.91) was observed between cfTIM and cfMAR, which indicates that the genetic variation within this species was very low. C. falcatum accession cfNAV and cf8436 represented a relatively wide diversity with similarity coefficient of 0.43 (Table 3). PIC values were calculated for each primer. Primer RPI1 gave highest PIC value of 0.849 and primer RPI15 gave the lowest PIC value of 0.489 with an average 0.705 PIC value per primer. Ten accessions were grouped into two major clusters based on the RAPD analysis at a cut-off value of 0.75 (Fig. 1). Both cluster consisted of five (cfNAV, cfPAR, cfTIM, cfMAR, and cfVES) and three accessions (cfKAM, cfCHA and cfMAD) respectively, indicating the close relation among them. Geographically distant C. falcatum accession cfGAN collected from Gandevi had a level of resemblance with Coimbatore accession cf8436 presented on different clade of bootstrapped dendrogram.
Table 2.
Numerical data as obtained from PCR amplification by RAPD primers among Colletotrichum falcatum accessions. (TL: Total Loci, ML: Monomorphic Loci, PL: Polymorphic Loci, %P: % Polymorphism, PIC: Polymorphic Information Content).
| Sr No | Primer | Sequence (5′-3′) | TL | ML | PL | % P | PIC |
|---|---|---|---|---|---|---|---|
| 1 | RPI1 | CCAGCTTAGG | 7 | 2 | 5 | 71.43 | 0.849 |
| 2 | RPI2 | CCGCCCAAAC | 3 | 1 | 2 | 66.67 | 0.625 |
| 3 | RPI5 | GGTTGTACCC | 3 | 0 | 3 | 100.00 | 0.541 |
| 4 | RPI6 | CCCGCTACAC | 5 | 2 | 3 | 60.00 | 0.778 |
| 5 | RPI9 | CTCACCGTCC | 6 | 2 | 4 | 66.67 | 0.822 |
| 6 | RPI10 | CCCGGCATAA | 3 | 0 | 3 | 100.00 | 0.631 |
| 7 | RPI11 | CCCGTTGGGA | 4 | 0 | 4 | 100.00 | 0.734 |
| 8 | RPI12 | TCTCCGCTTG | 6 | 2 | 4 | 66.67 | 0.814 |
| 9 | RPI14 | CTCCATGGGG | 3 | 0 | 3 | 100.00 | 0.653 |
| 10 | RPI15 | CCTCTCGACA | 2 | 0 | 2 | 100.00 | 0.489 |
| 11 | RPI17 | TGAGCCTCAC | 6 | 0 | 6 | 100.00 | 0.791 |
| 12 | RPI18 | TGAGCGGACA | 3 | 0 | 3 | 100.00 | 0.548 |
| 13 | RPI19 | ACCTGAACGG | 5 | 0 | 5 | 100.00 | 0.778 |
| 14 | RPI20 | TTGGCACGGG | 7 | 4 | 3 | 42.86 | 0.828 |
| 15 | RPI25 | GGCTGCAGAA | 4 | 0 | 4 | 100.00 | 0.692 |
| Total | – | 67 | 13 | 54 | 80.60 | – | |
| Average | – | 4.467 | 0.867 | 3.6 | 0.705 | ||
Table 3.
Dice’s similarity coefficient based on RAPD analysis in 10 Colletotrichum falcatum accessions.
| Accessions | cfNAV | cfVES | cfPAR | cfTIM | cfMAR | cfGAN | cfKAM | cfCHA | cfMAD | cf8436 |
|---|---|---|---|---|---|---|---|---|---|---|
| cfNAV | 1.00 | |||||||||
| cfVES | 0.84 | 1.00 | ||||||||
| cfPAR | 0.88 | 0.81 | 1.00 | |||||||
| cfTIM | 0.89 | 0.83 | 0.90 | 1.00 | ||||||
| cfMAR | 0.85 | 0.78 | 0.84 | 0.91 | 1.00 | |||||
| cfGAN | 0.60 | 0.65 | 0.64 | 0.63 | 0.67 | 1.00 | ||||
| cfKAM | 0.65 | 0.66 | 0.74 | 0.74 | 0.72 | 0.71 | 1.00 | |||
| cfCHA | 0.71 | 0.74 | 0.79 | 0.79 | 0.77 | 0.67 | 0.78 | 1.00 | ||
| cfMAD | 0.72 | 0.76 | 0.77 | 0.79 | 0.75 | 0.75 | 0.80 | 0.83 | 1.00 | |
| cf8436 | 0.43 | 0.52 | 0.49 | 0.49 | 0.49 | 0.57 | 0.59 | 0.59 | 0.61 | 1.00 |
Fig. 1.
