Abstract
Novel influenza viruses of the H7N9 subtype have infected 33 and killed nine people in China as of 10 April 2013. Their haemagglutinin (HA) and neuraminidase genes probably originated from Eurasian avian influenza viruses; the remaining genes are closely related to avian H9N2 influenza viruses. Several characteristic amino acid changes in HA and the PB2 RNA polymerase subunit probably facilitate binding to human-type receptors and efficient replication in mammals, respectively, highlighting the pandemic potential of the novel viruses.
Humans are rarely infected with avian influenza viruses, with the exception of highly pathogenic avian influenza A(H5N1) viruses, which have caused 634 infections and 371 deaths as of 12 March 2013 [1]. A few isolated cases of human infection with viruses of the H7N2, H7N3, and H7N5 subtypes have been reported, but none were fatal [2–11]. In 2003, in the Netherlands, 89 people were infected with an influenza virus of the H7N7 subtype that caused conjunctivitis and one fatality [5,7].
On 19 February 2013, an 87 year-old man in Shanghai developed a respiratory infection and died on 4 March, and on 27 February 2013, a 27 year-old pork seller in a Shanghai market became ill and died on 10 March. A 35 year-old woman in Chuzhou City in Anhui province (west of Shanghai), who had contact with poultry, became ill on 15 March 2013, and remains hospitalised in critical condition. There is no known epidemiological relationship among these three cases. A 38 year-old man in Hangzhou (Zhejiang province, south of Shanghai) became ill on 7 March 2013 and died on 27 March. All four cases presented with respiratory infections that progressed to severe pneumonia and breathing difficulties.
On 31 March 2013, the Chinese Centre for Disease Control and Prevention announced the isolation in embryonated eggs of avian influenza viruses of the H7N9 subtype (designated A/Shanghai/1/2013, A/Shanghai/2/2013, and A/Anhui/1/2013) from the first three cases. The sequences of the coding regions of all eight viral genes were deposited in the influenza sequence database of the Global Initiative on Sharing All Influenza Data (GISAID) on 31 March (Table 1). On 5 April 2013, the Hangzhou Center for Disease Control and Prevention deposited the haemagglutinin (HA), neuraminidase (NA), and matrix (M) gene sequences of A/Hongzhou/1/2013 virus (Table 1), which was isolated in cell culture from samples obtained from the 38 year-old man.
Table 1.
Origin of influenza A(H7N9) isolates included in the phylogenetic analysis, China, February–April 2013 (n=7)
| Segment ID | Segment | Isolate name | Collection date |
Originating laboratory | Submitting laboratorydate |
Submitter/Authors |
|---|---|---|---|---|---|---|
| EPI439488 | PB2 | A/Shanghai/1/2013 | 2013 | - | WHO Chinese National Influenza Center |
Lei yang |
| EPI439489 | PB1 | |||||
| EPI439490 | PA | |||||
| EPI439486 | HA | |||||
| EPI439491 | NP | |||||
| EPI439487 | NA | |||||
| EPI439493 | M | |||||
| EPI439494 | NS | |||||
| EPI439495 | PB2 | A/Shanghai/2/2013 | 2013 | - | ||
| EPI439501 | PB1 | |||||
| EPI439498 | PA | |||||
| EPI439502 | HA | |||||
| EPI439496 | NP | |||||
| EPI439500 | NA | |||||
| EPI439497 | M | |||||
| EPI439499 | NS | |||||
| EPI439504 | PB2 | A/Anhui/1/2013 | 2013 | - | ||
| EPI439508 | PB1 | |||||
| EPI439503 | PA | |||||
| EPI439507 | HA | |||||
| EPI439505 | NP | |||||
| EPI439509 | NA | |||||
| EPI439506 | M | |||||
| EPI439510 | NS | |||||
| EPI440095 | HA | A/Hangzhou/1/2013 | 2013-03-24 | Hangzliou Center for Disease Control and Prevention |
Hangzhou Center for Disease Control and Prevention |
Li,J; Pan,JC; Pu,XY; Yu,XF; Kou,Y; Zhou,YY |
| EPI440096 | NA | |||||
| EPI440097 | M | |||||
| EPI440682 | PB2 | A/Chicken/Shanghai /S1053/2013 |
2013-04-03 | Harbin Veterinary Research Institute |
Harbin Veterinary Research Institute |
Huihui kong |
| EPI440683 | PB1 | |||||
| EPI440681 | PA | |||||
| EPI440685 | HA | |||||
| EPI440678 | NP | |||||
| EPI440684 | NA | |||||
| EPI440680 | M | |||||
| EPI440679 | NS | |||||
| EPI440690 | PB2 | A/Environment/ Shanghai /S1088/2013 |
2013-04-03 | |||
| EPI440691 | PB1 | |||||
| EPI440689 | PA | |||||
| EPI440693 | HA | |||||
| EPI440686 | NP | |||||
| EPI440692 | NA | |||||
| EPI440688 | M | |||||
| EPI440687 | NS | |||||
| EPI440698 | PB2 | A/Pigeon/Shanghai /S1069/2013 |
2013-04-02 | |||
| EPI440699 | PB1 | |||||
| EPI440697 | PA | |||||
| EPI440701 | HA | |||||
| EPI440694 | NA | |||||
| EPI440700 | NA | |||||
| EPI440696 | M | |||||
| EPI440695 | NS |
We gratefully acknowledge the authors and laboratories for originating and submitting these sequences to the EpiFlu database of the Global Initiative on Sharing All Influenza Data (GISAID);these sequences were the basis for the reseaech presented here.
