Abstract
Background
Aberrant activation of the signal transducer and activator of transcription 3 (STAT3) is frequently seen in patients with gastric cancer (GC), and is generally associated with worse prognosis. HJC0152, a novel STAT3 inhibitor, has shown significant anti-tumor effects in several cancers, although its role in GC remains to be clarified.
Methods
The effect of HJC0152 on STAT3 signaling pathway and the biological behaviors of GC cells were evaluated through in vitro and/or in vivo experiments. Meanwhile, RNA sequence analysis was used to further explore its potential anti-tumor mechanisms.
Results
HJC0152 inhibited the expression of activated STAT3 and its downstream target genes (c-Myc and clyclinD1) in GC cells, and restrained tumor growth in vivo. HJC0152 treatment induced apoptosis in the STAT3 hyper-activated AGS and MKN45 cell lines, along with down-regulation of survivin and Mcl1, and up-regulation of cleaved-poly(ADP-ribose) polymerase. Moreover, HJC0152 markedly inhibited migration and invasion of these cells. Finally, RNA sequence analysis and protein expression analyses showed that in addition to STAT3 suppression, HJC0152 also exerts its anti-tumor effects at least partly via the mitogen-activated protein kinases pathway.
Conclusion
Our findings highlight that HJC0152 is a promising therapeutic agent for GC.
Keywords: gastric cancer, inhibitor, HJC0152, STAT3, MAPK
Introduction
Despite a decrease in incidence rate, gastric cancer (GC) is still one of the major causes of cancer-associated deaths worldwide.1–3 In addition, a significant proportion of GC patients die within 5 years of diagnosis due to rapid tumor progression even after clinical interventions, such as surgical treatment and/or chemotherapy.4–6 Patients at the advanced stage of GC often respond poorly to chemotherapy and suffer from serious side effects, resulting in a 5-year survival rate of <5%.2,7 Therefore, new drugs with high anti-tumor efficiency as well as minimal side effects are urgently needed.
The signal transducers and activators of transcription (STAT) proteins are important transcription factors involved in relaying extracellular stimuli and regulating multiple biological processes.8,9 Aberrant activation of STAT3 is observed in several diseases, including cancer. Constitutive activation and/or overexpression of STAT3 occurs in various hematological and solid tumors, and is associated with worse clinicopathological phenotypes and poor prognosis.10 Since STAT3 dysregulation drives tumor initiation and progression, it stands to reason that tumors with high STAT3 expression/activity can be controlled by STAT3-targeted therapy.11 STAT3 is hyper-activated in GC and associated with aggressive clinical features and poor outcomes12–15 making it an attractive therapeutic target.
Niclosamide is an effective inhibitor of STAT3 signaling, but is limited by poor aqueous solubility and bioavailability.16 HJC0152, its O-alkylamino-tethered derivative, is a more potent inhibitor compared with niclosamide, and significantly restrained in vitro and in vivo growth of breast cancer and head and neck squamous cell carcinoma (HNSCC) cells.17 Based on these findings, our study was designed to determine the potential anti-tumor effects of HJC0152 in GC. We also analyzed the effect of HJC0152 on the mitogen-activated protein kinases (MAPKs) signaling pathway, which is frequently dysregulated in cancers and plays a critical role in tumor progression.18–21
Materials and methods
Cell lines and culture conditions
Six GC cell lines (AGS, HGC-27, MKN28, MKN45, SGC7901, and BGC-823) were purchased from the Institute of Cellular Biology (Chinese Academy of Science, Shanghai, People’s Republic of China) and cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Rockville, MD, USA) supplemented with 10% FBS (Gibco) in a humidified 37°C incubator with 5% CO2.
Regents and antibodies
HJC0152 was obtained from Selleck Chemicals LLC (Houston, TX, USA) and dissolved in dimethyl sulfoxide (DMSO) to prepare a 20 µM stock solution. Primary antibodies against STAT3, p-STAT3 (Y705), p-STAT3 (S727), c-Jun N-terminal kinase (JNK), phospho-JNK, p38 MAPK, and p-p38 MAPK were bought from Abcam (Cambridge, UK) and those against p44/42 MAPK (extracellular signal-regulated kinase [ERK]1/2), p-p44/42 MAPK (p-ERK1/2), c-Myc, cyclinD1, survivin, Mcl1, and cleaved-poly(ADP-ribose) polymerase (c-PARP) from Cell Signaling Tech (Beverly, MA, USA). Finally, the anti-glyceraldehyde-3-phosphate dehydrogenase antibody and horseradish peroxidase-conjugated secondary antibodies were purchased from Proteintech (Hubei, People’s Republic of China).
Cell viability and colony formation assay
AGS (4×103 cells/well), MKN45 (4×103 cells/well), and HGC-27 (2×103 cells/well) cells were seeded in 96-well plates at suitable densities, and treated with 0, 5, 10, and 20 µM HJC0152 (in 1 µM DMSO). After incubation for 0, 24, 48, and 72 hours, the cells were stained using reagents of the Cell Counting Kit-8 (CCK8; Dojindo, Tokyo, Japan) as per the manufacturer’s instructions, and absorbance was measured at 450 nm. For colony formation assay, AGS and MKN45 cells were seeded in 6-well culture plates at the density of 8×102 cells/well and cultured with different concentrations of HJC0152 for ~14 days. The resulting colonies were fixed with 95% ethanol, stained with 0.1% crystal violet for 30 minutes, and the number of colonies was counted.
