Abstract
Cellulase producing bacteria were isolated from both soil and ward poultry, using CMC (carboxymethylcellulose) agar medium and screened by iodine method. Cellulase activity of the isolated bacteria was determined by DNS (dinitrosalicylic) acid method. The highly cellulolytic isolates (BTN7A, BTN7B, BMS4 and SA5) were identified on the basis of Gram staining, morphological cultural characteristics, and biochemical tests. They were also identified with 16S rDNA analysis. The phylogenetic analysis of their 16S rDNA sequence data showed that BTN7B has 99% similarity with Anoxybacillus flavithermus, BMS4 has 99% similarity with Bacillus megaterium, SA5 has 99% homology with Bacillus amyloliquefaciens and BTN7A was 99% similar with Bacillus subtilis. Cellulase production by these strains was optimized by controlling different environmental and nutritional factors such as pH, temperature, incubation period, different volumes of media, aeration rate and carbon source. The cellulase specific activity was calculated in each case. In conclusion four highly cellulolytic bacterial strains were isolated and identified and the optimum conditions for each one for cellulase production were determined. These strains could be used for converting plant waste to more useful compounds.
Keywords: Cellulase producing bacteria, Isolates, Optimization, 16S rDNA
1. Introduction
Cellulose is the most abundant biomass and most dominating agricultural waste on earth [36]. It is a polymer chain of glucose units connected by β-1, 4 linkages. Cellulose waste is a huge renewable bioresource produced by the photosynthetic process [15], [39]. It has a high potential for bioconversion to important bioproducts such as ethanol. The ability to obtain cheap ethanol will depend on the successful identification of novel cellulase producing strains [21]. More research activities are needed to obtain novel cellulases with hyper activity on pretreated biomass substrates by screening and sequencing novel classes of microorganisms and engineering cellulases with improved industrial qualities and by identifying proteins that can stimulate cellulases [38].
One of the requirements of the biological conversion of lignocellulosic wastes into industrial products is the use of cellulolytic and hemicellulolytic enzymes [18]. Microorganisms that can produce cellulase enzymes (cellulolytic microorganisms) can degrade cellulose. Fungi and bacteria isolated from soil secrete several enzymes which degrade lignocellulosic biomass [7]. These enzymes are commonly produced by some bacterial genera such as Cellulomonas, Pseudomonas [26] Bacillus, and Micrococcus [14] and fungi [32] that are widely used now in industrial applications. Cellulosic biomass hydrolysis requires successive action of three types of enzymes, which are cellobiohydrolase, endoglucanase carboxymethyl cellulase (CMCase), and β-glucosidases [5].
Scientific research efforts try to improve the hydrolysis process in an economical way. There are different factors which affect bacterial growth and enzyme production such as pH, temperature, aeration, incubation period, inoculum size and carbon source. The air pollution in Cairo is a matter of serious concern. One of the most notable sources of pollution is open-air waste-burning. A black cloud over Cairo has been noticed each year for many decades during harvest time where farmers burn leftover rice husks at the end of the growing season. The overall aim of this research was to produce more healthy air by converting plant wastes to economical products. Any bacterial strain has its own identity and has to be investigated for its optimum culture conditions for its best activity for the application purpose. On the other hand, economical high-efficient bacterial strains are not available unless paying their Know-How. As an initial step this article aimed to isolate indigenous bacterial strains efficient in cellulose degradation and maximize their activity using different nutrients and culture condition.
2. Materials and methods
2.1. Sample collection
Soil and ward poultry samples were collected from Beni-Suef and El-Sharkia Governorates, Egypt, and stored under sterile conditions at 4 °C until bacterial isolation.
2.2. Media
Luria-Bertani agar medium (LB) [4] was used for different physiological and molecular biology procedures. Bushnell Haas medium (BHM) [8] was used for isolation of cellulose hydrolytic bacteria after amendment with carboxymethylcellulose (CMC) as the sole carbon source.
2.3. Isolation and screening of cellulases producing bacteria
Ten grams of each soil or ward poultry sample were diluted with 90 ml saline solution in Erlenmeyer flasks. Isolation of bacteria was carried out on BHM medium using pour plate method [17], and morphologically different colonies were selected, purified on LB medium and kept at 4 °C for further study. Screening for cellulase producers was done on CMC agar medium. The purified bacterial isolates were inoculated on CMC plates and incubated at 37 ± 2 °C or 55 ± 2 °C for 24 h and then flooded with Gram's iodine for 3–5 min [19]. After incubation, a bacterial colony with a clearing zone, an indication of cellulase activity, was selected.