All submitters of data may be contacted directly via the GIASID website www.gisaid.org
All four human influenza A(H7N9) viruses are similar at the nucleotide and amino acid levels, suggesting a common ancestor. The HA gene of the novel viruses belongs to the Eurasian lineage of avian influenza viruses and shares ca. 95% identity with the HA genes of low pathogenic avian influenza A(H7N3) viruses isolated in 2011 in Zhejiang province (south of Shanghai) (Figure 1, Table 2). The NA gene of the novel viruses is ca. 96% identical to the low pathogenic avian influenza A(H11N9) viruses isolated in 2010 in the Czech Republic (Figure 1, Table 2).
Figure 1.
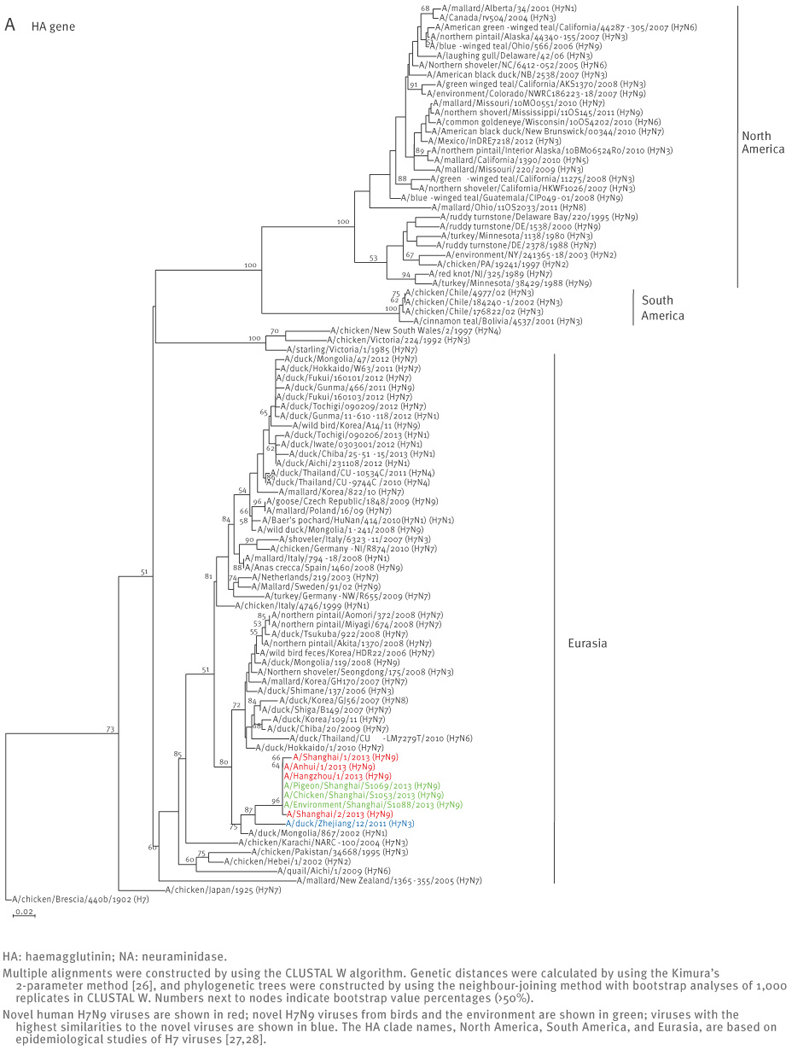
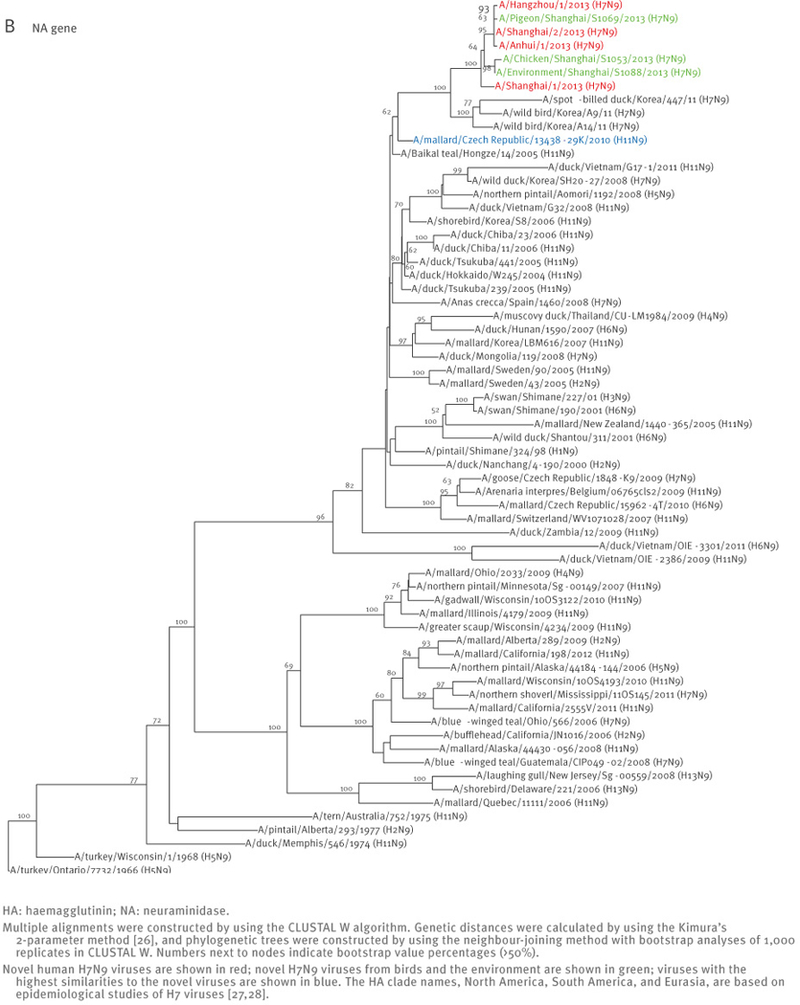
Phylogenetic analysis of the haemagglutinin (A) and neuraminidase (B) genes of the novel influenza A(H7N9) viruses, China, February - April 2013 (n=7)
Table 2.
Nucleotide identity of novel influenza A(H7N9) virus genes and their closest relative, China, February - April 2013
| Viral gene | Closest influenza virus relative | Nucleotide identity (%) |
