Abstract
Abundant, low prices and a highly reduced nature make glycerol to be an ideal feedstock for the production of reduced biochemicals and biofuels. Escherichia coli has been paid much attention as the platform of microbial cell factories due to its high growth rate (giving higher metabolite production rate) and the capability of utilizing a wide range of carbon sources. However, one of the drawbacks of using E. coli as a platform is its mixed metabolite formation under anaerobic conditions. In the present study, it was shown that ethanol could be exclusively produced from glycerol by the wild type E. coli, while d-lactic acid could be exclusively produced from glucose by pflA.cra mutant, where the glucose uptake rate could be increased by this mutant as compared to the wild type strain. It was also shown that the growth rate is significantly reduced in pflA.cra mutant for the case of using glycerol as a carbon source due to redox imbalance. The metabolic regulation mechanisms behind the fermentation characteristic were clarified to some extent.
Abbreviations: AcCoA, acetyl-coenzyme A; ATP, adenosine triphosphate; cAMP, cyclic adenosine monophosphate; cAMP-Crp, cAMP receptor protein; DCW, dry cell weight; DHA, dihydroxyacetone; GAPDH, -glyceraldehyde-3-phosphate dehydrogenase; GDH, glutamate dehydrogenase; KH2PO4, potassium dihydrogen phosphate; KOH, potassium hydroxide; LB, Luria Bertani; LDH, lactate dehydrogenase; M9, type of minimal media; MgSO4, magnesium sulfate; NAD+, nicotinamide adenine dinucleotide; NADH, reduced form of nicotinamide adenine dinucleotide; NaCl, sodium chloride; Na2HPO4, sodium phosphate; NaOH, sodium hydroxide; OD, optical density; OAA, oxaloacetic Acid; PEP, phosphoenolpyruvic acid; Pfl, pyruvate formatelyase; PTS, phospho-transferase system; PYR, pyruvate; PEP, phosphoenol pyruvate; TCA, tri-carboxylic acid; UV, ultra violet
Keywords: Glycerol, Glucose, Biofuel, Escherichia coli, pflA.cra mutant, Ethanol production
1. Introduction
Present global energy requirements are fulfilled primarily via fossil fuel combustion, and thus the world is dependent on a non-renewable resource for its energy needs [10]. The increasing economic growth and prosperity have been accelerated worldwide with an increasing demand on energy mostly generated from fossil fuels. This has brought the rapid global warming caused by the emission of green-house gases such as CO2, resulting in a disastrous climate change, where this problem is becoming crucial. Currently the importance of an alternative energy source has become even more necessary, not only due to the expected depletion of the limited fossil fuel stock, but also for a much safer and better environment, and there has also been an increasing interest worldwide in seeking alternative sources of energy [7], [11], [16], [17], [32], [34].
It is, therefore, important to consider the alternative renewable sources for energy and chemical production. In particular, biomass-oriented fuels and chemicals seem to be most promising. Although extensive investigations have, therefore, been made on biofuel and biochemical production from biomass resources, the main problem is the economic feasibility [36]. It is, therefore, highly desirable to utilize the cheap raw materials for fermentative production of such products.
Since glycerol is the byproduct of biodiesel production [26], [36] it is preferred as a low cost and abundant substrate for the production of bio-chemicals and biofuels [3]. Glycerol has a highly reduced nature when compared to other sugars such as glucose, xylose, etc., which indicates that glycerol may be more useful for the production of succinate, ethanol, lactate, and diols [36].
Among biofuels, bioethanol has been extensively investigated, even in an industrial scale in Brazil and USA, where ethanol can be produced by fermentation of food stocks, such as corn, sugarcanes and sugar beets. In 1925, Henry Ford had quoted ethyl alcohol, ethanol, as ‘the fuel of the future’ [2]. Although Saccharomyces cerevisiae has been extensively used for ethanol fermentation due to ethanol tolerance [14], [18] and it can utilize hexose sugars such as glucose, it does not have the capability of utilizing pentose sugars and others; therefore, the major drawback is the narrow range of its capability of assimilating carbon sources.
On the other hand, most bacteria such as Escherichia coli can assimilate a broad range of carbon sources including hexoses, pentoses, and others. E. coli is a gram-negative, facultative anaerobic and non-sporulating bacterium [33] and it is the most widely used prokaryotic system which produced heterologous proteins for the industrial production of bacterial metabolites by batch and fed-batch operations [15], [37]. E. coli has been regarded as the workhorse of modern biotechnology [12] for the potential microbial production of biofuels and bio-chemicals.