2.4. Cellulase activity assay
Cellulase activity was determined using (3,5-dinitrosalicylic acid) DNS according to [25]. The selected bacterial isolates were grown in LB broth medium for 24 hrs at 37 ± 2 °C or 55 ± 2 °C, and then centrifuged at 13,000 rpm for 5 min. One ml of the supernatant (enzyme solution) was mixed with 1 ml of CMC solubilized in phosphate buffer (1%) and incubated at 37 ± 2 °C or 55 ± 2 °C for 30 min under shaking (120 rpm). One ml dinitrosalicylic (DNS) acid reagent was added and the mixture was boiled for 5 min; then, the absorbancy was measured at 540 nm. One unit of enzyme activity was defined as µmol substrate consumed or product formed per minute.
Total protein concentration was determined according to [6] and absorbance was measured at 595 nm. Different concentrations of Bovine serum albumin (BSA) were conducted for plotting standard curve according to Lowry et al.’s method [24]. The highest isolates for cellulase production were selected for further studies for identification and optimization. Cellulase specific activity was calculated using the following equation:
Cellulase activity under various conditions (such as temperature, pH value, aeration rate, incubation period and carbon source) was measured taking cellulase activity of the control as 100% (in the absence of any factors). Data are presented as mean ± standard error for triplicate.
2.5. Identification of the bacterial isolates
2.5.1. Biochemical characterization
Bacterial isolates were identified using Bergey's Manual of Determinative Bacteria [20]. The identification was carried out using morphological characteristics and biochemical tests such as Gram stain, endospore stain, starch hydrolysis, citrate utilization, nitrate reduction, carbohydrate fermentation, Vogas Proskauer and catalase production.
2.5.2. Molecular identification by 16S rDNA sequencing
Bacterial genomic DNA was extracted using the method presented by Ostuki et al. [27]. Selected isolates were grown overnight on LB agar. A loopful of culture was resuspended in 100 µl of sterile distilled water and vortexed well, incubated at 95 °C for 20 min and then vortexed, and incubated on ice for 10 min. Centrifugation was done for 10 min at 10,000 rpm (Microcentrifuge Sigma 215k, USA).
The PCR reaction mixture was as follows: 12.5 μl Master Mix, 1.5 μl DNA, 0.75 μl universal 16S reverse primer (1492R)(GGTTACCTTGTTACGACTT), 0.75 μl 16S universal forward primer (8F) (AGAGTTTGATCATGGCTCAG) and water free nuclease up to 25 μl.
PCR amplification was performed in a gradient thermal cycler Amplitronyx™ (NYXTECHNIK, UK). Different annealing temperatures were tested (from 56 to 46 °C). The Best annealing temperature was one minute at 48 °C where it produced only one high intense DNA ampilicon. The optimum program was one cycle at 95 °C for five minutes, and then 35 cycles were performed as follows: two minutes at 95 °C for denaturation, one minute at 48 °C for annealing, four minutes at 72 °C for elongation, and then 20 min at 72 °C for final extension. The reaction mixtures were held at 4 °C until used. The obtained PCR products were sent to Macrogen, Korea, for sequencing. Nucleotide sequence was then compared to available data in GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.6. Effect of temperature on bacterial growth
Cultures of selected bacterial isolates corresponding to 0.01 OD620 were inoculated in 100 ml Erlenmeyer flasks containing 20 ml LB medium and incubated under shaking (120 rpm) for 24 h at different temperatures (25, 37, 45 and 55 °C). After incubation the absorbance was measured at 620 nm against a blank (uninoculated LB broth) using a Spectronic 21 spectrophotometer (Bauch and Lomb, New York, USA).
2.7. Optimization of cellulase production
2.7.1. pH value
Four pH values (3.0, 5.0, 7.0 and 9.0) were tested to select the optimum pH for cellulase production. Twenty ml of LB medium in 100 ml conical flasks were inoculated with overnight bacterial culture to OD620 0.01 and incubated for 24 h at 37 ± 2 °C for BMS4, BTN7A and SA5 or at 55 ± 2 °C for BTN7B under shaking (120 rpm).