In vivo tumor modeling and treatment
The animal studies were performed in accordance with the Animal Management Rules of the Ministry of Health of the People’s Republic of China and were approved by the Institutional Animal Care and Use Committee of Zhejiang University (approval ID: SYXK (ZHE) 2005–0072). MKN45 cells (6×106) were subcutaneously inoculated into 4–6 weeks old female BALB/c nude mice that were randomized into the control and HJC0152 groups (n=5 each). The mice were accordingly injected with either 100 µL PBS or 7.5 mg/kg HJC0152 dissolved in 100 µL PBS intraperitoneally twice weekly for 21 days. The tumor volume (mm3; length × width2/2) and body weights of the mice were monitored twice weekly. At the end of treatment, the animals were euthanized, and their tumors were excised, weighed, and processed for further analysis.
H&E staining and immunohistochemistry (IHC)
The resected tumors were fixed with formalin and embedded in paraffin, and cut into 4 µm sections. The tissues were de-paraffinized and rehydrated, and then stained with H&E. IHC staining against Ki-67 (Servicebio, Wuhan, People’s Republic of China) was performed as previously described.22
Annexin V/propidium iodide (PI) staining
Apoptosis was evaluated using the Annexin V-fluorescein isothiocyanate (FITC)/PI Detection Kit (BD Biosciences, San Jose, CA, USA). AGS and MKN45 cells were seeded in 6-well plates and treated with different concentrations of HJC0152 for 24 hours. The cells were then harvested, re-suspended, and incubated with Annexin V-FITC (5 µL) and PI (5 µL) at 4°C for 15 minutes. Flow cytometric analysis was performed on FACScan (BD Biosciences) within 1 hour of staining.
Transwell assay
Cell migration and invasion assays were performed in Tran-swell® plates containing 8-µm pore polycarbonate filters (Corning Costar, Corning, NY, USA) either coated with Matrigel (BD Biosciences) or left uncoated. Cells (5×104) pre-treated with HJC0152 (0 or 20 µM in 1 µM DMSO) for 8 hours were suspended in serum-free medium and seeded into the upper chambers, and the lower chambers were filled with 700 µL of RPMI-1640 containing 10% FBS. After incubation for 18 hours, the cells that had penetrated into the lower chamber through the membrane were fixed with 95% ethanol, and stained with 0.1% crystal violet. The stained cells were observed under a light microscope at 200× magnification.
Western blotting
Western blotting was performed as described previously.22 The immune-reactive protein bands were visualized using an enhanced chemiluminescence Western blotting kit (Bio-Rad, Hercules, CA, USA) as per the manufacturer’s instructions.
RNA sequencing
RNA sequencing (RNA-Seq) was performed as described previously.22
Statistical analysis
All data are presented as the mean ± SD of three independent experiments. The treatment and control groups were compared using Student’s t-test (GraphPad Prism 5.01; San Diego, CA, USA). P-values <0.05 were considered statistically significant.
Results
HJC0152 inhibits STAT3 phosphorylation in GC cells
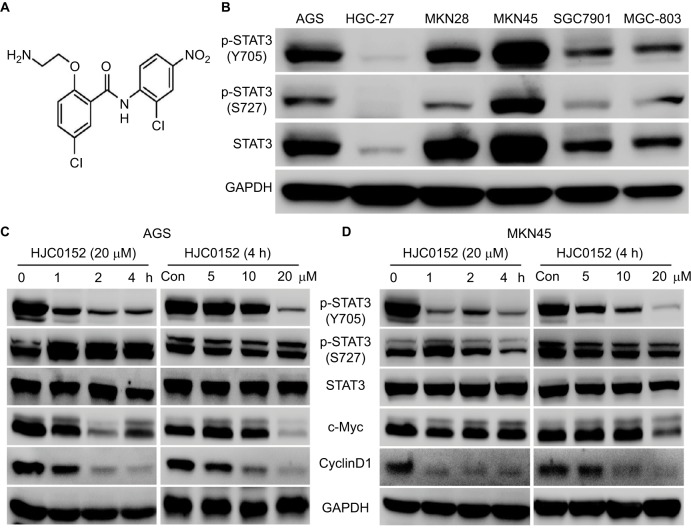
The chemical structure of HJC0152 is shown in Figure 1A. The expression status of total STAT3 and p-STAT3 (Y705 and S727) was evaluated in 6 GC cell lines (AGS, HGC-27, MKN28, MKN45, SGC7901, and BGC-823). As shown in Figure 1B, the AGS, MKN28, and MKN45 cell lines showed higher levels of p-STAT3 compared with the rest. Based on these results, the AGS, MKN45, and HGC-27 cell lines were selected for subsequent experiments. HJC0152 inhibited p-STAT3 (Y705) levels in a time- and dose-dependent manner in the AGS and MKN45 cell lines, but had no significant effect on p-STAT3 (Y705) and total STAT3 expression levels (Figure 1C,D). In addition, HJC0152 significantly down-regulated c-Myc and cyclinD1, the downstream targets of STAT3. However, these effects were not significant in HGC-27 cells (Figure S1A). These results demonstrated that HJC052 down-regulated STAT3 phosphorylation and its proliferation-related target genes in STAT3 overexpressed GC cells.
Figure 1.
HJC0152 inhibits the phosphorylation of STAT3 in GC cells.
Note: (A) Chemical structure of HJC0152. (B) Expression of STAT3 and p-STAT3 (Y705 and S727) in six GC cell lines (AGS, HGC-27, MKN28, MKN45, SGC7901, and BGC-823). Western blots showing the levels of p-STAT3 and downstream targets in AGS (C) and MKN45 (D) cells treated with 20 µM HJC0152 for varying durations (0, 1, 2, and 4 hours) or with different concentrations of HJC0152 (0, 5, 10, and 20 µM) for 4 hours. GAPDH was used as internal control.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GC, gastric cancer; STAT3, signal transducer and activator of transcription 3.
