Abstract
Background
Lung carcinogenesis is related to silencing of tumor suppressor genes and activation of oncogenes. The aim was to investigate the significance of death-associated protein kinase (DAPK) methylation in non-small-cell lung cancer (NSCLC) through a meta-analysis.
Methods
A detailed literature search was made in PubMed, Embase, and Web of Science databases. All analysis was performed with Review Manager 5.2.
Results
In total, 28 studies with a total of 2,148 patients were involved. The frequency of DAPK promoter hypermethylation was 40.50% in NSCLC, significantly higher than in nonmalignant lung tissue; the pooled OR was 5.69, P<0.00001. Additionally, DAPK promoter hypermethylation was significantly correlated with poor overall survival in patients with NSCLC. However, there was no significant difference found while comparing the rate of DAPK promoter hypermethylation in adenocarcinoma and squamous cell cancer. The rate of DAPK promoter hypermethylation was similar between stage III/IV and stage I/II. In addition, the data showed that DAPK promoter hypermethylation was not associated with smoking behavior in patients with NSCLC.
Conclusion
DAPK promoter hypermethylation is correlated with risk of NSCLC and is a potential biomarker for prediction of poor prognosis in patients with NSCLC.
Keywords: DAPK, NSCLC, biomarker, methylation, adenocarcinoma, squamous cell cancer, drug target
Background
Lung cancer is the second most commonly diagnosed malignancy in men and the third most commonly diagnosed malignancy in women worldwide.1 Lung cancer can be classified two major histological groups: small cell lung cancer and non-small-cell lung cancer (NSCLC). NSCLC accounts for more than 80% of all lung cancers, whereas 15%–20% is small cell lung cancer.2,3 NSCLC includes adenocarcinoma (ADC), squamous cell carcinoma (SCC), and large-cell carcinoma, within them, ADC accounts for 40%, SCC for 25%–30%, and large-cell carcinoma for 10%–15%.4,5 Lung carcinogenesis is related to silencing of tumor suppressor genes and activation of oncogenes. Accumulating data indicate that hypermethylation in CpG-rich promoter regions of many suppressor genes can contribute to the development and progression of a variety of cancers.6,7
Deiss and Kimchi discovered a large group of new genes by using “technical knock-out (TKO) and rescue” screen.8 Those genes function as positive mediators of cell death pathways, therefore they were named “Death-Associated Protein or DAP genes.9 One of the genes isolated by the TKO approach encoded a calcium calmodulin regulated serine/threonine kinase and was named DAP kinase (DAPk1 or DAPK).10,11 Further investigation indicated that DAPK plays an important role in apoptotic and autophagic cell death,12–14 tumor progression suppression, and metastasis suppression.15–19 Last two decades, a number of studies showed that DAPK loss by its promoter hypermethylation was associated with the development and progression of NSCLC. However, the results from individual studies were inconsistent due to small size of samples. In the present study, 28 relevant studies were pooled, and a meta-analysis was performed to evaluate the clinicopathological significance of DAPK promoter hypermethylation in NSCLC.
Methods
Search strategy and selection criteria
The following electronic databases were screened for relevant articles without any language restrictions: PubMed (1966–2018), Embase (1980–2018), Web of Science (1945–2018), Cochrane Library Database (1972–2018). We used the following keywords: “DAPK methylation”, “NSCLC”, “Non-small-cell lung cancer”, and “lung cancer”. A search of PubMed yielded 65 articles, Embase yielded 101, and Web Science yielded 138 articles. The included criteria were as follows: (1) the association between DAPK methylation and the clinicopathological significance of NSCLC; (2) the association of DAPK and prognosis in patients with NSCLC. After screening by titles and abstracts, 38 relevant articles were included for full text review. The following exclusion criteria were used: (1) the same population or overlapping database; (2) conference abstracts containing insufficient data reviews, editorials, letters, case reports, and expert opinion; (3) the studies utilized cell lines. After evaluation, 28 articles fulfilled the entry criteria of this meta-analysis. The detailed information of 28 relevant articles was listed in Table 1.
Table 1.