UPGMA cluster analysis of 10 Colletotrichum falcatum accessions with Dice’s similarity coefficient of RAPD.
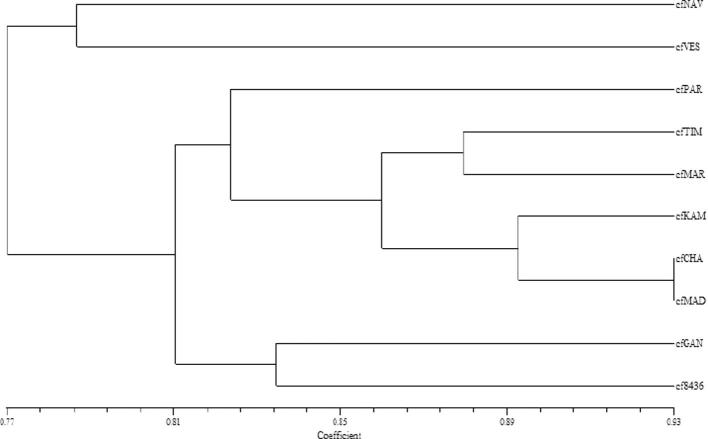
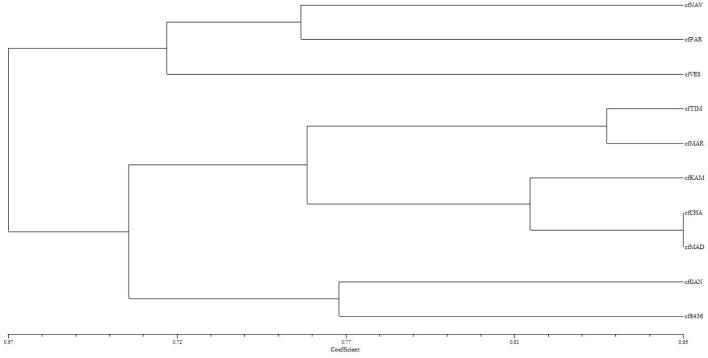
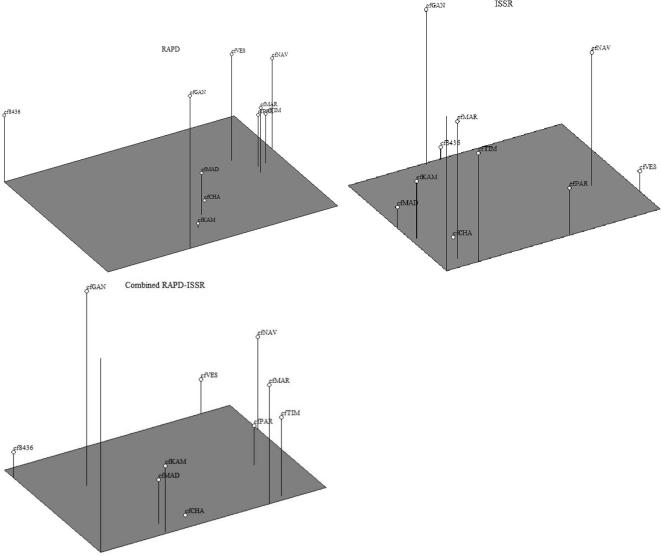
ISSR amplifications using 21 primers yielded total 119 scorable loci, of which 38 monomorphic loci (Table 4). The number of scorable bands for ISSR markers varied from 2 (SSR822) to 8 (ISSR807, ISSR823 and ISSR15). Highest number of bands (57) was obtained primer ISSR835, while only 16 bands were obtained by primers ISSR822. Out of 21 ISSR primers, four primers (ISSR830, ISSR845, ISSR4 and ISSR15) showed the 100% per cent polymorphism. Urena-Padilla et al. characterized Colletotrichum species causing strawberry by using RAPD and ISSR markers and they found variation at 40 putative genetic loci in this fungus [25]. The highest genetic similarity coefficient (0.93) was observed between cfCHA and cfMAD, which indicates that the genetic variability within this species was very low. C. falcatum genotypes cfVES and cfGAN represented a comparatively wide diversity with similarity coefficient of 0.73 (Table 5). The highest PIC value obtained with primer ISSR807 were of 0.849 and the lowest PIC value observed was 0.469 for primer ISSR822, with mean PIC value per primer was 0.767. The UPGMA dendrogram clearly indicated the one large cluster of six accessions and second with two accessions (cfGAN, cf436) situated with similarity matrix of 0.81 (Fig. 2). In these ten accessions cfNAV and cfVES were placed separately and do not fall into any cluster with similarity coefficient of 0.78. The existence of the diverse isolate cfGAN warrants extensive study with other molecular approaches. Further, to outline the genetic similarities between the C. falcatum accessions, data obtained from both the primers were combined to generate genetic diversity (Table 7). Dendrogram generated with combined data revealed three major clusters: I, II and III with 3, 5 and 2 accessions, respectively (Fig. 3). The Cluster I had three accessions cfNAV, cfVES and cfPAR, while Cluster III had only two accessions cfGAN and cf8436. Rest of the accessions were fall under major Cluster II. Similarity coefficient calculated from combined RAPD-ISSR data were ranged from 0.58 to 0.86 (Table 6). The results showed that cfCHA (Chalthan) and cfMAD (Madhi) were closely related accessions with the highest similarity index (0.86). On flip side, geographically