HJC0152 suppresses the growth of GC cells in vitro
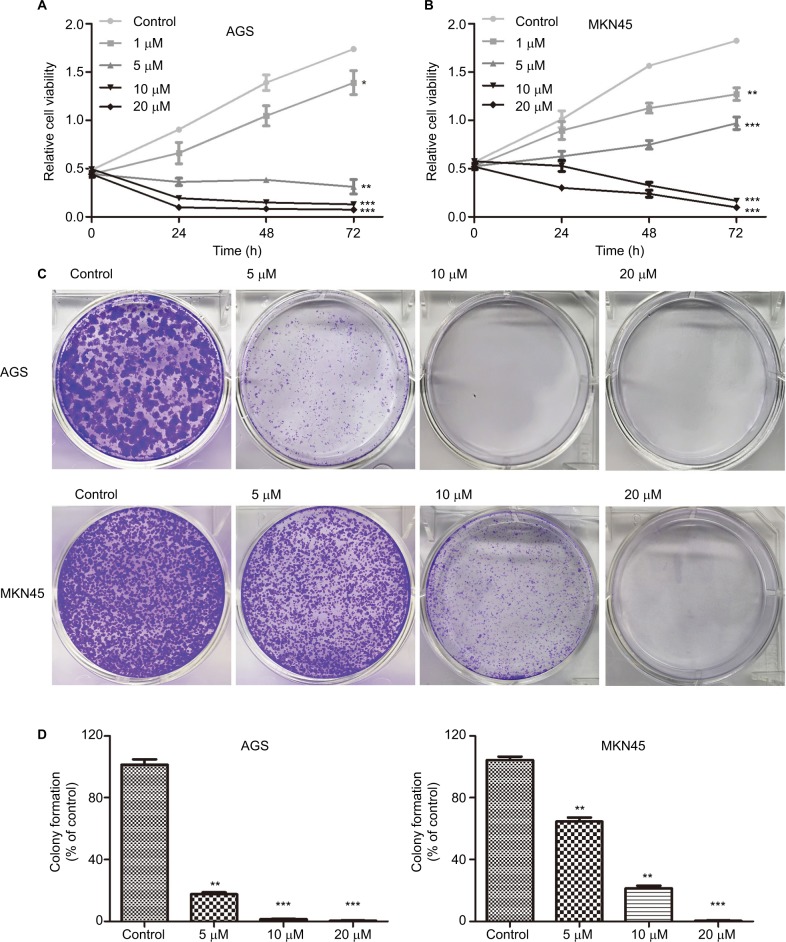
The effect of HJC0152 on the growth of GC cells was evaluated using the CCK8 assay. As shown in Figure 2A, B, HJC0152 treatment significantly inhibited the growth of AGS and MKN45 cells compared with the untreated controls. Additionally, HJC0152-treated AGS and MKN45 cells formed smaller and fewer colonies compared with controls in a dose-dependent manner (Figure 2C, D). In HGC-27 cells, however, HJC0152 only inhibited their growth at relatively high concentrations (Figure S1B). These results indicated that HJC0152 exerted a potent inhibitory effect on the growth of GC cells by targeting STAT3.
Figure 2.
HJC0152 inhibits GC cell growth.
Note: (A, B) Viability of AGS and MKN45 cells treated with different concentrations of HJC0152 (0, 1, 5, 10, and 20 µM) for 0, 24, 48, and 72 hours was determined by CCK8 assay. (C) Representative pictures of crystal violet-stained colonies formed by the AGS (upper) and MKN45 (lower) cells were treated with HJC0152 (0, 5, 10, and 20 µM) for ~14 days. (D) Percentage colony counts of the HJC0152-treated AGS (left) and MKN45 (right) cells relative to the DMSO controls. Data were obtained from three independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs control.
Abbreviations: CCK8, Cell Counting Kit-8; DMSO, dimethyl sulfoxide; GC, gastric cancer.
HJC0152 inhibits tumor growth in vivo
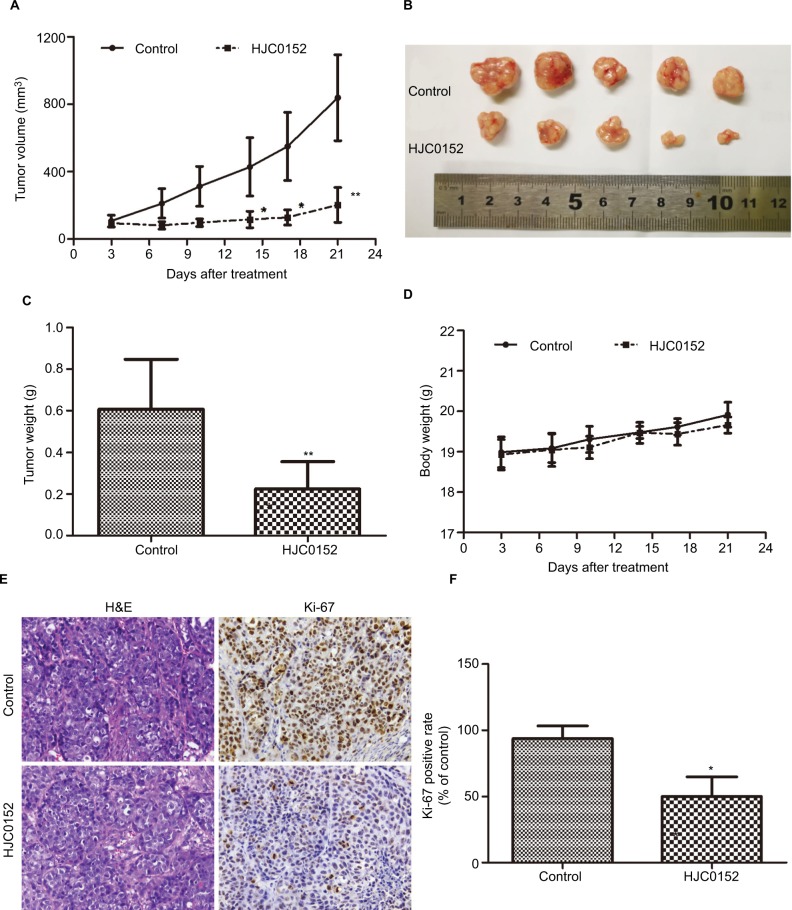
To determine the anti-tumor effect of HJC0152 in vivo, nude mice were subcutaneously inoculated with MKN45 cells and treated with HJC0152 (7.5 mg/kg) or PBS. As exhibited in Figure 3A–C, the tumor volumes (P<0.001) and tumor weight (P<0.01) were significantly lower in the HJC0152-treated group compared with the control group. Furthermore, the mice of both groups had similar body weights indicating that HCJ0152 had no parent side effects (Figure 3D). The tumor tissues were analyzed histo-pathologically by H&E staining, and the proportion of Ki-67 positive cells was significantly reduced in the HJC0152-treated mice compared with the control mice (Figure 3E, F). Taken together, HJC0152 retarded tumor growth in vivo by inhibiting the proliferative capacity of the tumor cells.