2.7.2. Incubation period
This factor was studied by inoculating 20 ml of LB medium (pH 7) with overnight bacterial culture to OD620 0.01 and incubation at 37 ± 2 or 55 ± 2 °C for different periods (24 and 48 hr) on a rotary shaker (120 rpm).
2.7.3. Medium ratio
Different volumes (10, 20, 30 and 40 ml) of LB medium were distributed into 100 ml Erlenmeyer flasks and inoculated with overnight bacterial culture to OD620 0.01. The flasks were incubated at 37 ± 2 or 55 ± 2 °C for 24 h on a rotary shaker (120 rpm).
2.7.4. Aeration rate
A one hundred ml Erlenmeyer flask containing 20 ml LB broth medium was inoculated with culture of overnight bacterial isolates to OD620 0.01 and then incubated in static or with shaking at 120 rpm for 24 hrs at 37 ± 2° or 55 ± 2 °C.
2.7.5. Carbon sources
Different carbon sources (glucose, sucrose, lactose, mannitol, cellobiose, cellulose, and CMC) alone and in combination were used in Bushnell Haas Medium (BHM) [8]. The medium was inoculated with overnight bacterial culture to OD620 0.01 and incubated for 24 h at 37 ± 2 °C /or 55 ± 2 °C.
3. Results and discussion
Due to the importance of cellulolytic microorganisms in biofuel production, several research groups are interested in isolation of many cellulase producing strains such as Anoxybacillus flavithermus [13]; Bacillus subtilis [29] and Bacillus amyloliquefaciens [33]. The present studies aimed to isolate novel strains with highly level of cellulase producing abilities.
3.1. Isolation and identification of cellulolytic microorganisms
In this study, fifty four bacterial cultures were isolated from soil and ward poultry according to their cellulase productivity. All strains were Gram positive, spore formers and showed different reactions for biochemical tests. Among the 54 isolates, four isolates named SA5, BMS4, BTN7A and BTN7B, showed high cellulase activity. SA5 was isolated from a ward poultry sample, while BMS4, BTN7A and BTN7B were obtained from soil samples. Table 1 shows the biochemical characteristics of the four promising isolates. The obtained results indicate that these isolates belong to the Genus Bacillus.
Table 1.
Biochemical characteristics for the identification of selected isolates.
| Biochemical test | Isolate code |
|||
|---|---|---|---|---|
| BTN7A | SA5 | BMS4 | BTN7B | |
| Gram stain | + | + | + | + |
| Cell shape | B | B | B | B |
| Endospore stain | + | + | + | + |
| Oxygen requirements | F | F | F | F |
| Motility test | − | − | − | + |
| Catalase test | + | + | + | + |
| Starch hydrolysis | + | + | + | + |
| Citrate utilization | − | − | − | − |
| Methyl red | − | + | + | + |
| Vogas Proskauer | + | + | − | − |
| Nitrate reduction | ||||
| NH3 | − | − | − | − |
| NO2 | − | − | − | − |
| NO3 | + | + | + | + |
| Carbohydrates fermentation | ||||
| Glucose | − | − | A | A |
| Lactose | − | − | − | − |
| Mannitol | − | − | − | A |
| Arabinose | − | − | − | − |
| Growth at 55 °C | − | − | − | + |
| 6.5% NaCl growth | + | + | + | − |
F = Facultative anaerobes, A = Acid, B = Bacilli.
According to previous study suggesting that, 16S rDNA sequencing is an accurate method for species identification and distinguishing between closely related bacterial species [3], therefore, the selected isolates were further analyzed by 16S rDNA sequencing and submitted to GenBank with the accession numbers KC438369, KC429572, KC438368 and KC429571. The phylogenetic analysis of their 16S rDNA sequences according to the available data in NCBI indicates that BTN7A shows 99% homology with B. subtilis subsp. subtilis, BTN7B has 99% similarity with A. flavithermus WK1, BMS4 has 99% identity to B. megaterium, and SA5 has 99% homology with B. amyloliquefaciens subsp. Plantarum, the data are summarized in Table 2.
Table 2.
Isolates accession number and their sequence similarities based on 16SrDNA.