distinct isolates cfNAV (Navsari) and cf8436 (Coimbatore) were most diverse accessions with lowest similarity index 0.58. PCA is one of the most useful statistical tools for screening multivariate data with significantly high correlation. The PCA plots of RAPD, ISSR and RAPD-ISSR combined markers was comparable to the cluster analysis obtained in UPGMA dendrogram (Fig. 4). The RAPD-based PCA revealed that the cfGAN and cf8436 accessions were located separately in the 3D PCA plot. Similarly ISSR markers based PCA revealed that the results are consistent with clusters obtained in ISSR dendrogram, indicating considerable genetic variability among C. falcatum accessions.
Table 4.
Numerical data as obtained from PCR amplification by ISSR primers among Colletotrichum falcatum accessions. (TL: Total Loci, ML: Monomorphic Loci, PL: Polymorphic Loci, %P: % Polymorphism, PIC: Polymorphic Information Content).
| Sr No | Primer | Sequence (5′-3′) | TL | ML | PL | % P | PIC |
|---|---|---|---|---|---|---|---|
| 1 | ISSR807 | AGAGAGAGAGAGAGAGT | 8 | 3 | 5 | 62.50 | 0.849 |
| 2 | ISSR822 | TCTCTCTCTCTCTCTCA | 2 | 1 | 1 | 50.00 | 0.469 |
| 3 | ISSR823 | TCTCTCTCTCTCTCTCC | 8 | 2 | 6 | 75.00 | 0.827 |
| 4 | ISSR830 | TGTGTGTGTGTGTGTGG | 7 | 0 | 7 | 100.00 | 0.804 |
| 5 | ISSR835 | AGAGAGAGAGAGAGAGYC | 7 | 4 | 3 | 42.86 | 0.848 |
| 6 | ISSR845 | CTCTCTCTCTCTCTCTRG | 5 | 0 | 5 | 100.00 | 0.776 |
| 7 | ISSR848 | CACACACACACACACARG | 7 | 1 | 6 | 85.71 | 0.829 |
| 8 | ISSR857 | ACACACACACACACACYG | 6 | 2 | 4 | 66.67 | 0.822 |
| 9 | ISSR859 | TGTGTGTGTGTGTGTGRC | 6 | 2 | 4 | 66.67 | 0.785 |
| 10 | ISSR860 | TGTGTGTGTGTGTGTGRA | 4 | 1 | 3 | 75.00 | 0.746 |
| 11 | ISSR873 | GACAGACAGACAGACA | 6 | 4 | 2 | 33.33 | 0.823 |
| 12 | ISSRG1 | GCTGCATCGGAGATAGGGAA | 4 | 2 | 2 | 50.00 | 0.702 |
| 13 | ISSRG9 | CAGCAGCGAGAAGTTTAGCA | 3 | 1 | 2 | 66.67 | 0.664 |
| 14 | ISSRG10 | CGGAGCTCCTATCATTCCAA | 6 | 3 | 3 | 50.00 | 0.759 |
| 15 | ISSR1 | AGCAGCAGCAGCAGCGA | 7 | 3 | 4 | 57.14 | 0.826 |
| 16 | ISSR2 | AGCAGCAGCAGCAGCGG | 4 | 2 | 2 | 50.00 | 0.728 |
| 17 | ISSR4 | AGCAGCAGCAGCAGCGC | 4 | 0 | 4 | 100.00 | 0.715 |
| 18 | ISSR12 | GTGTGTGTGTGTGTGT | 4 | 1 | 3 | 75.00 | 0.687 |
| 19 | ISSR14 | GTGTGTGTGTGTGTCT | 7 | 4 | 3 | 42.86 | 0.804 |
| 20 | ISSR15 | GTGTGTGTGTGTGTAT | 8 | 0 | 8 | 100.00 | 0.835 |
| 21 | ISSR19 | GCAGAGAGAGAGAGAGA | 6 | 2 | 4 | 66.67 | 0.798 |
| Total | - | 119.00 | 38 | 81 | 68.07 | - | |
| Average | - | 5.667 | 1.809 | 3.857 | 0.767 | ||
Table 5.