Main characteristics of included studies
| Study | Year | Country | Sample size (M/T) | DAPK methylation rate (%) | Histology
|
Stage (TNM)
|
Smoking status
|
Method | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCT | AC | SCC | I+II | III+IV | + | − | ||||||
|
| ||||||||||||
| Ali et al28 | 2017 | India | 133/160 | 83.13 | 49/70 | – | – | – | – | – | – | MSP |
| Jin et al29 | 2016 | China | 120/199 | 60.30 | – | 55/95 | 65/104 | – | – | 94/145 | 26/54 | MSP |
| Guo et al30 | 2015 | China | 35/202 | 17.33 | – | 15/111 | 20/91 | 16/100 | 5/27 | – | – | MSP |
| Kontic et al31 | 2012 | Serbia | 11/47 | 23.40 | – | 4/18 | 7/29 | 10/35 | 2/18 | 1/11 | 11/44 | MSP |
| Fujii et al32 | 2012 | Japan | 6/46 | 13.04 | 0/25 | – | – | – | – | – | – | MSP |
| Zhang et al33 | 2011 | China | 120/200 | 61.00 | 23/200 | – | – | – | – | – | – | MSP |
| Zhang et al34 | 2010 | China | 11/78 | 14.10 | 3/78 | – | – | – | – | – | – | MSP |
| Jin et al35 | 2010 | China | 88/150 | 58.67 | 15/150 | – | – | – | – | – | – | MSP |
| Peng et al36 | 2010 | China | 48/82 | 58.54 | 0/25 | – | – | – | – | – | – | MSP |
| Niklinska et al37 | 2009 | Japan | 22/61 | 36.07 | – | – | – | – | – | – | – | MSP |
| Han et al38 | 2009 | USA | 8/14 | 57.14 | 4/20 | – | – | – | – | – | – | MSP |
| Licchesi et al39 | 2008 | USA | 7/19 | 36.8 | 0/46 | MSP | ||||||
| Katayama et al40 | 2007 | Japan | – | – | – | – | 2/8 | 8/26 | – | – | MSP | |
| Yanagawa et al41 | 2007 | Japan | 26/101 | 25.74 | 8/101 | 14/62 | 12/39 | 20/75 | 6/26 | 20/73 | 6/28 | MSP |
| Liu et al42 | 2007 | China | 40/122 | 32.79 | – | 25/72 | 15/50 | 17/54 | 17/54 | 28/81 | 12/41 | MSP |
| Belinsky et al43 | 2007 | USA | 22/72 | 30.56 | 5/25 | – | – | – | – | – | – | MSP |
| Fischer et al44 | 2007 | Germany | – | – | – | – | – | – | – | – | MSP | |
| Kim et al45 | 2005a | Korea | 23/72 | 31.94 | 4/72 | – | – | – | – | – | – | MSP |
| de Fraipont et al46 | 2005 | France | – | – | – | – | – | – | 18/121 | 1/4 | MSP | |
| Safar et al47 | 2005 | USA | 12/32 | 37.50 | 6/32 | – | – | 16/48 | 8/57 | – | – | MSP |
| Russo et al48 | 2005 | USA | 22/49 | 44.90 | 1/27 | – | – | – | – | – | – | MSP |
| Kim et al49 | 2005b | Korea | – | – | 13/42 | 7/17 | 9/34 | 9/27 | – | – | MSP | |
| Fujiwara et al50 | 2005 | Japan | 10/91 | 10.99 | 5/100 | – | – | 6/60 | 4/31 | 9/43 | 10/38 | MSP |
| Divine et al51 | 2005 | USA | 72/206 | 34.95 | – | – | – | – | – | 16/45 | 45/125 | MSP |
| Lu et al19 | 2004 | USA | – | – | – | – | – | – | – | – | MSP | |
| Toyooka et al52 | 2003 | USA | 14/38 | 36.84 | 1/15 | 8/20 | 6/18 | – | – | – | – | COBRA |
| Soria et al53 | 2002 | USA | – | – | – | – | – | – | 13/89 | 4/11 | MSP | |
| Zöchbauer-Müller et al54 | 2001 | Australia | 20/107 | 18.69 | 6/104 | 7/45 | 9/43 | 17/82 | 3/25 | 18/98 | 2/9 | MSP |
Abbreviations: ADC, adenocarcinoma; COBRA, combined bisulfite restriction analysis; DAPK, death-associated protein kinase; M, number of NSCLC with methylation; MSP, methylation-specific PCR; NCT, normal control tissue; NSCLC, non-small-cell lung cancer; SCC, squamous cell cancer; T, total number of NSCLC.
Data extraction and study assessment
Two reviewers (YZ and JW) extracted data from selected studies independently by using a standardized data extraction form including the following items: first author’s name, year of publication, country, number of patients, histology, stage of NSCLC, smoking status of patients with NSCLC, method for methylation detection. Any disagreement was discussed and reached a consensus for all issues.