|---|---|---|
| PB2 | A/brambling/Beijing/16/2012(H9N2) | 99 |
| PB1 | A/chicken/Jiangsu/Q3/2010(H9N2) | 98 |
| PA | A/brambling/Beijing/16/2012(H9N2) | 99 |
| HA | A/duck/Zhejiang/12/2011(H7N3) | 95 |
| NP | A/chicken/Zhejiang/611/2011(H9N2) | 98 |
| NA | A/mallard/Czech Republic/13438-29K/2010(H11N9) | 96 |
| M | A/chicken/Zhejiang/607/2011(H9N2) | 98 |
| NS | A/chicken/Dawang/1/2011(H9N2) | 99 |
HA:haemagglutinin; M:matrix gene;NA:neuraminidase;NP:nucleoprotein;NS:non-structural;PA:RAN polymerase acidic subunit;PB1:RNA polymerase basic subunit 1;PB2:RAN polymerase basic subunit 2.
The sequences of the remaining viral genes are closely related (>97% identity) to avian influenza A(H9N2) viruses, which recently circulated in poultry in Shanghai, Zhejiang, Jiangsu, and neighbouring provinces of Shanghai (Table 2, Figure 2). These findings strongly suggest that the novel influenza A(H7N9) viruses are reassortants that acquired their H7 HA and N9 NA genes from avian influenza viruses, and their remaining genes from recent influenza A(H9N2) poultry viruses (Figure 1, Figure 3, Table 2).