3.2. Effect of temperature on bacterial growth
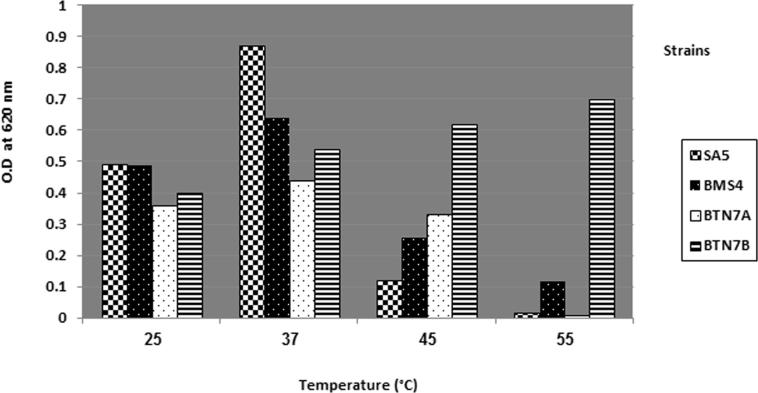
Data shown in Fig. 1 indicate that the optimum growth temperature of SA5, BMS4 and BTN7A was 37⁰C while for BTN7B optimum growth was achieved at 55 °C. The thermal stability of cellulase produced from thermostable A. flavithermus BTN7B may be used as potent industrial applications.
Figure 1.
Effect of different temperatures on growth.
3.3. Optimization of cellulase production
Factors affecting cellulase production were optimized for the selected bacterial isolates.
3.3.1. Effect of different pH on cellulase production
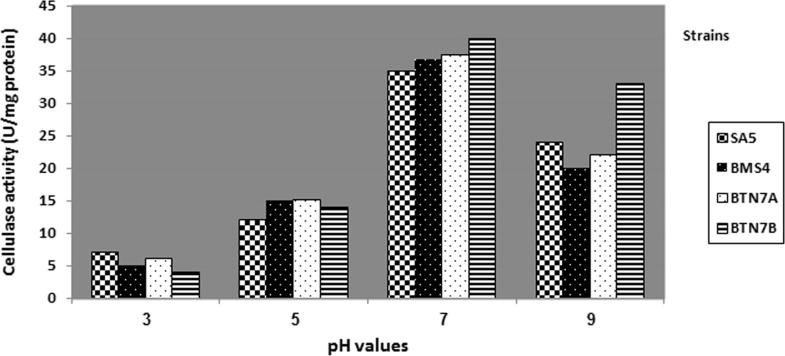
Fig. 2 reveals that the minimum cellulase production was detected at pH 3 for all selected bacterial isolates: B. megaterium BMS4, B. subtilis BTN7A, B. amyloliquefaciens SA5 and A. flavithermus BTN7B, while the optimum cellulase production was at pH 7 for all strains tested. These results are in accordance with the previous studies on different strains of Bacillus spp. [14]; B. subtilis [2], [9], and [23], B. megaterium [34] and B. amyloliquefaciens DL-3 [33]. In contrast, to the previous work [30], it is observed that the maximum cellulolytic activities of Bacillus circulans and B. megaterium were at pH 8.0, and the present data were in agreement with other studies stated that most bacterial enzymes show optimal activity between pH 6 and 8 [10], [28].
Figure 2.
Cellulase activity of selected strains growing at different pH values.
3.3.2. Effect of incubation period on cellulase production
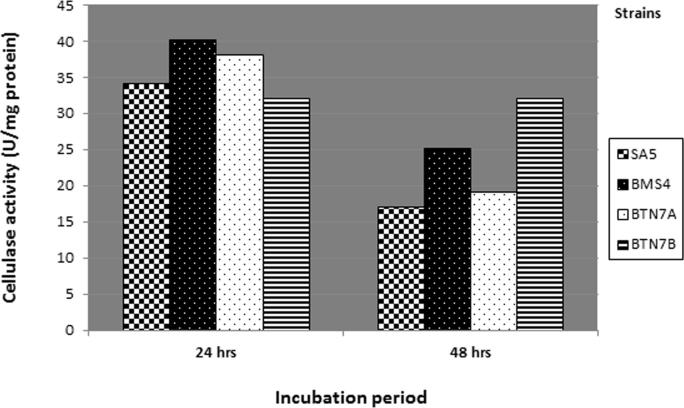
Results in Fig. 3 show that the maximum productivity of cellulase was achieved after 24 h of incubation and decreased after 48 h for B. amyloliquefaciens SA5 (34 U/mg), B. subtilis BTN7A (38 U/mg) and B. megaterium BMS4 (40 U/mg), while A. flavithermus BTN7B showed the same productivity (32 U/mg) after 24 and 48 h.