Biofuel and biochemical production by recombinant E. coli has been paid much attention due to its high growth rate (contributing to the productivity) and a broad range of carbohydrate utilization [22]. In the present study, therefore, the fermentative utilization of glycerol by E. coli is considered as compared to glucose.
One of the drawbacks of using E. coli, is the mixed metabolite production (such as formate, lactate, acetate, succinate, and ethanol) under micro-aerobic or anaerobic conditions, where it lowers the yield of the target metabolite and gives burden for the downstream processing. It is highly desirable to produce a single metabolite in practice. In fact, this may be attained by the specific pathway mutation. For example, pfl gene knockout mutation allows the exclusive D-lactate production from glucose in E. coli to be cultivated under anaerobic condition [38].
Another important factor for the useful metabolite production is the substrate uptake rate. In particular, it is highly desirable to increase the glycolytic flux to yield higher pyruvate formation, where pyruvate is the starting metabolite for a variety of target metabolite formations. The carbon flow in E. coli is controlled by Global regulator Cra (catabolite repressor/activator) [21], [29]. Its control mechanism is cAMP-independent [5], where Cra represses the expressions of the sugar uptake genes such as ptsHI, and the glycolysis genes such as pfkA, pykF, zwf, edd, eda as well as TCA cycle gene acnB, while it activates the gluconeogenic genes such as fbp, ppsA, pckA, the glyoxylate pathway gene aceA and the TCA cycle genes such as icdA and acnA [28]. The set of such genes implies that Cra activates the gluconeogenic pathway genes and represses the glycolysis genes. This implies that the glycolytic flux may be increased by cra gene knockout [30], [35], where acetate is overproduced in cra mutant, since the expression of aceA and icdA is repressed [30]. This problem may be avoided in pfl mutant, cultivated under anaerobic conditions, since AcCoA formation is blocked by this mutation.
In the present investigation, we considered the single metabolite production such as ethanol (biofuel) production by the wild type E. coli, and D-lactic acid (biochemical) production by pflA.cra mutant using either glucose or glycerol as a carbon source.
2. Materials and methods
2.1. Strains used
The strains used in the present study were E. coli BW25113 (lacIqrrnBT14 ΔlacZwJ16 hsdR514 ΔaraBADAH33ΔrhaBADLD78), and its pflA.cra double gene knockout mutant, where the mutant was constructed based on the method of Datsenko and Wanner [4].
The gene knockout mutant was constructed at Keio University, and open to the public as KEIO collection [1]. The double-gene knockout mutant was constructed in the similar method. The basic knockout strategy is to replace a cra gene of the pflA gene knockout mutant (kanamycin resistant) with a selectable antibiotic (ampicillin) resistant gene (amp) that is generated by PCR using primers with homology extensions. After selection, the resistant gene can be eliminated using a helper plasmid. The mutant was verified by comparing the length of the PCR amplified fragments with the expected length from the genome database.
2.2. Media compositions
Micro-aerobic batch culture was carried out using M9 synthetic medium containing the following components: 48 mM Na2HPO4, 22 mM KH2PO4, 10 mM NaCl and 30 mM (NH4)2SO4. The carbon source was either glucose (10 g/L) or glycerol (20 g/L) for the micro-aerobic batch culture. The following components were filter sterilized and then added (per liter) with 1 ml of 1 M MgSO4, 1 ml of 0.1 mM CaCl2, 1 ml of 1.0 mg/L Vitamin B1 and 10 ml of trace element solution containing (per liter): 0.55 g CaCl2·2H2O, 1.67 g FeCl3·6H2O, 0.10 g MnCl2·4H2O, 0.17 g ZnCl2, 0.043 g CuCl2·2H2O, 0.06 g CoCl2·2H2O and 0.06 g Na2MoO4·2H2O.