Figure 2.
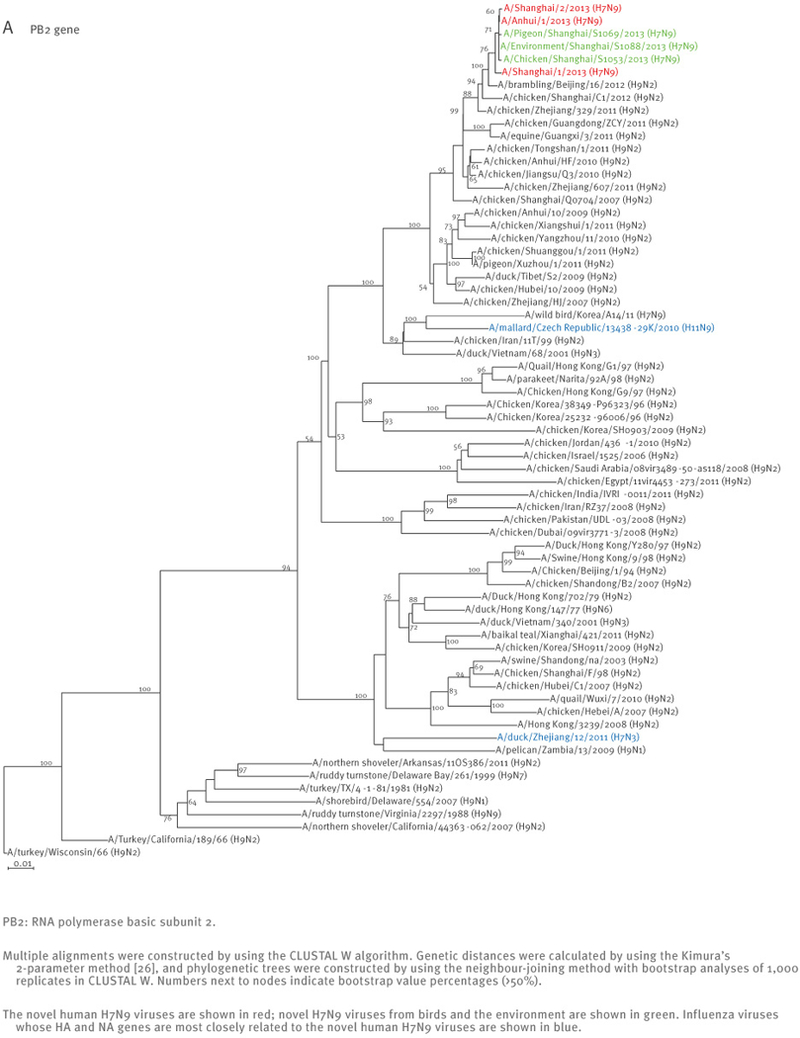
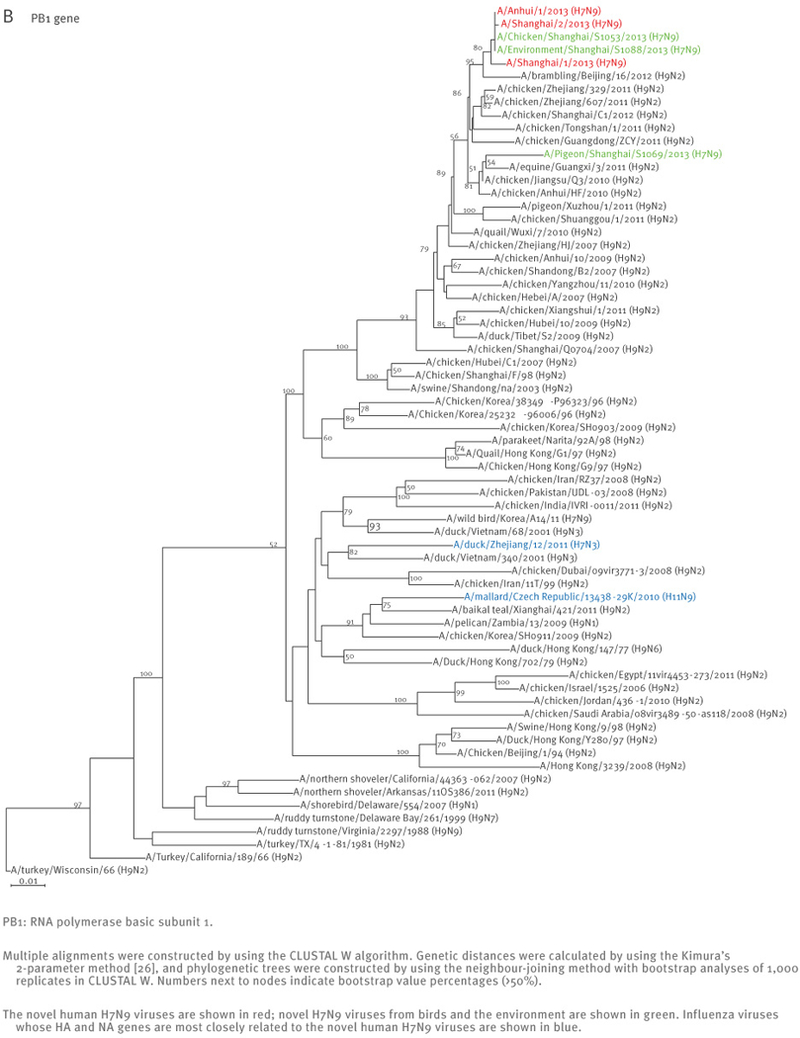