Figure 3.
Cellulase activity of selected strains growing on LB at different incubation periods.
The current work indicates that the enzyme production decreased, as the incubation time exceeds the optimum period, and the explanation of these results has been previously discussed by Haq et al. [12] who suggested that a decrease in enzymatic activity with increasing incubation time may be due to the depletion of nutrients and production of other by-products in the fermentation medium, and also Ariffin et al. [1], found that depletion of nutrients in the medium causes bacterial stress that result in inactivation of enzyme secretion. In contrast to the previous observation, the thermostable cellulases from A. flavithermus BTN7B show optimal activity after 48 h in agreement with Verma et al. [37] who reported that thermophilic Bacillus reaches its maximum activity after 48 h.
3.3.3. Effect of shaking on cellulase production
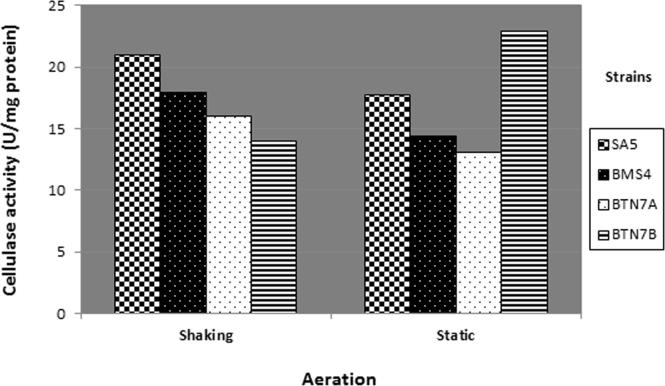
The effect of shaking on cellulase production of selected strains is presented in Fig. 4. The obtained results reveal that incubation of B. megaterium BMS4, B. subtilis BTN7A, and B. amyloliquefaciens SA5 under shaking conditions gave more cellulase activity than static incubation. In contrast to this finding, A. flavithermus BTN7B produced more cellulase in the static state as compared with shaking. Previous studies explain similar result as agitation speed is an important factor which governs the dissolved oxygen level in the culture broth that affects cell growth of cellulase producing microorganisms [16]. However, higher agitation speed has been shown to inhibit cellulase activity [16], [22].
Figure 4.
Cellulase activity of selected strains growing static and under shaking incubation.
3.3.4. Effect of medium volume on cellulase production
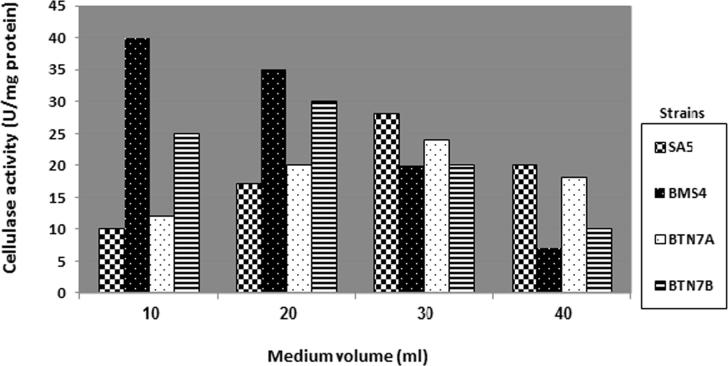
The results shown in Fig. 5 demonstrate the effect of different volumes of LB broth medium (10, 20, 30 and 40 ml) on cellulase activity. Optimum cellulase production varies from one strain to another. B. subtilis BTN7A and B. amyloliquefaciens SA5 produced high levels of enzyme at 30 ml LB (3: 7 medium: air). However, the maximum enzyme productivity of B. megaterium BMS4 was at 10 ml LB (1:9 medium:air) and A. flavithermus BTN7B shows optimum production at 20 ml LB (2:8 medium:air).
Figure 5.
Cellulase activity of selected strains growing in different volumes of medium.