Figure 3.
HJC0152 inhibits the growth of MKN45 xenografts in vivo.
Note: (A) Tumor growth curves in different groups. (B) Representative pictures of tumor xenografts. (C) Tumor weights of different groups measured at the end of the experiment. (D) Body weight of HJC0152-treated and control groups. (E) Representative pictures of HE- and Ki-67-stained xenograft tissues. Magnification ×200. (F) The positive rate of Ki-67 in different groups. Three independent experiments were performed. *P<0.05, **P<0.01 vs control.
HJC0152 induces apoptosis in GC cells
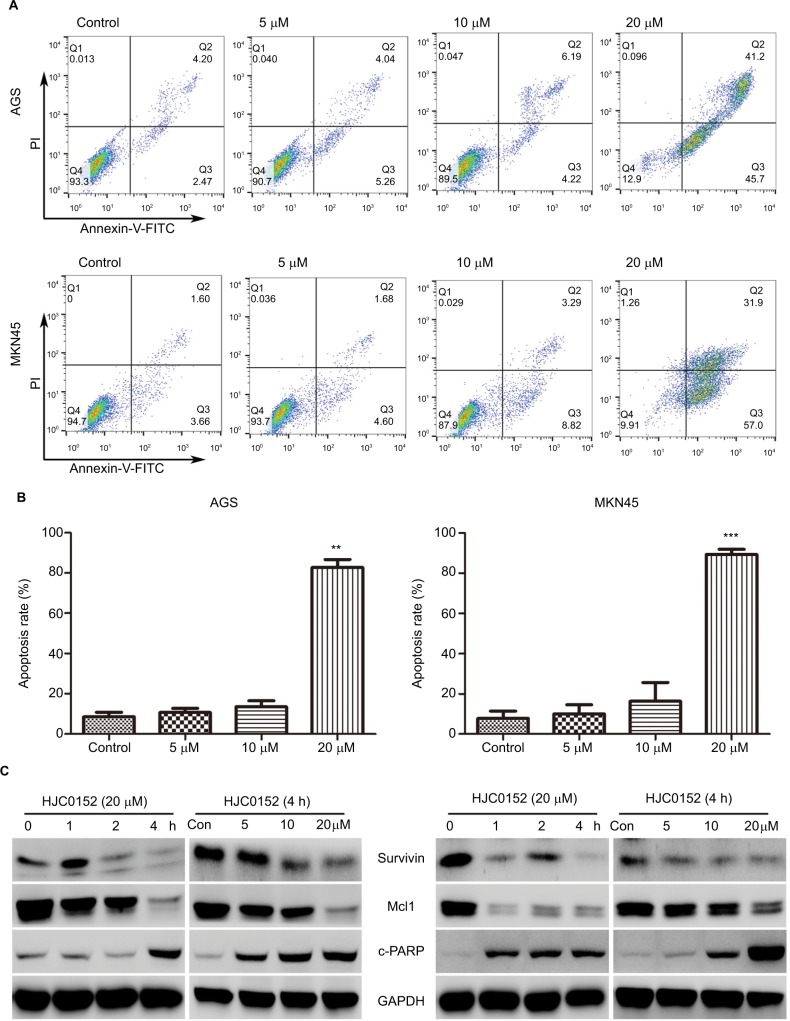
The apoptosis rates in the AGS and MKN45 cells were analyzed after the 24 hours HJC0152 treatment using the Annexin V-FITC/PI double staining assay. As shown in Figure 4A, B, the percentage of apoptotic cells was significantly increased in the 20 µM HJC0152-treated group compared with the controls, in both AGS (P<0.001) and MKN45 (P<0.001) cells. In addition, the expression of apoptosis-related survivin and Mcl1 proteins, which are regulated by the STAT3 signaling pathway, was also analyzed. HJC0152 treatment markedly reduced expression levels of both proteins, and significantly increased that of the pro-apoptotic c-PARP in GC cells in a time- and dose-dependent manner (Figure 4C).
Figure 4.