Statistical analysis
ORs with 95% confidence intervals (CIs) were calculated by using a fixed or random effect model depending on heterogeneity (a fixed effect model for I2<50%, a random effect model for I2>50%). The analysis was performed to compare DAPK promoter hypermethylation between NSCLC and normal tissue, DAPK promoter hypermethylation in different stage of NSCLC, DAPK promoter hypermethylation in different histology type of NSCLC, as well as in smoker and nonsmoker patients with NSCLC. All P-values were two sided. P-value less than 0.05 was considered statistically significant. Funnel plots were used for detection of publication bias. All analysis was performed with Review Manager 5.2.
Results
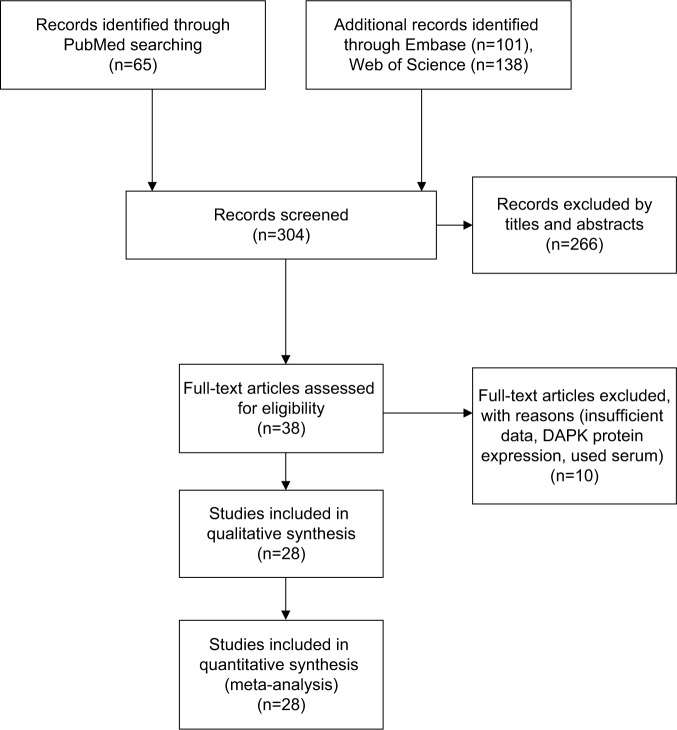
In total, 28 studies were included in the present study after screening 304 articles by two reviewers (Figure 1). The following items were collected from each study: first author, published year, country, histology of NSCLC, and DAPK hypermethylation status, smoking status as well as patient prognosis (Table 1).
Figure 1.
Schematic flow diagram for selection of included studies.
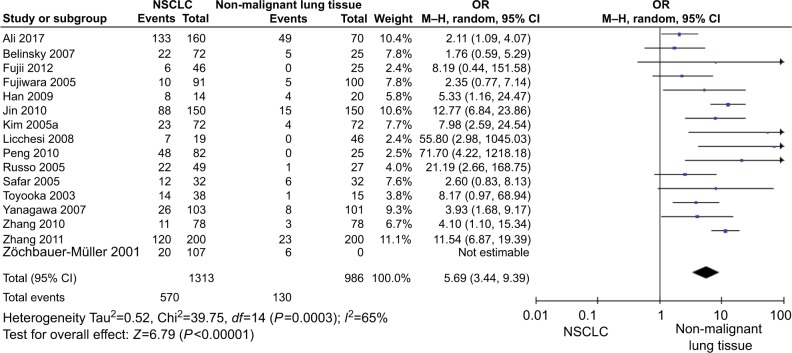
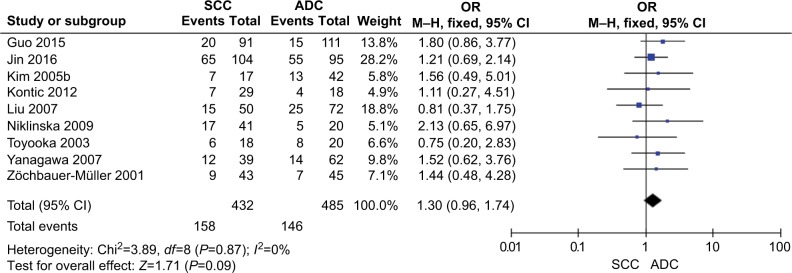
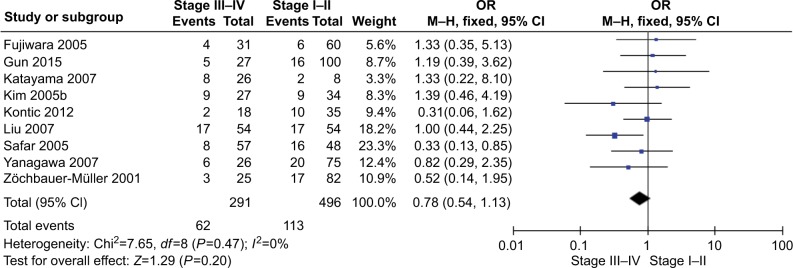
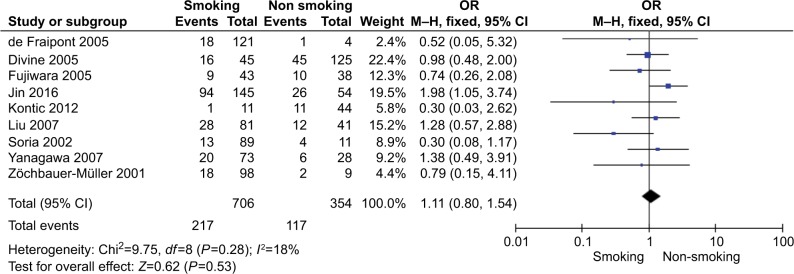
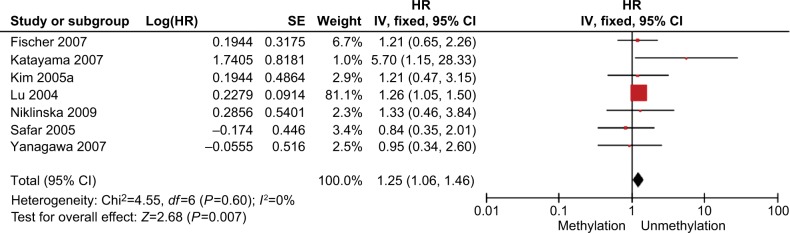
The total number of NSCLC tumor from 28 studies is 2148, 870 of them were with DAPK promoter hypermethylated, the rate was 40.50%. Whereas the promoter hyper-methylation rate from individual study ranged from 10.99% to 83.13% (Table 1). The frequency of DAPK promoter hypermethylation was significantly higher in NSCLC than in non-malignant lung; and the pooled OR was 5.69 with 95% CI 3.44–9.39, Z=6.79, P<0.00001 (Figure 2). DAPK promoter methylation was similar between SCC and ADC; the pooled OR was 1.30 with 95% CI 0.96–1.74, Z=1.71, P=0.09, I2=0% (Figure 3). In addition, DAPK methylation was not significantly correlated with stages of NSCLC; OR was 0.78 with 95% CI 