2.3. Culture conditions
Micro-aerobic batch cultivation was carried out in Erlenmeyer flasks at 150 rpm where the temperature was maintained at 37 °C. The inoculum was prepared by 100 μl of transferring cells from a glycerol stock to 50 ml test tube containing 10 ml of LB medium. Then the culture was incubated for 6 h and 0.25 ml of culture broth was then transferred to a 125 ml Erlenmeyer conical flask containing 25 ml of M9 medium. Then that culture was incubated for 12 h in a shaking incubator at 37 °C controlling the stirring speed at 150 rpm. After that, the total 25 ml culture was transferred to a 500 ml Erlenmeyer conical flask containing 250 ml of M9 medium to continue micro-aerobic batch culture at the same condition, where the initial pH was set at 7.0. The triplicate samples were taken during cultivations.
2.4. Measurement of biomass concentrations
Cell concentration was measured by the optical density (OD) of the culture broth (at λ = 600 nm) with a UV spectrophotometer (Shimadzu Co, Japan). After the batch culture, E. coli cells from 250 ml of culture were harvested by centrifugation and dried in a laboratory oven at 60 °C. After confirming that the weight of the dried material was constant, the dry cell weight per liter of culture was calculated from the measured weight. It was then converted to dry cell weight (DCW) per liter based on the OD600 – DCW relationship (1 OD600 ≈ 0.3 g/L) [19], [27].
2.5. Measurement of metabolite concentrations
Glucose was measured according to Nelson’s modification of Somogy’s method (1944) [24] where the color intensity was measured (at λ = 500 nm) in a UV Spectrophotometer (Shimadzu Co, Japan) and compared with standard d-glucose.
Ethanol concentration was measured by the gas chromatography (GC) using GC-2010 Plus (Shimadzu Co, Japan) using Rtx-1301 column (30 m length × 0.32 mm internal diameter × 10 μm film thickness), where nitrogen gas was used as the carrier gas and flame ionization detection (FID) was used at the temperature of 200 °C. The oven temperature was initially maintained at 100 °C for 1 min.
2.6. Statistical analysis
Triplicate measurements were done in all the cases during the observation and assessment of bacterial growth and metabolite production. Data were captured into Microsoft Excel Software, version 2010 which was used to calculate means and standard deviations. Student's t-test was applied to confirm that the observed changes were statistically significant.
3. Results and discussion
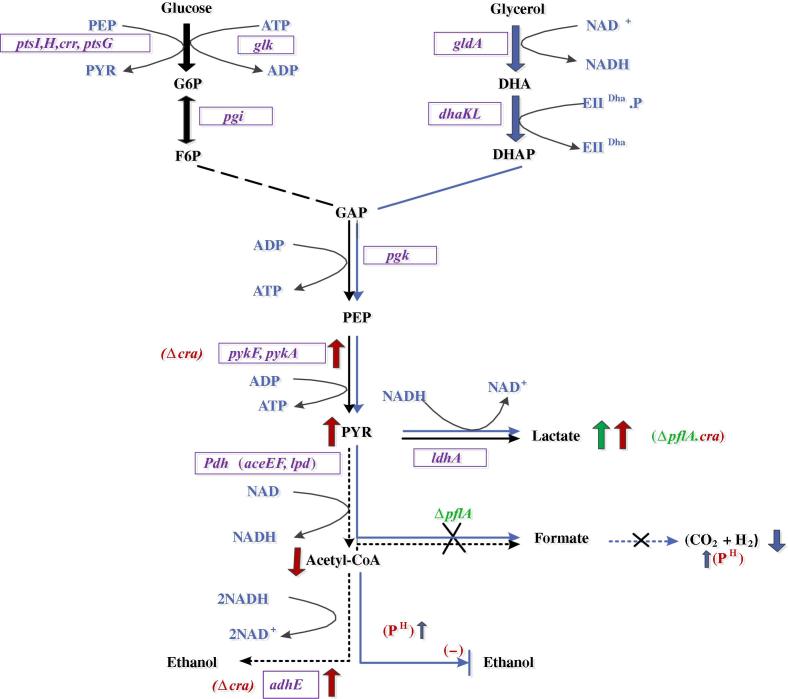
One of the promising features of using glycerol as a carbon source is its highly reduced nature of carbon atoms, which gives an advantage for the production of reduced chemicals and fuels as compared to the case of using sugars. As illustrated in Fig. 1, the number of reducing equivalents for the conversion of glycerol to pyruvate is twice that of glucose (and other sugars as well).
Figure 1.
Metabolic pathways of E. coli cultivated under microaerobic condition using glucose and glycerol.