HJC0152 induces apoptosis in GC cells
Note: (A) AGS (upper) and MKN45 (lower) cells were treated with HJC0152 (0, 5, 10, and 20 µM) for 24 hours, and stained with Annexin-FITC/PI. (B) Histograms showing the percentage of apoptotic cells in AGS (left) and MKN45 (right) cells. Data were presented as the mean ± SD of three independent experiments (C) Western blots showing the levels of apoptosis-related proteins in AGS and MKN45 cells treated with 20 µM HJC0152 for varying time durations (0, 1, 2, and 4 hours) or treated with different concentrations of HJC0152 (0, 5, 10, and 20 µM) for 4 hours. GAPDH served as the internal control. Data were collected from three independent experiments. **P<0.01, ***P<0.001 vs control.
Abbreviations: FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GC, gastric cancer; PI, propidium iodide.
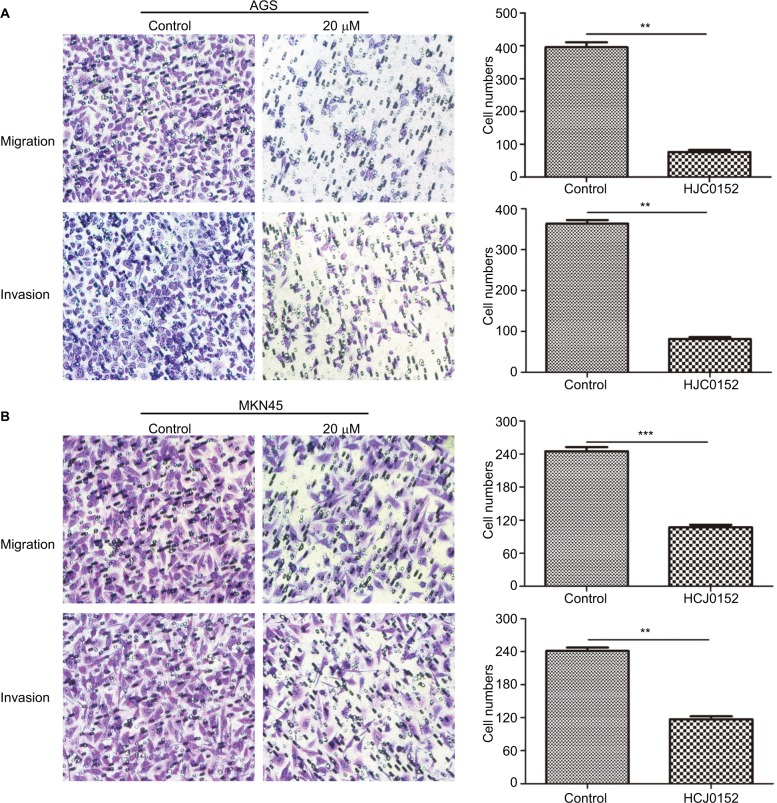
HJC0152 inhibits the migration and invasion capacity of GC cells
Transwell migration and invasion assays were performed to evaluate the effect of HJC0152 on GC cell mobility. As shown in Figure 5A, B, the percentage of migrated and invaded cells were significantly decreased in HJC0152-treated groups (20 µM) compared with that in the control (P<0.01). Thus, HJC0152 suppresses the migration and invasion capacities of AGS and MKN45 cells.
Figure 5.
HJC0152 inhibited migration and invasion of GC cells.
Note: Representative pictures of transwell chambers showing the migration (uncoated inserts) and invasion capacity (Matrigel-coated inserts) of AGS (A) and MKN45 (B) cells. Magnification ×200. The average number of migrating and invading cells was calculated from three independent experiments. **P<0.01, ***P<0.001 vs control.
Abbreviation: GC, gastric cancer.
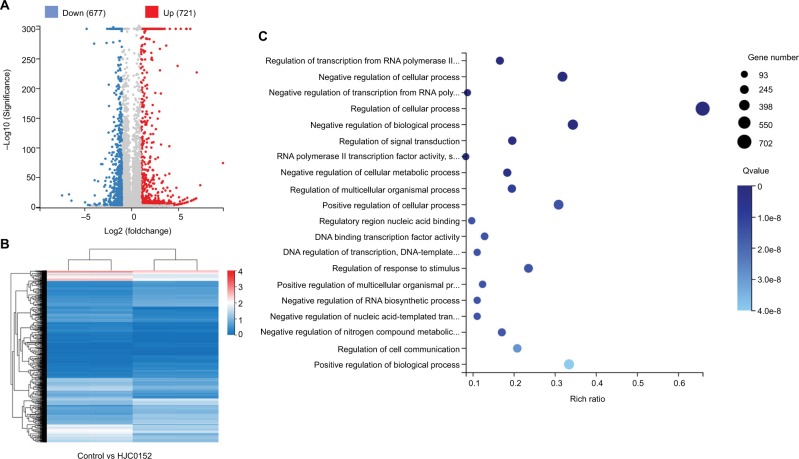
HJC0152 treatment alters multiple cancer-associated biological processes
To determine the potential mechanisms underlying HJC0152 action in the GC cells, we compared the gene expression profiles of the control and treated cells using RNA-Seq. All samples showed high inter-consistency (Figure S2). A total of 1,398 differentially expressed genes (DEGs) were identified between the control and HJC0152-treated AGS cells, of which 721 were up-regulated and 677 down-regulated (Figure 6A, B). The detailed information of the DEGs is summarized in Tables S1 and S2. Gene ontology analysis23 revealed that the DEGs were enriched in various biological functions like regulation of cell processes (Figure 6C and Table S3). Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis24 indicated that a wide range of cancer-related biological pathways of DEGs were affected by HJC0152 exposure, including FoxO, Jak-STAT, Wnt, tumor necrosis factor, mammalian target of rapamycin, and MAPK signaling pathways (Table 1).