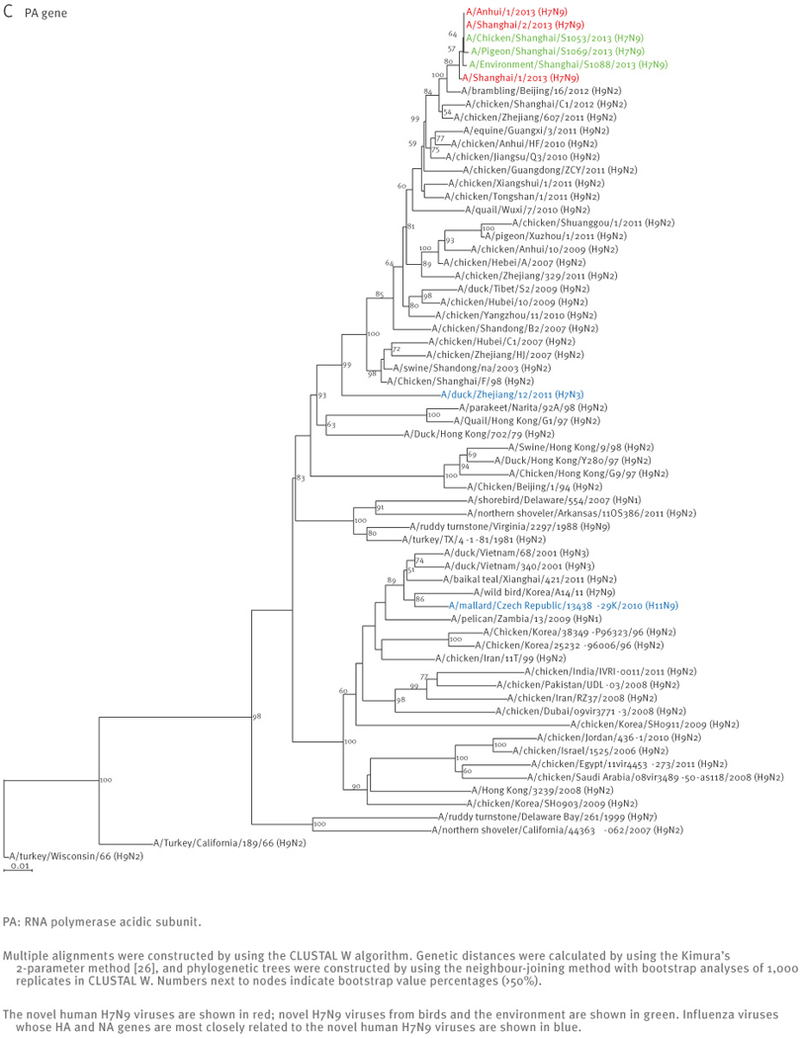
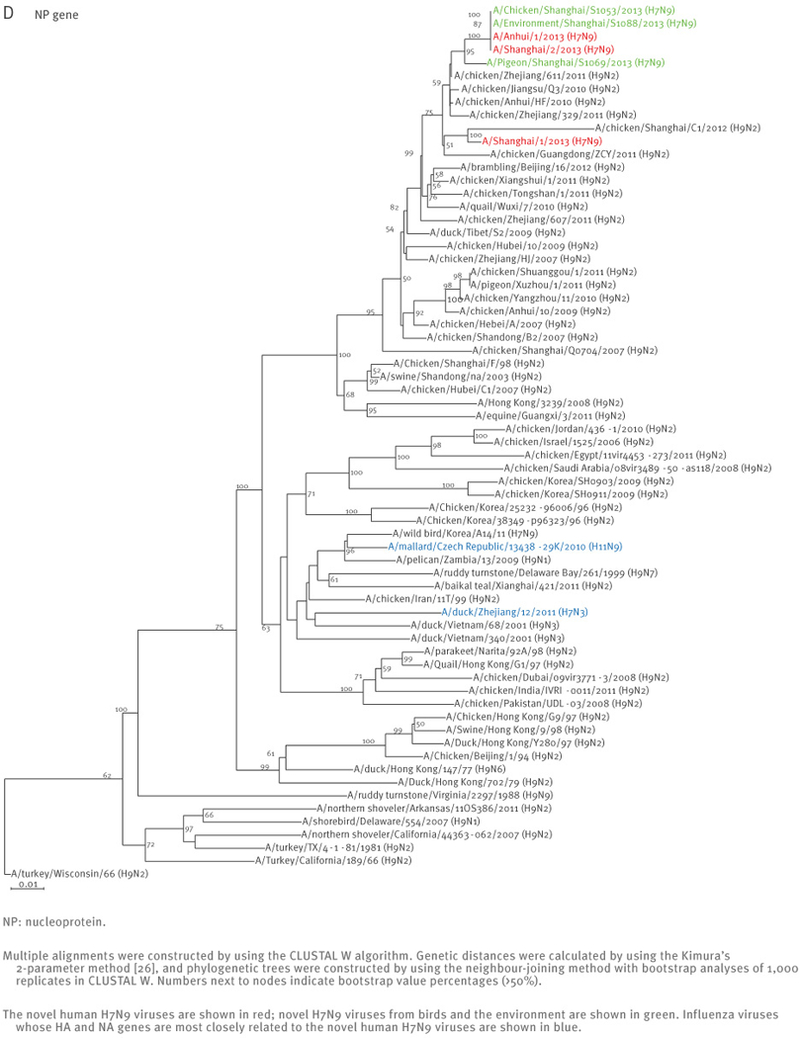
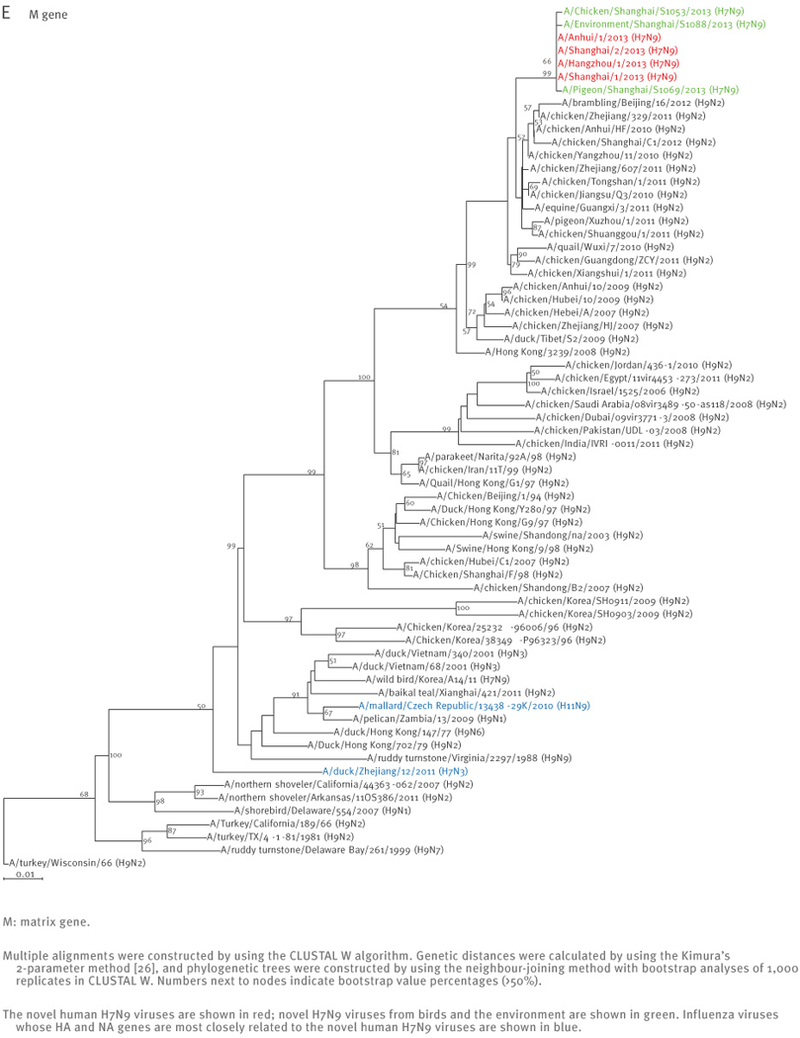