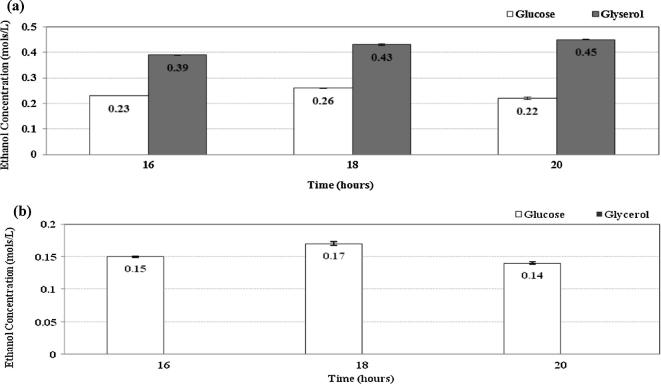
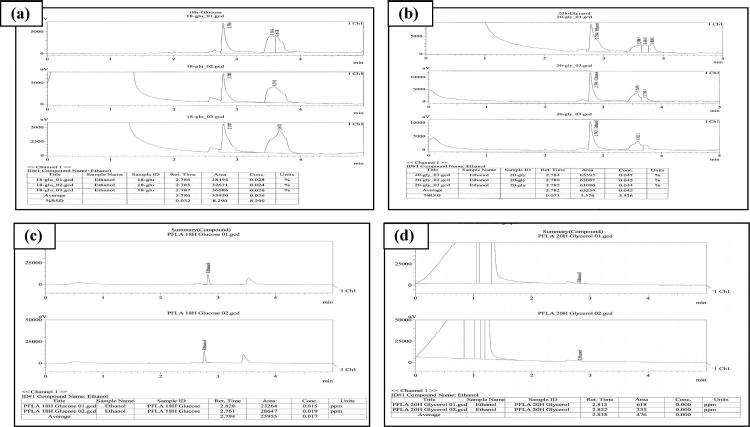
The ethanol production was compared (Fig. 2a) for the case of using glycerol as compared to that of using glucose with respect to time in the wild type E. coli. As illustrated in Fig. 2a, the ethanol yield is nearly doubled for each case of using glycerol (P < 0.01 for all cases) as compared to the case of using glucose (18 and 20 h, respectively), where the corresponding GC data are given in Fig. 7. The maximum theoretical yield may be computed for the case using glucose:
| (1) |
while the following equation may be considered for the case of using glycerol:
| (2) |
where half of glucose is lost as CO2 due to its oxidation state as shown in Eq. (1), while glycerol can co-produce both ethanol and formate (or ethanol and hydrogen).
Figure 2.
Comparison of ethanol formation in wild type E. coli (a), and pflA.cra mutant (b) cultivated under microaerobic conditions using glucose and glycerol as a carbon source.
Figure 7.
Chromatogram of ethanol produced at 18 h using glucose (a and c) and 20 h using glycerol (b and d) as carbon source during batch culture by E. coli BW25113 and its pflA.cra mutant.
In E. coli, glycerol is converted first to dihydroxyacetone phosphate (DHAP) in the glycolysis, where glycerol dehydrogenase (GLDH) encoded by gldA can convert glycerol to dihydroxyacetone (DHA), producing a reducing equivalent, and then DHA is converted to DHAP by DHA kinase (DHAK) encoded by dhaKLM using PEP as the phosphate donor under anaerobic or micro-aerobic conditions [9]. Therefore, 2 mols of reducing equivalents are formed at GLPDH and glycerol 3-phosphate dehydrogenase (GAPDH) in the case of using glycerol as a carbon source, instead of one mole of NADH production at GAPDH in the case of using glucose (Fig. 1). Namely, the conversion of glycerol to pyruvate generates twice the amount of reducing equivalents as compared to the case of converting glucose to the pyruvate [6]. This permits, for example, the co-production of ethanol and formic acid (or ethanol and hydrogen), which would result in doubling the overall product yield as compared to the case of using glucose for the production of ethanol, where half of the sugar is lost as CO2 due to the oxidation state of glucose. The higher production of reducing equivalents allows the wild type E. coli to produce ethanol exclusively. Although formate may be also formed from pyruvate, this may be converted to hydrogen and CO2 by formate-hydrogen lyase (Fhl) under acidic conditions [23].