Figure 6.
DEGs affected by HJC0152 treatment.
Note: (A) Scatter plot of DEGs. Red and blue dots represent the down- and up-regulated genes, respectively. (B) Hierarchical clustering of global gene expression pattern of the HJC0152-treatment and control cells. Gradient color barcode at top right indicates fold change of expression (log2). (C) The 20 most enriched pathways (with lowest Q value) based on GO analysis. Rich factor is the ratio of DEGs to all genes in this pathway.
Abbreviations: DEG, differentially expressed genes; GO, Gene Ontology.
Table 1.
Important pathway and genes affected by HJC0152 treatment
| Pathway | Counts | Dysregulation of genes | P-value |
|---|---|---|---|
|
| |||
| FoxO signaling pathway | 24 | FOXO6,CDKN1A,CDKN2B,CDKN2D,KLF2,PLK2,PLK3,PLK3,EGFR,FOXO3,SIRT1,IRS1,PIK3CB,PIK3R2, BCL6,SGK1,SKP2,BNIP3,SOS1,SETD7,CCNG2,NXPE3,HOMER1,RALGPS1 | 0.00037241 |
| Jak-STAT signaling pathway | 26 | CDKN1A,IL24,TOM1L2,EGFR,IFNL1,IFNE,IL3RA,IL3RA,IL6R,IL10RA,IL13RA2,LIF,MCL1,MYC,PDGFA,PD GFB,PIK3CB,PIM1,PIK3R2,IL17D,SOS1,STAT5B,SOCS1,SOCS3,IL27RA,RALGPS1 | 0.000718774 |
| Wnt signaling pathway | 27 | FRAT1,LOC100996598,CTBP2,DKK1,FZD2,TES,JUN,JUN,NRROS,MYC,NFATC2,NFATC4,CXXC5,MA NSC1,PRKCA,PRKCB,VANGL2,Table 1X,WNT9A,FOSL1,WNT5B,VANGL1,AXIN2,DIXDC1,FHDC1,RO CK2,DGCR2 | 0.001921694 |
| TNF signaling pathway | 21 | MAP3K8,CILP2,DAB2IP,MLKL,FOS,CXCL1,JUN,JUNB,LIF,MMP3,CCDC88C,PIK3CB,PIK3R2,PTGS2,BCL 3,TNFAIP3,TRAF3,TRAF5,NLRC5,MAP3K14,SOCS3 | 0.001731405 |
| mTOR signaling pathway | 26 | LOC100996598,FLCN,FZD2,GRB7,IRS1,NRROS,PIK3R2,PIK3R2,DDIT4,MANSC1,PRKCA,PRK CB,MOSPD1,SGK1,SKP2,SLC3A2,SOS1,WNT9A,PRR5L,WNT5B,SESN2,DIXDC1,ATP6V1E2,F NIP1,RALGPS1,DGCR2 | 0.003190345 |
| MAPK signaling pathway | 40 | PLA2G4B,ANGPTL7,DUSP10,MAP3K8,CSF1R,RASGEF1B,DDIT3,DDIT3,DUSP1,DUSP2,DUSP6,EFNA2, EGFR,EPHA2,FGF1,FLNB,FLNC,ANGPTL2,FOS,NR4A1,HSPA1L,HSPA6,HSPA8,JUN,JUND,SAMD5,CCD C88C,MYC,THAP4,PDGFA,PDGFB,PGF,PRKCA,CACHD1,CACHD1,PTPRR,SOS1,CACNB1, MAP3K14,RALGPS1 | 0.003195837 |
Abbreviations: MAPK, mitogen-activated protein kinases; mTOR, mammalian target of rapamycin; STAT, signal transducer and activator of transcription.
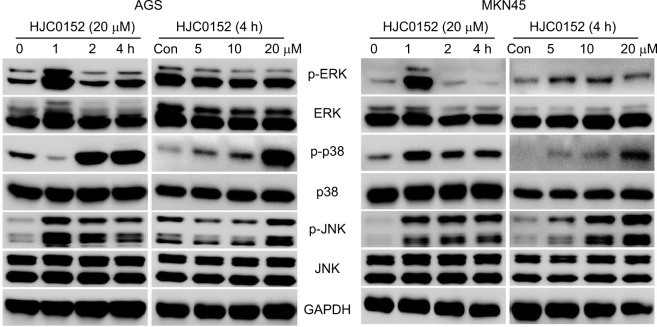
HJC0152 regulates the activation of MAPK signaling pathway in GC cells
RNA-Seq and bioinformatics analyses showed that in addition to STAT signaling pathway, HJC0152 affected many other signaling pathways as well, especially the MAPK pathway (P<0.01, Table S4). To further determine the effect of HJC0152 on the MAPK pathway, we analyzed the expression and activation status of the pathway mediators. As shown in Figure 7A, B, HCJ0152 exposure significantly increased the expression of the phosphorylation of p38 and JNK in a time- and dose-dependent manner relative to the total p38 and JNK levels in both AGS and MKN45 cells. However, HCJ0152 only slightly affected ERK phosphorylation and had no effect on total ERK expression levels. Taken together, these results suggested that p38/JNK MAPK activation was involved in HJC0152-induced apoptosis in GC.
Figure 7.
The effects of HJC0152 on MAPK pathway mediators.
Notes: Western blots showing the expression levels of p-ERK, ERK, p-p38, p38, p-JNK, and JNK in protein samples of the AGS (left) and MKN45 (right) cells treated with 20 µM HJC0152 of varying time duration (0, 1, 2, and 4 hours) or with different concentrations of HJC0152 (0, 5, 10, and 20 µM) for 4 hours. GAPDH was used as the internal control. Western blot analysis was used to evaluate. Three independent experiments were performed.