Dice’s similarity coefficient based on ISSR analysis in 10 Colletotrichum falcatum accessions.
| Accessions | cfNAV | cfVES | cfPAR | cfTIM | cfMAR | cfGAN | cfKAM | cfCHA | cfMAD | cf8436 |
|---|---|---|---|---|---|---|---|---|---|---|
| cfNAV | 1.00 | |||||||||
| cfVES | 0.78 | 1.00 | ||||||||
| cfPAR | 0.82 | 0.79 | 1.00 | |||||||
| cfTIM | 0.79 | 0.75 | 0.84 | 1.00 | ||||||
| cfMAR | 0.77 | 0.74 | 0.79 | 0.88 | 1.00 | |||||
| cfGAN | 0.79 | 0.73 | 0.77 | 0.80 | 0.81 | 1.00 | ||||
| cfKAM | 0.76 | 0.74 | 0.82 | 0.88 | 0.86 | 0.82 | 1.00 | |||
| cfCHA | 0.79 | 0.76 | 0.85 | 0.86 | 0.84 | 0.78 | 0.88 | 1.00 | ||
| cfMAD | 0.75 | 0.74 | 0.80 | 0.85 | 0.85 | 0.82 | 0.90 | 0.93 | 1.00 | |
| cf8436 | 0.78 | 0.77 | 0.79 | 0.78 | 0.76 | 0.83 | 0.83 | 0.85 | 0.84 | 1.00 |
Fig. 2.
UPGMA cluster analysis of 10 Colletotrichum falcatum accessions with Dice’s similarity coefficient of ISSR.
Table 7.
Genetic polymorphism among 10 Colletotrichum falcatum accessions as revealed by RAPD, ISSR and RAPD-ISSR analyses.
| Method | Number of genotypes | Mean Bands | Mean Monomorphic Bands | Mean Polymorphic Bands | % Polymorphism |
|---|---|---|---|---|---|
| RAPD | 10 | 4.47 | 0.87 | 3.60 | 80.60 |
| ISSR | 10 | 5.67 | 1.81 | 3.86 | 68.07 |
| RAPD-ISSR | 10 | 5.17 | 1.42 | 3.75 | 74.73 |
Fig. 3.
UPGMA cluster analysis of 10 Colletotrichum falcatum accessions with Dice’s similarity coefficient of RAPD and ISSR.
Table 6.
Dice’s similarity coefficient based on RAPD-ISSR combined analysis in 10 Colletotrichum falcatum accessions.
| Accessions | cfNAV | cfVES | cfPAR | cfTIM | cfMAR | cfGAN | cfKAM | cfCHA | cfMAD | cf8436 |
|---|---|---|---|---|---|---|---|---|---|---|
| cfNAV | 1 | |||||||||
| cfVES | 0.72 | 1 | ||||||||
| cfPAR | 0.75 | 0.71 | 1 | |||||||
| cfTIM | 0.74 | 0.68 | 0.79 | 1 | ||||||
| cfMAR | 0.70 | 0.66 | 0.72 | 0.84 | 1 | |||||
| cfGAN | 0.64 | 0.65 | 0.65 | 0.67 | 0.71 | 1 | ||||
| cfKAM | 0.61 | 0.63 | 0.72 | 0.76 | 0.75 | 0.76 | 1 | |||
| cfCHA | 0.65 | 0.67 | 0.75 | 0.76 | 0.75 | 0.69 | 0.80 | 1 | ||
| cfMAD | 0.63 | 0.67 | 0.70 | 0.76 | 0.75 | 0.77 | 0.83 | 0.86 | 1 | |
| cf8436 | 0.58 | 0.64 | 0.62 | 0.60 | 0.61 | 0.76 | 0.74 | 0.74 | 0.75 | 1 |
Fig. 4.