In the case of pflA.cra mutant, much less amounts of ethanol are produced from glucose, while no detectable ethanol could be produced from glycerol as shown in Fig. 2b. The reason for the later phenomenon may be due to the growth defect caused by the redox imbalance for the case of using glycerol.
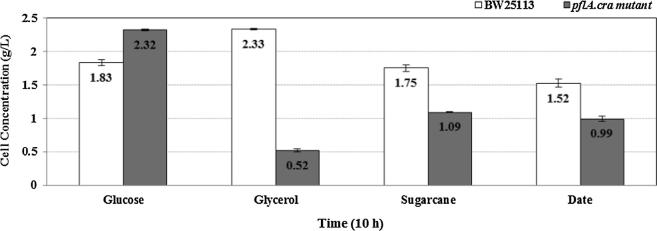
The comparison of the cell growth measured (Fig. 3) in terms of g DCW at 10 h of the batch cultivation, which indicates that the cell concentrations of the wild type E. coli (BW25113) and its pflA.cra mutant were 1.83 ± 0.04 g/L and 2.32 ± 0.01 g/L, respectively when glucose was used as a carbon source. Fig. 3 also shows that cell concentrations were 2.33 ± 0.01 g/L and 0.52 ± 0.02 g/L for the wild type and pflA.cra mutant, respectively, in the case of using glycerol as a carbon source.
Figure 3.
Comparison of the cell growth of the wild type E. coli and its pflA.cra mutant cultivated under microaerobic conditions using glucose and glycerol.
The cell concentration of the wild type using glycerol as a carbon source is similar (Fig. 3) to that of pflA.cra mutant using glucose as a carbon source. This may be explained as follows: that is, the cell growth rate is in general correlated with the specific ATP production rate. In the case of wild type using glycerol as a carbon source, the phosphate of PEP is utilized at DHAK for the phosphorylation of DHA to yield DHAP, and therefore, one mole of ATP is formed at phosphoglucokinase (Pgk) in the glycolysis from one mole of glycerol to yield pyruvate. On the other hand, in the case of pflA.cra mutant using glucose as a carbon source, one mole of PEP is used for the phosphotransferase system (PTS) to phosphorylate glucose, and the net ATP production from glucose to pyruvate is two moles, where one ATP is consumed at phosphofructokinase (Pfk), while 2 mols of ATP is produced at Pgk and one mole at pyruvate kinase (Pyk) (another mole is used for PTS). Since the number of carbons in glucose is two times that of glycerol, the same amount of ATP is formed from the same weight of carbon sources, resulting in the similar growth rate as shown in Fig. 3.
In the case of using glycerol as a carbon source, its assimilation rate is low, where the glycerol assimilation pathway genes appear to be expressed at low levels as inferred by the kinetics of glycerol fermentation [6]. This may be considered to avoid the accumulation of DHAP, otherwise it may cause the toxic methylglyoxal production. Slow glycerol uptake causes fructose 1,6-bisphosphate (FBP) concentration to be decreased, where FBP controls the glycolysis and anaplerotic pathways via enzyme levels and transcriptional regulations. Namely, FBP allosterically activates Pyk and PEP carboxylase (Ppc), while FBP also inhibits Cra activity, and the transcriptional regulation is made via Cra [13]. The decrease in FBP level causes the deactivation of Ppc and Pyk, resulting in lower succinate production through Ppc, and the lower glycolytic flux through Pyk. In the later case, the pyruvate is pulled off through Pfl and ADH to produce ethanol for the redox balancing, and thus the pyruvate concentration is also lower, and D-lactate production is lower due to its relatively lower affinity (higher Km value) of LDH to pyruvate. These may force the wild type E. coli to produce a single ethanol production from glycerol under micro-aerobic condition.
The growth defect of pflA.cra mutant for the case of using glycerol may be due to redox imbalance, causing shortage of NAD+. A similar situation may be also seen for pfl and ldhA mutations [39].