0.54–1.13, Z=1.29, P=0.20, I2=0% (Figure 4). The rate of DAPK methylation was not associated with smoking behavior in patients with NSCLC; OR was 1.11 with 95% CI 0.80–1.54, Z=0.62, P=0.53, I2=18% (Figure 5). However, DAPK promoter hypermethylation was significantly associated with poor prognosis in patients with NSCLC; HR was 1.25 with 95% CI 1.06–1.46, Z=2.68, P=0.007, I2=0% (Figure 6).
Figure 2.
Forest plot for DAPK promoter hypermethylation in NSCLC and non-malignant lung tissue.
Notes: The squares represent the weight of individual study in the meta-analysis, the line width indicates the corresponding 95% CI, the diamond represents the pooled OR, and the width of diamond indicates 95% CI.
Abbreviations: DAPK, death-associated protein kinase; M–H, Mantel–Haenszel; NSCLC, non-small-cell lung cancer.
Figure 3.
Forest plot for DAPK promoter hypermethylation in SCC and ADC.
Notes: The squares represent the weight of individual study in the meta-analysis, the line width indicates the corresponding 95% CI, the diamond represents the pooled OR, and the width of diamond indicates 95% CI.
Abbreviations: ADC, adenocarcinoma; DAPK, death-associated protein kinase; M–H, Mantel–Haenszel; NSCLC, non-small-cell lung cancer; SCC, squamous cell carcinoma.
Figure 4.
Forest plot for DAPK promoter hypermethylation in NSCLC stage III/IV and stage I/II.
Notes: The squares represent the weight of individual study in the meta-analysis, the line width indicates the corresponding 95% CI, the diamond represents the pooled OR, and the width of diamond indicates 95% CI.
Abbreviations: DAPK, death-associated protein kinase; M–H, Mantel–Haenszel; NSCLC, non-small-cell lung cancer.
Figure 5.
Forest plot for DAPK promoter hypermethylation in NSCLC patients with smoking and non-smoking behavior.
Notes: The squares represent the weight of individual study in the meta-analysis, the line width indicates the corresponding 95% CI, the diamond represents the pooled OR, and the width of diamond indicates 95% CI.