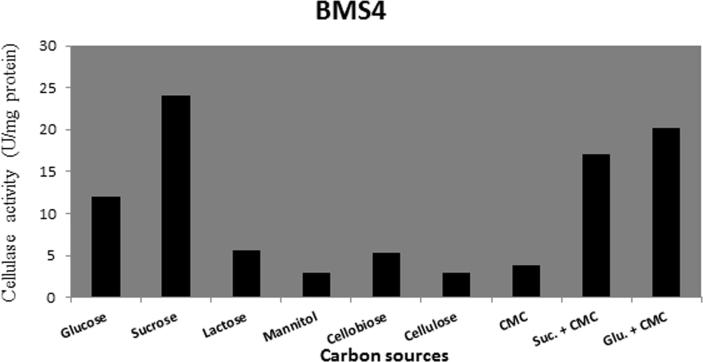
3.3.5. Effect of different carbon sources on cellulase production
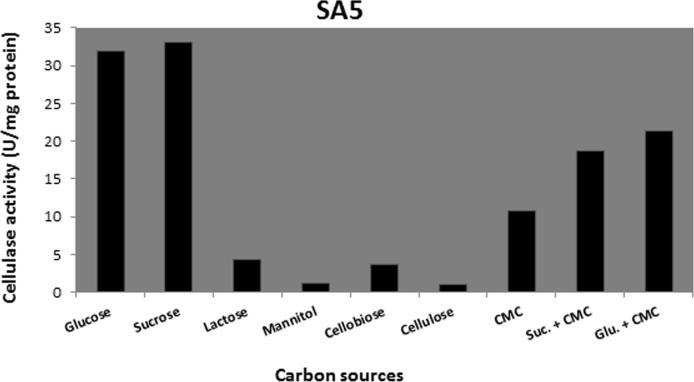
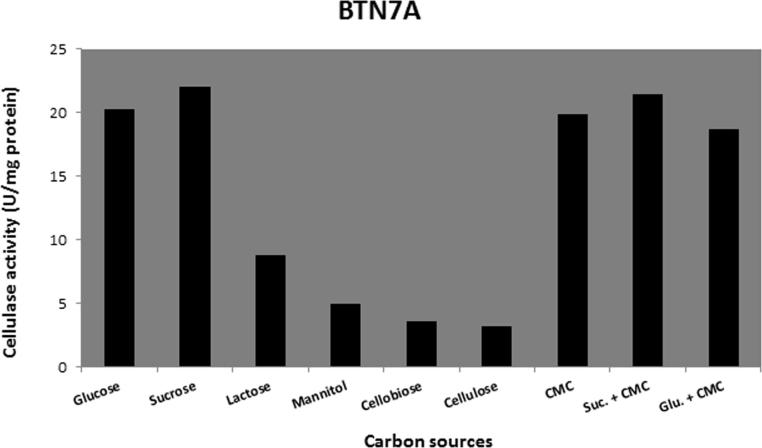
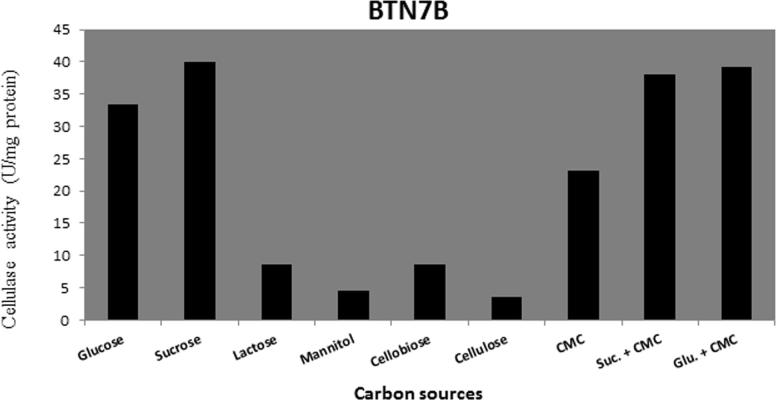
Different carbon sources (glucose, sucrose, lactose, mannitol, cellobiose, cellulose, CMC sucrose + CMC, glucose + CMC) were tested for their effects on cellulase production. Data present in Figure 6, Figure 7, Figure 8, Figure 9 show that sucrose is the best sole source of carbon for cellulase production where cellulase activity was 33 U/mg for B. amyloliquefaciens SA5, 22 U/mg for B. subtilis BTN7A, 24 U/mg for B. megaterium BMS4 and 40 U/mg for A. flavithermus BTN7B.