Three-dimensional plot of principal component analysis of Colletotrichum falcatum accessions based on RAPD, ISSR and combined (RAPD - ISSR) analysis.
4. Discussion
Although work on morphological, biochemical, pathogenic characterization of C. falcatum isolates from various region of India has been attempted extensively, studies on genetic diversity are scanty. Studies at the molecular level are essential to understand genetic diversity and relationships among accessions. In our previous study we have characterized these South Gujarat C. falcatum accessions on the basis of morphology and virulence pattern [16]. Since, the virulence assays are subject to ecological variations and sometimes leading to disease escape and misinterpretation. So, polymorphism or molecular diversity can be used to depict any discrepancy among isolates of loci, regardless of frequency. In present study, one of the major objectives was to determine intra-population genetic variation using RAPD and ISSR in C. falcatum accessions from South Gujarat region. Although simple and rapid but possibility of inconsistency with RAPD marker, we also carried out ISSR analysis as an independent and combined means to make accurate and specific characterization.
The inherent genetic variability in current study on C. falcatum isolates was apparent from the RAPD, ISSR and combined RAPD-ISSR profiles and the dendrogram showed they were separated unambiguously. It was noted that all primers tested were reliable and produced apparent amplification profiles with appropriate PCR condition. The genetic diversity among C. falcatum accessions based on RAPD and ISSR markers showed a correlation between genetic and geographical distribution which is similar to research presented by Ratanacherdchai et al. [26]. The clustering of C. falcatum accessions proved the suitability of RAPD and ISSR in detecting allele characteristic of C. falcatum genotypes. The percent of polymorphism ranged from 42.86% to 100% and 33.33% to 100% in RAPD and ISSR respectively. Our findings are in accordance with the research presented by Kumar Narender who reported only 95% polymorphism in thirteen C. falcatum accessions with RAPD markers [9], [27]. Another study conducted on standard accessions with 40 RAPD on Cf01, Cf02, Cf03, Cf07, Cf08 and Cf09 accessions by Suman et al. [28] markers found 78.6% polymorphism which we found with only 15 primers. Though, RAPDs cover the whole genome for amplification, ISSR amplifies the regions between two microsatellites, but the average PIC of both RAPD and ISSR marker systems was higher and almost analogous. In present study average PIC recorded for RAPD and ISSR marker system were 0.705 (range = 0.489–0.849) and 0.767 (range = 0.469–0.849) and 8 out of 15 RAPD primers and 12 out of 21 ISSR primers exceeded this value, respectively. The higher values of PIC recorded in this study indicated the power of these selected primers for discrimination genetic diversity of the C. falcatum accessions. Furthermore the relatively low similarity coefficients in present study among C. falcatum accessions from different sugarcane cultivars based on RAPD and ISSR analysis provides additional support for genetic variability between them. In present work, data from combination of RAPD and ISSR marker systems has been analysed to estimate the genetic relatedness between C. falcatum accessions which revealed that the degree of genetic diversity was relatively higher with broad gene pool. This might be due to the frequent genetic changes and sexual recombination increases genotype diversity in populations. This high level of variability in C. falcatum is challenge for the breeders to breed for red rot resistance hence information about the genetic variability in each region is the basis for resistance breeding programmes. This proves that although red rot disease caused by C. falcatum is very old problem, continues research is requires on this pathogen.
5. Conclusion
There were differences in the numbers of RAPD and ISSR primer loci produced by PCR amplification, which indicates differences in sequence composition of C. falcatum genome. Although these molecular techniques would help in characterization molecular analysis needs to be further validated with advanced markers like SCAR, APC and SNPs to get a précised and comprehensive results on existing variability of red rot isolates, which can help to improve sugarcane red rot resistance and varietal deployment approach.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Barber CA. Madras Department of Agriculture Bulletin 2; 1901. p. 181–93.