Abbreviations: M–H, Mantel–Haenszel; NSCLC, non-small-cell lung cancer.
Figure 6.
Forest plot for the association of DAPK promoter hypermethylation and the overall survival of NSCLC patients.
Notes: The squares represent the weight of individual study in the meta-analysis, the line width indicates the corresponding 95% CI, the diamond represents the pooled OR, and the width of diamond indicates 95% CI.
Abbreviations: DAPK, death-associated protein kinase; NSCLC, non-small-cell lung cancer; SE, standard error.
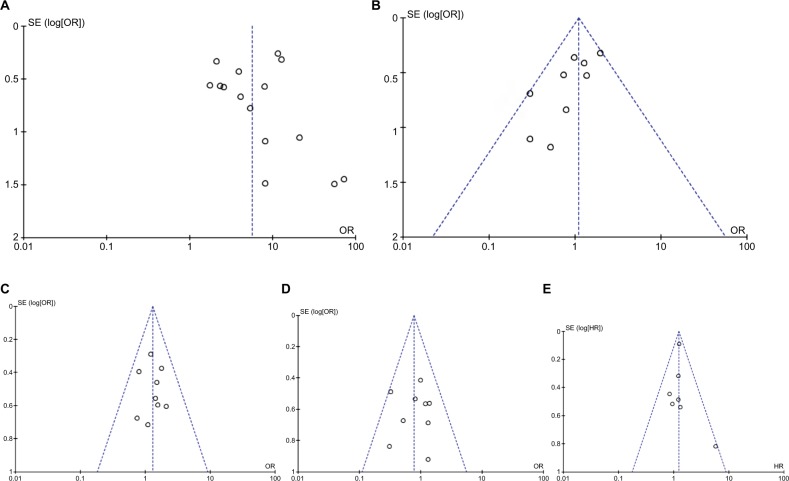
The quality of each study was assessed using the New-castle Ottawa Quality Assessment Scale (NOQAS). This scale for non-randomized case controlled studies and cohort studies was used to allocate a maximum of nine points for the quality of selection, comparability, exposure, and outcomes for study participants. Of the studies, 15 scored eight points, 11 scored seven points, and two scored six points (data not shown). Hence, the studies were of a relatively high quality. A sensitivity analysis, in which one study was removed at a time, was conducted to assess the result stability. The pooled ORs were not significantly changed, indicating the stability of our analyses (data not shown). The funnel plots were largely symmetric (Figure 7), suggesting there were no publication biases in the meta-analysis of DAPK promoter hypermethylation and clinicopathological features.
Figure 7.
Funnel plot for publication bias.
Notes: (A) DAPK promoter hypermethylation in NSCLC and non-malignant lung tissue; (B) DAPK promoter hypermethylation in SCC and ADC; (C) DAPK promoter hypermethylation in NSCLC stage III/IV and stage I/II. (D) DAPK promoter hypermethylation in NSCLC patients with smoking and non-smoking behavior; (E) the association of DAPK promoter hypermethylation and the overall survival of NSCLC patients. Area of the circle represents the weight of individual study.
Abbreviations: ADC, adenocarcinoma; DAPK, death-associated protein kinase; NSCLC, non-small-cell lung cancer; SCC, squamous cell carcinoma; SE, standard error.
Discussion
Aberrant methylation in tumor suppressor genes has been linked to carcinogenesis. Hypermethylation is the predominant mechanism to make tumor suppressor genes silent by promoter inactivation. DAPK gene is located on chromosome 9q34.1. It encodes a proapoptotic protein involved in apoptosis initiated by THN-α, IFN-γ, Fas, and TRAIL. DAPK promoter methylation has been observed in about 30 types of tumor including NSCLC. Moreover, aggressiveness of malignant tumors has been associated with the methylation of the promoter region of the DAPK gene and loss of DAPK expression.20 A number of studies evaluated the methylation rate in NSCLC, which ranged from 10.99% to 83.33% due to small size of samples. We pooled 28 studies including 2,148 NSCLC patients, 870 of them were with DAPK gene promoter hypermethylated; hypermethylation rate was 40.50%, 5.69 times higher than the one in non-malignant lung tissue. Therefore, DAPK promoter hypermethylation was correlated with the risk of NSCLC. Previous evidence indicated that the expression of DAPK was partially restored by treatment with a demethylation agent, 5′-aza-2′-deoxycitidine.15 DAPK could be a potential target for development of new strategy of treatment.