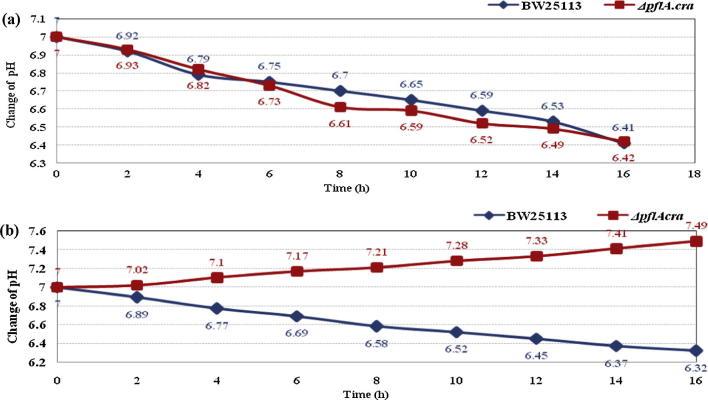
Fig. 4 shows the changes of culture pH during batch culture containing glucose and glycerol for E. coli BW25113 and its pflA.cra mutant. Fig. 4a indicates that both strains produced acids, causing pH to continuously decrease during cultivation, in the case of using glucose as a carbon source. In pflA.cra mutant, the pH downshift may be caused by the exclusive d-lactic acid production, while multiple acids may have been formed in the wild type strain.
Figure 4.
Comparison of the pH changes during the batch culture of E. coli BW25113 and its pflA.cra mutant containing glucose (a) and glycerol (b) as carbon sources.
On the other hand, the pH changing patterns are quite different for the case of using glycerol as a carbon source as shown in Fig. 4b. In the case of wild type strain, multiple acids are formed with less amounts, causing a decrease in pH level during cultivation, where it can ferment glycerol in a pH-dependent manner [8] and formate may be converted to CO2 and H2 under acidic condition [23], [31]. The strange phenomenon may be seen for the case of pflA.cra mutant, where pH level keeps increasing irrespective of the growth defect. The reason for this is not clear at this stage.
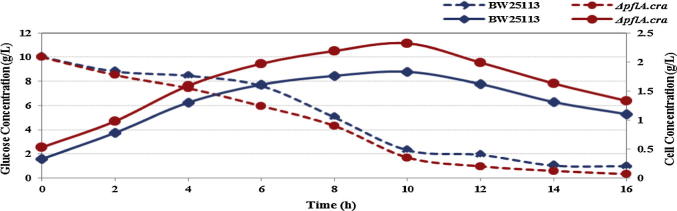
Under anaerobic or microaerobic conditions, the improvement of substrate uptake rate or the glycolytic flux directly increases the metabolite production from pyruvate. Fig. 5 shows the comparison of the glucose consumption for the wild type and its pflA.cra mutant. It indicates that the glucose consumption rate is faster in the case of pflA.cra mutant as compared to the wild type [20], [35].
Figure 5.
The glucose consumption and the cell growth of the wild type and its pflA.cra mutant cultivated using glucose as a carbon source under micro aerobic condition.
The conventional approach for ethanol production by E. coli is to introduce the heterologous ethanol-forming pathway such as pyruvate decarboxylase (PDC) gene (pdc) and alcohol dehydrogenase II (ADHII) gene (adhB) from Zymomonas mobilis into the chromosome of pfl gene (called as KO11) [25]. Many recombinant E. coli strains have been developed since then. In particular, it is essential to disrupt pfl gene for the production of ethanol from pyruvate, where NADH produced at GAPDH is reoxidized at ADH, resulting in redox balanced. Without introducing pdc and adhB genes, E. coli exclusively produces d-lactate from glucose with redox balancing between GAPDH and LDH in a similar way to ethanol production with the heterologous pathway [38], [39]. This implies that the present result can also be applied to the conventional ethanol producer such as KO11 and its related strains in terms of redox balance with different metabolite productions.
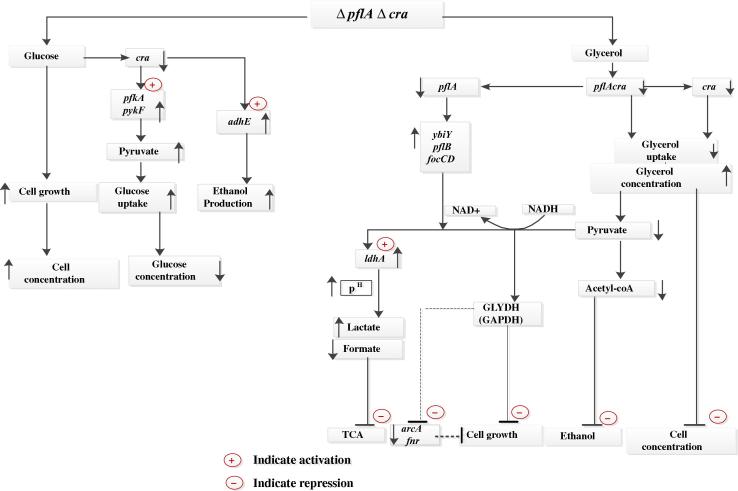
The possible metabolic regulation mechanism of pflA.cra mutant in our study is briefly illustrated in Fig. 6.