Figure 6.
Cellulase activity of SA5 strain growing on different carbon sources.
Figure 7.
Cellulase activity of BTN7A strain growing on different carbon sources.
Figure 8.
Cellulase activity of BTN7B strain growing on different carbon sources.
Figure 9.
Cellulase activity of BMS4 strain growing on different carbon sources.
The best carbon source for cellulase production obtained in this study was sucrose. Our results did not agree with those of Saraswati et al. [31] who reported that the best carbon source for optimum cellulase production of B. subtilis isolated from cow dung was lactose. Teodoro and Martins [35] found that maltose served as the best carbon source for Bacillus sp. However, Deka et al. [11] reported that CMC plays a significant role in cellulase production by Bacillus sp. The optimum carbon source differs from research work to another; this may be due to the tested strain, carbon sources or isolation environment.
4. Conclusions
Soil contains a variety of cellulolytic bacteria many of which have not been isolated. We isolated four strains with high cellulase activity and characterized them by biochemical tests and on the basis of 16S rDNA. Isolates were identified as B. amyloliquefaciens SA5, B. subtilis BTN7A, B. megaterium BMS4 and A. flavithermus BTN7B. Cellulase production was optimized for each strain. The optimum conditions for B. amyloliquefaciens SA5 and B. subtilis BTN7A, were at pH 7, under shaking conditions, at ratio 3:7 media to air, after 24 h incubation time and sucrose used as a sole carbon source. However, for B. megaterium BMS4 were at pH 7, under shaking conditions, at ratio 1:9 of media to air, after 24 h of incubation time and sucrose used as a sole carbon source and for A. flavithermus BTN7B were at pH 7, under static conditions, at ratio 2:8 media to air, after 24 or 48 h of incubation time and sucrose used as a sole carbon source.
The four strains can be used in various industrial applications using these optimized conditions. Further studies with large scale culture are needed, as well as the optimization of other parameters such as inoculum size, presence of inducers and medium additives.
Acknowledgments
We are very grateful to Dr. Pete Chandrangus, Microbiology Department, Cornell University, Ithaca NY 14853, USA, for his help in correcting the language of this article. This work was supported by Microbial Genetics Department, National Research Centre, Dokki, Giza, Egypt.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Ariffin H., Abdullah N., Umikalsom M.S., Shirai Y., Hassan M.A. Int. J. Eng. Technol. 2006;3:47–53. [Google Scholar]
- 2.Bagudo A.I., Argungu A.U., Aliero A.A., Suleiman S., Kalpana S. Int. J. Modern Cell. Mol. Biol. 2014;3(1):1–9. [Google Scholar]
- 3.Basavaraj I.P., Shivasharan C.T., Kaliwal B.B. Int. J. Curr. Microbiol. Appl. Sci. 2014;3(5):59–69. [Google Scholar]
- 4.Bertani G. J. Bacteriol. 1952;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhat M.K. Biotechnol. Adv. 2000;18(5):355–383. doi: 10.1016/s0734-9750(00)00041-0. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M.M. Annal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 7.Bruce T., Martinez I.B., Maia Neto O., Vicente A.C., Kruger R.H., Thompson F.L. Microbial Ecol. 2010;60:840–849. doi: 10.1007/s00248-010-9750-2. [DOI] [PubMed] [Google Scholar]
- 8.Bushnell L.D., Haas H.F. J. Bacteriol. 1941;41:653. doi: 10.1128/jb.41.5.653-673.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chundakkadu K. Bioresour. Technol. 1998;69:231–239. [Google Scholar]
- 10.Daniel R.M., Peterson M.E., Danson M.J. Biochem. J. 2010;425(2):353–360. doi: 10.1042/BJ20091254. [DOI] [PubMed] [Google Scholar]
- 11.Deka D., Bhargavi P., Sharma A., Goyal D., Jawed M., Goyal A. Enzyme Res. 2011;2011:1–8. doi: 10.4061/2011/151656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haq I.U., Hameed K., Shahzadi M.M., Javed S.A., Qadeer M.A. Int. J. Bot. 2005;1:19–22. [Google Scholar]
- 13.Ibrahim A.S.S., EL-Diwany A.I. Austr. J. Basic Appl. Sci. 2007;1:473–478. [Google Scholar]
- 14.G. Immanuel, R.D. Hanusha, P. Prema, A. Palavesam, 3(1) (2006) 25–34.