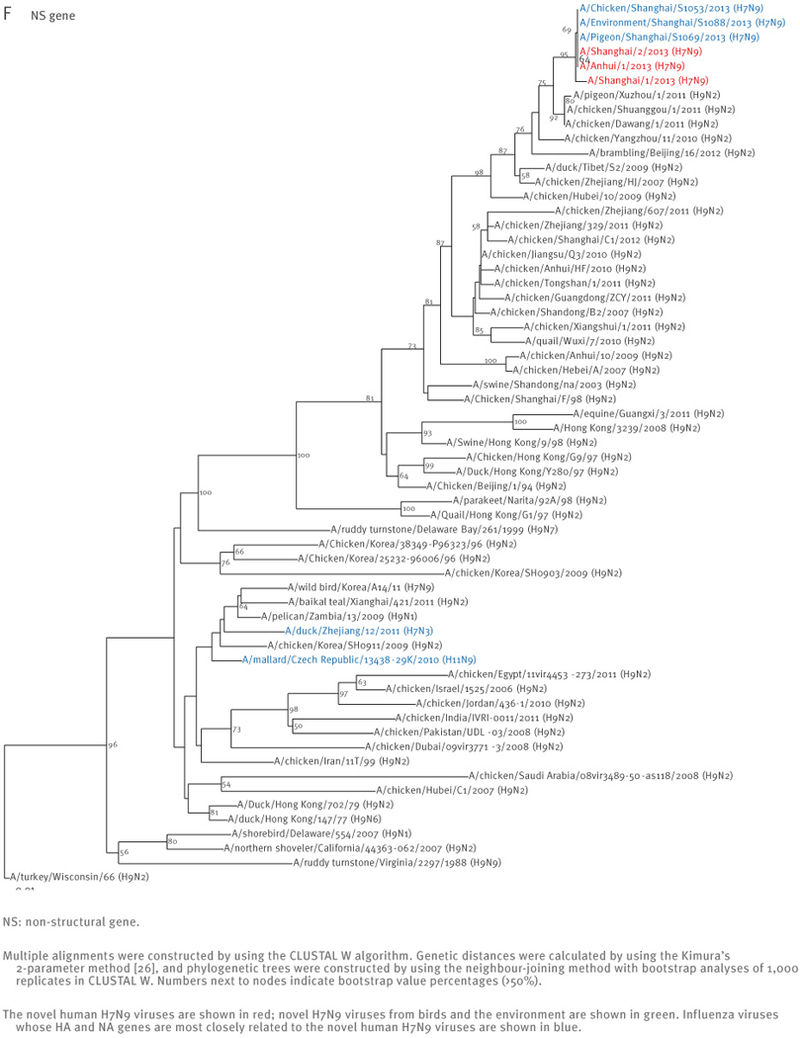
Phylogenetic analysis of the six remaining genes of the novel influenza A(H7N9) viruses, China, February – April, 2013 (n=7)
Figure 3.