Figure 6.
Overall regulation mechanism of pflA.cra mutant.
Further study of the effects of different supplements and conditions in their (E. coli BW25113 and its pflA.cra mutant) growth is needed to identify their efficiency as ethanol producers, where optimization of pH, nutrients, temperature, incubation time can influence metabolite (ethanol) production capacity [8].
Biofuel and biochemical production by recombinant E. coli, has been paid much attention due to its high growth rate (contributing to the productivity) and a broad range of fermentative utilization of glycerol compared to glucose [22], might be helpful to formulate and develop the industrial production of alternative energy sources.
To make it a functional form for industrial biodiesel production, this is the base study to utilize glycerol, as it is an abundant, inexpensive carbon source [3], where current interest is increasing worldwide in seeking alternative sources of energy [7], [11], [16], [17], [32], [34] for a much safer and better environment.
4. Conclusion
In this study, two (E. coli BW25113 and its pflA.cra mutant) strains were used to aim single metabolite (ethanol) production under microaerobic condition using glycerol or glucose as a carbon source. These two strains were considered to measure biomass concentration, culture pH change, growth curve analysis on different kinds and concentration of carbon sources in minimal media to select the best candidates for metabolite (ethanol) production that might be further used.
Authors’ contributions
This work is a product of the intellectual effort of the whole team; and that all members have contributed in various degrees to the analytical methods used, to the research concept, to the experiment design and to the manuscript preparation.
Conflict of interest
No conflict of interest influenced in this research.
Acknowledgement
This research was partially supported by Department of Genetic Engineering and Biotechnology, University of Chittagong, Bangladesh.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K., Tomita M., Wanner B.L., Mori H. Mol. Syst. Biol. 2006 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandel A.K., Chan E.S., Rudravaram R., Narasu M.L., Rao L.V., Ravindra P. Biotechnol. Mol. Biol. Rev. 2007;2(1):14–32. [Google Scholar]
- 3.Da Silva G.P.M., Mack J. Biotechnol. Adv. 2009;27:30–39. doi: 10.1016/j.biotechadv.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Datsenko K.A., Wanner B.L. Proc. Natl. Acad. Sci. U.S.A. 2001;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dessein A., Schwartz M., Ullmann A. Mol. Gen. Genet. 1978;162:83–87. doi: 10.1007/BF00333853. [DOI] [PubMed] [Google Scholar]
- 6.Dharmadi Y., Murarka A., Gonzalez R. Biotechnol. Bioeng. 2006;94:821–829. doi: 10.1002/bit.21025. [DOI] [PubMed] [Google Scholar]
- 7.Dien B.S., Cotta M.A., Jeffries T.W. Appl. Microbiol. Biotechnol. 2003;63:258–266. doi: 10.1007/s00253-003-1444-y. [DOI] [PubMed] [Google Scholar]
- 8.Doi Y., Ikegami Y. J. Bacteriol. 2014;196:2472–2480. doi: 10.1128/JB.01512-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durnin G., Clomburg J., Yeates Z., Alvarez P.J.J., Zygourakis K., Campbell P., Gonzalez R. Biotechnol. Bioeng. 2009;103:148–161. doi: 10.1002/bit.22246. [DOI] [PubMed] [Google Scholar]
- 10.Hansen A.C., Zhang Q., Lyne P.W.L. Biores. Technol. 2005;96:277–285. doi: 10.1016/j.biortech.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Herrera S. Nat. Biotechnol. 2006;24:755–760. doi: 10.1038/nbt0706-755. [DOI] [PubMed] [Google Scholar]