- 15.Jarvis M. Nature. 2003;426:611–612. doi: 10.1038/426611a. [DOI] [PubMed] [Google Scholar]
- 16.Jo K.I., Lee Y.J., Kim B.K. Biotech. Bioproc. Eng. 2008;13(2):182–188. [Google Scholar]
- 17.Johnson L.F., Curl E.A., Bond J.H., Fibourg H.A. Burgess Minneapolis; 1959. Methods of Studying Soil Microflora and Plant Disease Relationships. [Google Scholar]
- 18.Jourdier E., Ben Chaabane F., Poughon L., Larroche C., Monot F. Chem. Eng. Trans. 2012;27:313–318. [Google Scholar]
- 19.Kasana R.C., Salwan R., Dhar H., Dutt S., Gulati A.A. Curr. Microbiol. 2008;57:503–507. doi: 10.1007/s00284-008-9276-8. [DOI] [PubMed] [Google Scholar]
- 20.Krieg N.R., Holt J.G. second ed. Williams & Wilkins; Baltimore, London: 1986. Bergey's Manual of Systematic Bacteriology. [Google Scholar]
- 21.Lee S.M., Koo Y.M. J. Microbiol. Biotechnol. 2001;11:229–233. [Google Scholar]
- 22.Lejeune R., Baron G.V. Appl. Micro. Biotech. 1995;43(2):249–258. [Google Scholar]
- 23.Li W., Zhang W.W., Yang M.M., Chen Y.L. Mol. Biotechnol. 2008;40:195–201. doi: 10.1007/s12033-008-9079-y. [DOI] [PubMed] [Google Scholar]
- 24.Lowry O.H., Rosenbrough N.J., Farr A.L., Randall R.J. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Miller G.L. Anal. Chem. 1959;31:426–428. [Google Scholar]
- 26.Nakamura K., Kppamura K. J. Ferm. Tech. 1982;60(4):343–348. [Google Scholar]
- 27.Ostuki K., Guayeurus T.V., Cx A., Vicent P. Mem. Inst. Oswaldo Cruz. 1997;92:107–108. [Google Scholar]
- 28.Oyeleke S.B., Oduwole A.A. J. Microbiol. Res. 2009;3(4):143–146. [Google Scholar]
- 29.Rahna K.R., Divya J., Balasaravanan T. J. Micro. Biotech. Food Sci. 2013;2(6):2383–2386. [Google Scholar]
- 30.Sangbriba R.U., Duan C.J., Tang J.L. Bioresour. Technol. 2006;14:2727–2733. [Google Scholar]
- 31.Saraswati B., Ravi K.M., Mukesh Kumar D.J., Balashanmugam P., Bala Kumaran M.D., Kalaichelvan P.T. Appl. Sci. Res. 2012;4(1):269–279. [Google Scholar]
- 32.Shin C.S., Lee J.P., Lee J.S., Park S.C. Appl. Biochem. Biotechnol. 2000;84:237–245. doi: 10.1385/abab:84-86:1-9:237. [DOI] [PubMed] [Google Scholar]
- 33.Shuchi S., Moholkar V.S., Goyal A. ISRN Microbiol. 2013;13:1–7. doi: 10.1155/2013/728134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shumaila S., Saira A., Rehman Abdul. Pakistan J. Zool. 2013;45(6):1655–1662. [Google Scholar]
- 35.Teodoro C.E.S., Martins M.L.L. Braz. J. Microbiol. 2000;4:298–302. [Google Scholar]
- 36.Tomme P., Warren R.A.J., Gilkes N.R. Advance Micro. Phys. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 37.Verma V., Alpika V., Akhilesh K. Adv. Appl. Sci. Res. 2012;3(1):171–174. [Google Scholar]
- 38.Wilson D.B. Curr. Opin. Biotechnol. 2009;20:295–299. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y.H.P., Lynd L.R. Biotech. Bioeng. 2004;88(7):797–824. doi: 10.1002/bit.20282. [DOI] [PubMed] [Google Scholar]