We did not find significant association of DAPK promoter hypermethylation with tumor histology, smoking, and stage. Although TNM staging system still remained the most powerful tool for medical decision making, it is difficult to accurately predict the prognosis for individual patient. The 5-year survival rate for patients with stage I NSCLC is about 65%–80%,21 therefore a more accurate tool, independent from TNM stage, is very important to predict prognosis in those patients. Our finding indicated that DAPK was correlated to worse survival in our meta-analysis, supporting the importance of epigenetic gene regulation in NSCLC progression and prognosis. Loss of apoptotic functions would compromise cell death induced by unrepaired DNA damage.22 In addition, DAPK promoter hypermethylation is associated with metastatic status. Taken together, DAPK promoter hypermethylation leads to worse prognosis in patients with NSCLC. DAPK hypermethylation is a potential predictor of survival in patients with NSCLC.
Given the important role of smoking in the development of lung cancer and the fact that DNA methylation is an early event in carcinogenesis,23 several biomarker such as Wnt inhibitory factor-1 (Wif1), Phosphatase and tensin homologue deleted on chromosome 10 (PTEN), and TP53 were associated with smoking behavior.24–27 However, no correlation was found between DAPK promoter hypermethylation and the smoking behavior in the present study. Further confirmation needs to be finished in future when more relative studies are available.
Our findings should be interpreted in view of certain limitations. First, most of the included studies were retrospective, 26 out of 28 were of sufficient quality (NOQAS ≥ 7). Hence, the studies were of a relatively high quality. Although the possibility of selection, sample, and publication bias could not be excluded, no obvious bias was detected by the funnel plots. Second, present findings were based on individual unadjusted ORs and further confirmation needs to be finished by evaluation adjusted with other potential risk factors.
Conclusion
In summary, present findings suggested that DAPK promoter hypermethylation was correlated with the risk of NSCLC; and DAPK is a promising drug target for development of new therapy strategy. Additionally, DAPK promoter hyper-methylation was a potential predictor of poor prognosis in patients with NSCLC.
Data sharing statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work. The corresponding author had full access to all data and the final responsibility for the decision to submit the article for publication. All authors read and approved the final manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Allen C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3(4):524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reck M, Popat S, Reinmuth N, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii27–iii39. doi: 10.1093/annonc/mdu199. [DOI] [PubMed] [Google Scholar]
- 5.Thakur MK, Wozniak AJ. Spotlight on necitumumab in the treatment of non-small-cell lung carcinoma. Lung Cancer. 2017;8:13–19. doi: 10.2147/LCTT.S104207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10(7):687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 7.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 8.Deiss LP, Kimchi A. A genetic tool used to identify thioredoxin as a mediator of a growth inhibitory signal. Science. 1991;252(5002):117–120. doi: 10.1126/science.1901424. [DOI] [PubMed] [Google Scholar]
- 9.Kimchi A. DAP genes: novel apoptotic genes isolated by a functional approach to gene cloning. Biochim Biophys Acta. 1998;1377(2):F13–F33. doi: 10.1016/s0304-419x(98)00002-x. [DOI] [PubMed] [Google Scholar]
- 10.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9(1):15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 11.Gozuacik D, Kimchi A. DAPk protein family and cancer. Autophagy. 2006;2(2):74–79. doi: 10.4161/auto.2.2.2459. [DOI] [PubMed] [Google Scholar]
- 12.Kögel D, Reimertz C, Düssmann H, Mech P, Scheidtmann KH, Prehn JH. The death associated protein (DAP) kinase homologue Dlk/ZIP kinase induces p19ARF- and p53-independent apoptosis. Eur J Cancer. 2003;39(2):249–256. doi: 10.1016/s0959-8049(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 13.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157(3):455–468. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Kissil JL, Feinstein E, Cohen O, et al. DAP-kinase loss of expression in various carcinoma and B-cell lymphoma cell lines: possible implications for role as tumor suppressor gene. Oncogene. 1997;15(4):403–407. doi: 10.1038/sj.onc.1201172. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann U, Celikkaya G, Hasemeier B, Länger F, Kreipe H. Promoter hypermethylation of the death-associated protein kinase gene in breast cancer is associated with the invasive lobular subtype. Cancer Res. 2002;62(22):6634–6638. [PubMed] [Google Scholar]