- 2.Alvi A., Iqbal J., Shah A., Pan Y. Pak J Bot. 2008;40:1419–1425. [Google Scholar]
- 3.Afghan S, Hussnain Z. Annual Report, Shakarganj Sugar Research Institute, Jhang; 2006.
- 4.Saksena P., Vishwakarma S.K., Tiwari A.K., Singh A., Kumar A. J Plant Prot Res. 2013;53:37–41. [Google Scholar]
- 5.Bharti A.K.Y.P., Sharma D.D.K., Singh S.K., Shukla D.N. Afr J Microbiol Res. 2014;8:1040–1049. [Google Scholar]
- 6.Malathi P., Viswanathan R., Sundar A.R., Prakasam N., Padmanaban P., Jothi R., Devi S.R., Poongothai M. Sug Cane Intern. 2010;28:47–52. [Google Scholar]
- 7.Srinivasan K, Chenulu V. In: Proc. 9th Cong. ISSCT, vol. 1; 1956. p. 1097–107.
- 8.Viswanathan R. Plant disease: Red rot of sugarcane, Anmol Publications; 2010.
- 9.Jhang T., Kumar Narender, Sharma Tilak Raj. J Phytopathol. 2011;159:260–267. [Google Scholar]
- 10.Mahmodi F., Kadir J., Puteh A., Pourdad S., Nasehi A., Soleimani N. Plant Pathol J. 2014;30:10. doi: 10.5423/PPJ.OA.05.2013.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva A., Savi D., Gomes F., Gos F., Silva G., Glienke C. Eur J Plant Pathol. 2017;147:731–748. [Google Scholar]
- 12.Williams J.G., Kubelik A.R., Livak K.J., Rafalski J.A., Tingey S.V. Nucl Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganie S.H., Upadhyay P., Das S., Sharma M.P. Plant Gene. 2015;4:83–99. doi: 10.1016/j.plgene.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menzies J., Bakkeren G., Matheson F., Procunier J., Woods S. Phytopathology. 2003;93:167–175. doi: 10.1094/PHYTO.2003.93.2.167. [DOI] [PubMed] [Google Scholar]
- 15.Zietkiewicz E., Rafalski A., Labuda D. Genomics. 1994;20:176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]
- 16.Prittesh P., Amaresan N., Rushabh S., Krishnamurthy R., Bhasker V. J Plant Dis Prot. 2016;123:273–277. [Google Scholar]
- 17.Raeder U., Broda P. Let Appl Microbiol. 1985;1:17–20. [Google Scholar]
- 18.Dice L.R. Ecology. 1945;26:297–302. [Google Scholar]
- 19.Nei M., Li W.-H. Proc Nat Acad Sci. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khaleghi E., Sorkheh K., Chaleshtori M.H., Ercisli S. 3 Biotech. 2017;7:71. doi: 10.1007/s13205-017-0669-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Guerber J.C., Liu B., Correll J.C., Johnston P.R. Mycologia. 2003;95:872–895. [PubMed] [Google Scholar]
- 22.Mesquita A, Faleiro F, De Paula T, Ragagnin V, Moreira M, de Barros E. Fitopatolog. Brasil. 1998;23:58-61.
- 23.Talhinhas P., Sreenivasaprasad S., Neves-Martins J., Oliveira H. Phytopathology. 2002;92:986–996. doi: 10.1094/PHYTO.2002.92.9.986. [DOI] [PubMed] [Google Scholar]
- 24.Thottappilly G., Mignouna H., Onasanya A., Abang M., Oyelakin O., Singh N. Afr Crop Sci J. 1999;7:195–205. [Google Scholar]
- 25.Ureña-Padilla A., MacKenzie S., Bowen B., Legard D. Phytopathology. 2002;92:1245–1252. doi: 10.1094/PHYTO.2002.92.11.1245. [DOI] [PubMed] [Google Scholar]
- 26.Ratanacherdchai K., Wang H.-K., Lin F.-C., Soytong K. Afr J Microbiol Res. 2010;4:076–083. [Google Scholar]
- 27.Madan V., Mandal B., Ansari M., Srivastava A., Soni N., Solomon S., Agnihotri V. Sug Cane Int. 2000:5–8. [Google Scholar]
- 28.Suman A., Lal S., Shasany A., Gaur A., Singh P.W. J Microbiol Biotechnol. 2005;21:1135–1140. [Google Scholar]