Abbreviations: ERK, extracellular signal-regulated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; JNK, c-Jun N-terminal kinase.
Discussion
Under normal physiological conditions, STAT3 phosphorylation is transient and lasts for only a relatively short period of time during signal transduction.9 Activated STAT3 undergoes dimerization and nuclear translocation, and then binds to and transcriptionally activates downstream target genes (eg, c-Myc, cyclinD1, Bcl-xl, Bcl-2, survivin, matrix metal-loproteinase (MMP)2, and MMP9) involved in multiple biological processes.25,26 Constitutive activation of STAT3 has been associated with the initiation and progression of various diseases, especially cancer.10 STAT3 overexpression has been observed in the head and neck, prostate, lung, breast, ovarian, and GCs,13,27–32 and is generally associated with worse prognosis, indicating its potential as a therapeutic target.
Although several inhibitors of STAT3 signaling have been identified and developed, none of them have been approved by Food and Drug Administration for clinical treatment. HJC0152 was first designed in 2012 as a derivative of niclosamide,16 and showed significantly improved aqueous solubility and more potent STAT3 suppression ability in vitro. Additionally, 25 mg/kg HJC0152 showed superior anti-tumor effect on breast cancer xenografts compared with 75 mg/kg niclosamide without significant side effects. A recent study showed that HNSCC cell lines expressing high levels of p-STAT3 (Y705) were more sensitive to HJC0152 compared with the low-expressing cell lines. HJC0152 inhibited STAT3 signaling pathway as well as the miR-21/β-catenin axis leading to suppression of cell growth and invasion of HNSCC cells.17 These findings indicated the potential therapeutic utility of HJC0152 in various cancers.
In the present study, we explored the anti-tumor effect of HJC0152 in GC cells. We showed for the first time that HJC0152 significantly suppressed expression of p-STAT3 (Y705) and the activation of STAT3 target genes (c-Myc and cyclinD1) in dose-dependent manner in GC cells. Furthermore, HJC0152 markedly reduced the growth and colony formation capacity of AGS and MKN45 cells in vitro, and inhibited the growth of MKN45 xenografts in vivo with no apparent side effects. However, its suppressive effects were much worse in HGC-27 cells with lower expression levels of p-STAT3 (Y705). Besides, HJC0152 induced significant apoptosis at the concentration of 20 µM, and down-regulated the anti-apoptosis proteins (survivin and Mcl1) and up-regulated c-PARP in GC cells in vitro. Finally, the migration and invasion capacities of GC cells were dramatically reduced following HJC0152 treatment. Taken together, HJC0152 showed potent anti-tumor effects in GC cells both in vivo and in vitro.
The MAPK signaling pathway is critical in maintaining cellular homeostasis. The three best-studied members of this pathway are ERKs, p38, and JNKs MAPK.21 Aberrant activation of the MAPKs is closely associated with tumor development,19,21,33 and a number of compounds, therefore, have been identified or designed to target the MAPK pathway.34–37 Previous studies showed that niclosamide, the precursor of HJC0152, inhibited the expression of both the phosphorylated and total ERKs in a time-dependent manner in the human glioblastoma U-87 MG cell line,38 but not in the human breast cancer MCF-7 cell line.39 Another study indicated that niclosamide treatment down-regulated the expression of many genes related to the MAPK signaling pathway, using RNA-Seq analysis.40 Consistent with this, we identified several DEGs in GC cells following HJC0152 treatment, of which 40 were significantly enriched in the MAPK pathway. Furthermore, we verified that HJC0152 activated p38 and JNK in a time- and dose-dependent manner, although its effects on the expression of ERKs were not that promising. These data suggested that the MAPK signaling pathway was involved in the tumor suppressive effect of HJC0152 in GC.
Conclusion
In conclusion, this study demonstrates for the first time that HJC0152 effectively inhibits STAT3 activation in GC cells, and retards their growth in vitro and in vivo. Moreover, HJC0152 induced apoptosis of GC cells along with the inhibition of survivin and Mcl1, and activation of c-PARP time- and dose-dependently in p-STAT3 (Y705) overexpressing GC cells. Besides, RNA-Seq analysis and Western blotting revealed that HJC0152 time- and dose-dependently activated p38 and JNK MAPK. Taken together, our findings provide evidence for the therapeutic use of HJC0152 in GC and future investigations are warranted.