- 17.Lévy D, Plu-Bureau G, Decroix Y, et al. Death-associated protein kinase loss of expression is a new marker for breast cancer prognosis. Clin Cancer Res. 2004;10(9):3124–3130. doi: 10.1158/1078-0432.ccr-03-0213. [DOI] [PubMed] [Google Scholar]
- 18.Kim DH, Nelson HH, Wiencke JK, et al. Promoter methylation of DAP-kinase: association with advanced stage in non-small cell lung cancer. Oncogene. 2001;20(14):1765–1770. doi: 10.1038/sj.onc.1204302. [DOI] [PubMed] [Google Scholar]
- 19.Lu C, Soria JC, Tang X, et al. Prognostic factors in resected stage I non-small-cell lung cancer: a multivariate analysis of six molecular markers. J Clin Oncol. 2004;22(22):4575–4583. doi: 10.1200/JCO.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 20.Tang X, Khuri FR, Lee JJ, et al. Hypermethylation of the death-associated protein (DAP) kinase promoter and aggressiveness in stage I non-small-cell lung cancer. J Natl Cancer Inst. 2000;92(18):1511–1516. doi: 10.1093/jnci/92.18.1511. [DOI] [PubMed] [Google Scholar]
- 21.Bains MS. Surgical treatment of lung cancer. Chest. 1991;100(3):826–837. doi: 10.1378/chest.100.3.826. [DOI] [PubMed] [Google Scholar]
- 22.Buckingham L, Penfield Faber L, Kim A, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010;126(7):1630–1639. doi: 10.1002/ijc.24896. [DOI] [PubMed] [Google Scholar]
- 23.Feng Q, Hawes SE, Stern JE, et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17(3):645–654. doi: 10.1158/1055-9965.EPI-07-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo H, Zhou S, Tan L, Wu X, Wu Z, Ran R. Clinicopathological significance of WIF1 hypermethylation in NSCLC, a meta-analysis and literature review. Oncotarget. 2017;8(2):2550–2557. doi: 10.18632/oncotarget.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin G, Kim MJ, Jeon HS, et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung Cancer. 2010;69(3):279–283. doi: 10.1016/j.lungcan.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Tammemagi MC, Mclaughlin JR, Bull SB. Meta-analyses of p53 tumor suppressor gene alterations and clinicopathological features in resected lung cancers. Cancer Epidemiol Biomarkers Prev. 1999;8(7):625–634. [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali A, Kumar S, Kakaria VK, et al. Detection of promoter DNA methylation of APC, DAPK, and GSTP1 genes in tissue biopsy and matched serum of advanced-stage lung cancer patients. Cancer Invest. 2017;35(6):423–430. doi: 10.1080/07357907.2017.1309547. [DOI] [PubMed] [Google Scholar]
- 29.Jin Y, Xu P, Liu X, et al. Cigarette smoking, BPDE-DNA adducts, and aberrant promoter methylations of tumor suppressor genes (TSGs) in NSCLC from Chinese population. Cancer Invest. 2016;34(4):173–180. doi: 10.3109/07357907.2016.1156689. [DOI] [PubMed] [Google Scholar]
- 30.Guo M, Alumkal J, Drachova T, et al. CHFR methylation strongly correlates with methylation of DNA damage repair and apoptotic pathway genes in non-small cell lung cancer. Discov Med. 2015;19(104):151–158. [PubMed] [Google Scholar]
- 31.Kontic M, Stojsic J, Jovanovic D, et al. Aberrant promoter methylation of CDH13 and MGMT genes is associated with clinicopathologic characteristics of primary non-small-cell lung carcinoma. Clin Lung Cancer. 2012;13(4):297–303. doi: 10.1016/j.cllc.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujii M, Fujimoto N, Hiraki A, et al. Aberrant DNA methylation profile in pleural fluid for differential diagnosis of malignant pleural mesothelioma. Cancer Sci. 2012;103(3):510–514. doi: 10.1111/j.1349-7006.2011.02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang CY, Jin YT, Xu HY, et al. Relationship between promoter methylation of p16, DAPK and RAR beta genes and the clinical data of non-small cell lung cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011;28(1):23–28. doi: 10.3760/cma.j.issn.1003-9406.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303(1):21–28. doi: 10.1016/j.canlet.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Jin Y, Xu H, Zhang C, et al. Combined effects of cigarette smoking, gene polymorphisms and methylations of tumor suppressor genes on non small cell lung cancer: a hospital-based case-control study in China. BMC Cancer. 2010;10:422. doi: 10.1186/1471-2407-10-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng Z, Shan C, Wang H. Value of promoter methylation of RASSF1A, p16, and DAPK genes in induced sputum in diagnosing lung cancers. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2010;35(3):247–253. doi: 10.3969/j.issn.1672-7347.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 37.Niklinska W, Naumnik W, Sulewska A, Kozłowski M, Pankiewicz W, Milewski R. Prognostic significance of DAPK and RASSF1A promoter hypermethylation in non-small cell lung cancer (NSCLC) Folia Histochem Cytobiol. 2009;47(2):275–280. doi: 10.2478/v10042-009-0091-2. [DOI] [PubMed] [Google Scholar]