- 12.Himmel M.E., Adney W.S., Baker J.O., Elander R., McMillan J.D., Nieves R.A., Sheehan J.J., Thomas S.R., Vinzant T.B., Zhang M. Adv. Bioeth. Prod. Technol. A Perspect. 1997:1. [Google Scholar]
- 13.Kochanowski K., Volkmer B., Gerosa L., Haverkorn R.V., Schmidt B.R.A., Heinemann M. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1130–1135. doi: 10.1073/pnas.1202582110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laluce C. Crit. Rev. Biotechnol. 1991;11:149–161. [Google Scholar]
- 15.Lee S.Y. Trends Biotechnol. J. U.S.A. 1996;14(3):98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y., Tanaka S. Appl. Microbiol. Biotechnol. 2006;69:627–642. doi: 10.1007/s00253-005-0229-x. [DOI] [PubMed] [Google Scholar]
- 17.Lynd L.R. Annu. Rev. Energy Environ. 1996;21:403–465. [Google Scholar]
- 18.Lyons T.P., Kelsall D., Murtagh J. Nott. Univ. Press; Nottingham, United Kingdom: 1995. The Alcohol Textbook. [Google Scholar]
- 19.Marzan L.W., Shimizu K. Microb. Cell Fact. 2011;10:39. doi: 10.1186/1475-2859-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuoka Y., Shimizu K. Adv. Biosci. Biotechnol. 2013;4:455–468. [Google Scholar]
- 21.Moat A.G., Foster J.W., Spector M.P. fourth ed. Cold John Wiley & Sons; New York: 2002. [Google Scholar]
- 22.Munjal N., Mattam A.J., Pramanik D., Srivastava P.S., Yazdani S.S. Microb. Cell Fact. 2012;11:145. doi: 10.1186/1475-2859-11-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murarka A., Dharmadi Y., Yazdani S.S., Gonzalez R. App. Env. Microbiol. 2007;2:1124–1135. doi: 10.1128/AEM.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson N. J. Biol. Chem. 1944;153:375–380. [Google Scholar]
- 25.Ohta K., Beall D.S., Mejia J.P., Shanmugam K.T., Ingram L.O. Appl. Environ. Microbiol. 1991;57(4):893–900. doi: 10.1128/aem.57.4.893-900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagliaro M.R., Ciriminna H., Kimura M., Rossi C.D. Angew. Chem. Int. Ed. 2007;46:4434–4440. doi: 10.1002/anie.200604694. [DOI] [PubMed] [Google Scholar]
- 27.Peng L., Shimizu K. Enzyme Microb. Technol. 2006;38:512–520. [Google Scholar]
- 28.Perrenoud A., Sauer U. J. Bacteriol. 2005;187:3171–3179. doi: 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saier M.H., Ramseier T.M. J. Mol. Microbiol. Biotechnol. 1996;178:3411–3417. [Google Scholar]
- 30.Sarkar D., Shimizu K. Biochem. Eng. J. 2008;42:224–228. [Google Scholar]
- 31.Sawers R.G., Clark D.P. In: EcoSal-Escherichia coli and Salmonella: Cellular and Molecular Biology. Curtis R. III, editor. ASM Press; Washington, DC: 2004. [Google Scholar]
- 32.Schubert C. Nat. Biotechnol. 2006;24:777–784. doi: 10.1038/nbt0706-777. [DOI] [PubMed] [Google Scholar]
- 33.Singleton P. 5th ed. Wiley; 1999. Bacteria in Biology, Biotechnology and Medicine. pp. 444–454. [Google Scholar]
- 34.Wyman C.E. In: Handbook on Bioethanol: Production and Utilization. Wyman C.E., editor. Taylor and Francis; Washington, DC: 1996. pp. 1–18. [Google Scholar]
- 35.Yao R., Kurata H., Shimizu K. Adv. Biosci. Biotechnol. 2013;4:477–486. [Google Scholar]
- 36.Yazdani S.S., Gonzalez R. Curr. Opin. Biotechnol. 2007;18:213–216. doi: 10.1016/j.copbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Yee L., Blanch H.W. Biotechnology (NY) 1992;10(12):1550–1556. doi: 10.1038/nbt1292-1550. [DOI] [PubMed] [Google Scholar]
- 38.Zhu J., Shimizu K. Appl. Microbiol. Biotechnol. 2004;64:367–375. doi: 10.1007/s00253-003-1499-9. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J., Shimizu K. Metab. Eng. 2005;7:104–115. doi: 10.1016/j.ymben.2004.10.004. [DOI] [PubMed] [Google Scholar]