Supplementary Materials
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (no. 81201089 and 81272676) and by Natural Science Foundation of Zhejiang province, China (no. LY15H160026 and LY15H160012).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16(4):201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fidler IJ, Kripke ML. The challenge of targeting metastasis. Cancer Metastasis Rev. 2015;34(4):635–641. doi: 10.1007/s10555-015-9586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 8.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19(21):2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 9.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 10.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8(4):945–954. [PubMed] [Google Scholar]
- 11.Furtek SL, Backos DS, Matheson CJ, Reigan P. Strategies and approaches of targeting STAT3 for cancer treatment. ACS Chem Biol. 2016;11(2):308–318. doi: 10.1021/acschembio.5b00945. [DOI] [PubMed] [Google Scholar]
- 12.Yu S, Li G, Wang Z, et al. The prognostic value of pSTAT3 in gastric cancer: a meta-analysis. J Cancer Res Clin Oncol. 2016;142(3):649–657. doi: 10.1007/s00432-015-2023-1. [DOI] [PubMed] [Google Scholar]
- 13.Pan YM, Wang CG, Zhu M, et al. STAT3 signaling drives EZH2 transcriptional activation and mediates poor prognosis in gastric cancer. Mol Cancer. 2016;15(1):79. doi: 10.1186/s12943-016-0561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng JY, Sun D, Liu XY, Pan Y, Liang H. STAT-3 correlates with lymph node metastasis and cell survival in gastric cancer. World J Gastroenterol. 2010;16(42):5380–5387. doi: 10.3748/wjg.v16.i42.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Judd LM, Bredin K, Kalantzis A, Jenkins BJ, Ernst M, Giraud AS. STAT3 activation regulates growth, inflammation, and vascularization in a mouse model of gastric tumorigenesis. Gastroenterology. 2006;131(4):1073–1085. doi: 10.1053/j.gastro.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Yang Z, Ding C, et al. Discovery of O-alkylamino tethered niclosamide derivatives as potent and orally bioavailable anticancer agents. ACS Med Chem Lett. 2013;4(2):180–185. doi: 10.1021/ml3003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Wang S, Wu Y, et al. Suppression of the growth and invasion of human head and neck squamous cell carcinomas via regulating STAT3 signaling and the miR-21/β-catenin axis with HJC0152. Mol Cancer Ther. 2017;16(4):578–590. doi: 10.1158/1535-7163.MCT-16-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim EK, Choi EJ. Compromised MAPK signaling in human diseases: an update. Arch Toxicol. 2015;89(6):867–882. doi: 10.1007/s00204-015-1472-2. [DOI] [PubMed] [Google Scholar]
- 19.Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120(22):3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scuteri A, Galimberti A, Maggioni D, et al. Role of MAPKs in platinum-induced neuronal apoptosis. Neurotoxicology. 2009;30(2):312–319. doi: 10.1016/j.neuro.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410(6824):37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Rao X, Huang K, Jiang X, Wang H, Teng L. FH535 inhibits proliferation and motility of colon cancer cells by targeting Wnt/β-catenin signaling pathway. J Cancer. 2017;8(16):3142–3153. doi: 10.7150/jca.19273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye J, Fang L, Zheng H, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34(Web Server issue):W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Rogoff HA, Keates S, et al. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc Natl Acad Sci USA. 2015;112(6):1839–1844. doi: 10.1073/pnas.1424171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S. Regulation of metastases by signal transducer and activator of transcription 3 signaling pathway: clinical implications. Clin Cancer Res. 2007;13(5):1362–1366. doi: 10.1158/1078-0432.CCR-06-2313. [DOI] [PubMed] [Google Scholar]
- 27.Banerjee K, Resat H. Constitutive activation of STAT3 in breast cancer cells: a review. Int J Cancer. 2016;138(11):2570–2578. doi: 10.1002/ijc.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K. Stem cell marker (nanog) and Stat-3 signaling promote microRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31(2):149–160. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anglesio MS, George J, Kulbe H, et al. IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin Cancer Res. 2011;17(8):2538–2548. doi: 10.1158/1078-0432.CCR-10-3314. [DOI] [PubMed] [Google Scholar]
- 30.Takemoto S, Ushijima K, Kawano K, et al. Expression of activated signal transducer and activator of transcription-3 predicts poor prognosis in cervical squamous-cell carcinoma. Br J Cancer. 2009;101(6):967–972. doi: 10.1038/sj.bjc.6605212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mora LB, Buettner R, Seigne J, et al. Constitutive activation of STAT3 in human prostate tumors and cell lines: direct inhibition of STAT3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62(22):6659–6666. [PubMed] [Google Scholar]
- 32.Xu YH, Lu S. A meta-analysis of STAT3 and phospho-STAT3 expression and survival of patients with non-small-cell lung cancer. Eur J Surg Oncol. 2014;40(3):311–317. doi: 10.1016/j.ejso.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 33.Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010;1802(4):396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Temraz S, Mukherji D, Shamseddine A. Dual inhibition of MEK and PI3K pathway in KRAS and BRAF mutated colorectal cancers. Int J Mol Sci. 2015;16(9):22976–22988. doi: 10.3390/ijms160922976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gehringer M, Muth F, Koch P, Laufer SA. c-Jun N-terminal kinase inhibitors: a patent review (2010 – 2014) Expert Opin Ther Pat. 2015;25(8):849–872. doi: 10.1517/13543776.2015.1039984. [DOI] [PubMed] [Google Scholar]
- 36.Astolfi A, Iraci N, Manfroni G, Barreca ML, Cecchetti V. A comprehensive structural overview of p38α MAPK in complex with type I inhibitors. Chem Med Chem. 2015;10(6):957–969. doi: 10.1002/cmdc.201500030. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Y, Zhang G, Li G. Targeting MAPK pathway in melanoma therapy. Cancer Metastasis Rev. 2013;32(3-4):567–584. doi: 10.1007/s10555-013-9433-9. [DOI] [PubMed] [Google Scholar]
- 38.Cheng B, Morales LD, Zhang Y, Mito S, Tsin A. Niclosamide induces protein ubiquitination and inhibits multiple pro-survival signaling pathways in the human glioblastoma U-87 MG cell line. PLoS One. 2017;12(9):e0184324. doi: 10.1371/journal.pone.0184324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fonseca BD, Diering GH, Bidinosti MA, et al. Structure-activity analysis of niclosamide reveals potential role for cytoplasmic pH in control of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem. 2012;287(21):17530–17545. doi: 10.1074/jbc.M112.359638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prather GR, Maclean JA, II, Shi M, Boadu DK, Paquet M, Hayashi K. Niclosamide as a potential nonsteroidal therapy for endometriosis that preserves reproductive function in an experimental mouse model. Biol Reprod. 2016;95(4):76. doi: 10.1095/biolreprod.116.140236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.