- 38.Han W, Wang T, Reilly AA, Keller SM, Spivack SD. Gene promoter methylation assayed in exhaled breath, with differences in smokers and lung cancer patients. Respir Res. 2009;10:86. doi: 10.1186/1465-9921-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Licchesi JD, Westra WH, Hooker CM, Herman JG. Promoter hypermethylation of hallmark cancer genes in atypical adenomatous hyperplasia of the lung. Clin Cancer Res. 2008;14(9):2570–2578. doi: 10.1158/1078-0432.CCR-07-2033. [DOI] [PubMed] [Google Scholar]
- 40.Katayama H, Hiraki A, Fujiwara K, et al. Aberrant promoter methylation profile in pleural fluid DNA and clinicopathological factors in patients with non-small cell lung cancer. Asian Pac J Cancer Prev. 2007;8(2):221–224. [PubMed] [Google Scholar]
- 41.Yanagawa N, Tamura G, Oizumi H, et al. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58(1):131–138. doi: 10.1016/j.lungcan.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Gao W, Siegfried JM, Weissfeld JL, Luketich JD, Keohavong P. Promoter methylation of RASSF1A and DAPK and mutations of K-ras, p53, and EGFR in lung tumors from smokers and never-smokers. BMC Cancer. 2007;7:74. doi: 10.1186/1471-2407-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belinsky SA, Grimes MJ, Casas E, et al. Predicting gene promoter methylation in non-small-cell lung cancer by evaluating sputum and serum. Br J Cancer. 2007;96(8):1278–1283. doi: 10.1038/sj.bjc.6603721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer JR, Ohnmacht U, Rieger N, et al. Prognostic significance of RASSF1A promoter methylation on survival of non-small cell lung cancer patients treated with gemcitabine. Lung Cancer. 2007;56(1):115–123. doi: 10.1016/j.lungcan.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 45.Kim YT, Park SJ, Lee SH, et al. Prognostic implication of aberrant promoter hypermethylation of CpG islands in adenocarcinoma of the lung. J Thorac Cardiovasc Surg. 2005;130(5):1378–1378. doi: 10.1016/j.jtcvs.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 46.de Fraipont F, Moro-Sibilot D, Michelland S, Brambilla E, Brambilla C, Favrot MC. Promoter methylation of genes in bronchial lavages: a marker for early diagnosis of primary and relapsing non-small cell lung cancer? Lung Cancer. 2005;50(2):199–209. doi: 10.1016/j.lungcan.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 47.Safar AM, Spencer H, Su X, et al. Methylation profiling of archived non-small cell lung cancer: a promising prognostic system. Clin Cancer Res. 2005;11(12):4400–4405. doi: 10.1158/1078-0432.CCR-04-2378. [DOI] [PubMed] [Google Scholar]
- 48.Russo AL, Thiagalingam A, Pan H, et al. Differential DNA hyper-methylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin Cancer Res. 2005;11(7):2466–2470. doi: 10.1158/1078-0432.CCR-04-1962. [DOI] [PubMed] [Google Scholar]
- 49.Kim YT, Lee SH, Sung SW, Kim JH. Can aberrant promoter hyper-methylation of CpG islands predict the clinical outcome of non-small cell lung cancer after curative resection? Ann Thorac Surg. 2005;79(4):1180–1188. doi: 10.1016/j.athoracsur.2004.09.060. discussion 1180–1188. [DOI] [PubMed] [Google Scholar]
- 50.Fujiwara K, Fujimoto N, Tabata M, et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res. 2005;11(3):1219–1225. [PubMed] [Google Scholar]
- 51.Divine KK, Pulling LC, Marron-Terada PG, et al. Multiplicity of abnormal promoter methylation in lung adenocarcinomas from smokers and never smokers. Int J Cancer. 2005;114(3):400–405. doi: 10.1002/ijc.20761. [DOI] [PubMed] [Google Scholar]
- 52.Toyooka S, Toyooka KO, Miyajima K, et al. Epigenetic down-regulation of death-associated protein kinase in lung cancers. Clin Cancer Res. 2003;9(8):3034–3041. [PubMed] [Google Scholar]
- 53.Soria JC, Rodriguez M, Liu DD, Lee JJ, Hong WK, Mao L. Aberrant promoter methylation of multiple genes in bronchial brush samples from former cigarette smokers. Cancer Res. 2002;62(2):351–355. [PubMed] [Google Scholar]
- 54.Zöchbauer-Müller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61(1):249–255. [PubMed] [Google Scholar]