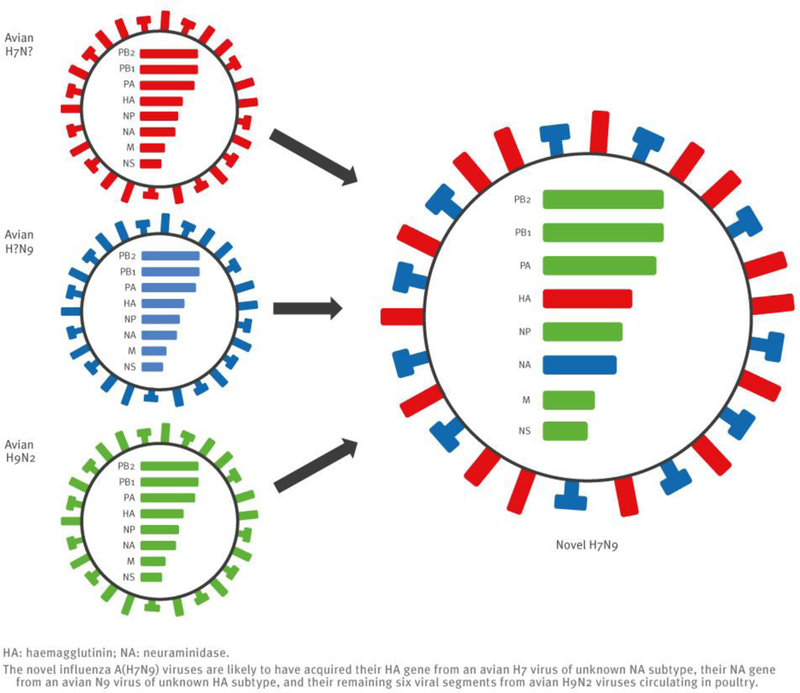
Schematic diagram of novel influenza A(H7N9) virus generation
At the nucleotide level, A/Shanghai/2/2013, A/Anhui/1/2013, and A/Hangzhou/1/2013 share more than 99% identity and differ by no more than three nucleotides per gene, even though they were isolated in different cities several hundred kilometres apart. On 7 April 2013, the Harbin Veterinary Research Institute deposited the full genome sequences of isolates from a pigeon (A/pigeon/Shanghai/S1069/2013), a chicken (A/chicken/Shanghai/S1053/2013), and an environmental sample (A/environment/Shanghai/S1088/2013) that were collected on 2 and 3 April from a Shanghai market (Table 1). All eight genes of these three isolates are similar to those of A/Shanghai/2/2013 and A/Anhui/1/2013 at the nucleotide level, except for the PB1 gene of A/pigeon/Shanghai/S1069/2013, which belongs to a different lineage than the PB1 of the other H7N9 isolates (Figures 1 and 2).
Interestingly, A/Shanghai/1/2013 and A/Shanghai/2/2013 differ by 52 nucleotides (for example, there are 13 nucleotide and nine amino acid differences in their HA sequences) even though these two cases were identified in the same city and at around the same time. These findings suggest that A/Shanghai/2/2013, A/Anhui/1/2013, A/Hangzhou/1/2013, as well as the viruses from the chicken and the environment, share a closely related source of infection, whereas A/Shanghai/1/2013 and A/pigeon/Shanghai/S1069/2013 are likely to have originated from other sources.
Highly pathogenic avian influenza viruses are characterised by a series of basic amino acids at the HA cleavage site that enable systemic virus spread. The HA cleavage sequence of the novel influenza A(H7N9) viruses possesses a single basic amino acid (EIPKGR*GL; *indicates the cleavage site), suggesting that these viruses are of low pathogenicity in avian species.
The amino acid sequence of the receptor-binding site (RBS) of HA determines preference for human- or avian-type receptors. At this site, A/Shanghai1/2013 encodes an S138A mutation (H3 numbering; Figure 4, Table 3), whereas A/Shanghai/2/2013, A/Anhui/1/2013, the two avian isolates, and the virus from the environmental sample encode G186V and Q226L mutations; any of these three mutations could increase the binding of avian H5 and H7 viruses to human-type receptors [12–14]. The finding of mammalian-adapting mutations in the RBS of these novel viruses is cause for concern. The A/Hangzhou/1/2013 isolate encodes isoleucine at position 226, which is found in seasonal influenza A(H3N2) viruses.
Figure 4.
Amino acid changes in the novel influenza A(H7N9) viruses that may affect their receptor-binding properties, China, February - April 2013 (n=7)
Table 3.
Selected characteristic amino acids of the novel influenza A(H7N9) viruses, China, February - April 2013 (n=7)
| Viral protein |
Amino acid position |
Shanghai/ 1/2013 |
Shanghai/ 2/2013 |
Anhui/ 1/2013 |
Hangzhou /1/2013 |
Chicken/ Shanghai/ S1053/2013 |
Environment/ Shanghai/ S1088/2013 |
Pigeon/ Shanghai/ S1069/2013 |
Human influenza viruses |
Avian influenza viruses |
Comments | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | 627 | K | K | K | Nd | E | E | E | K | E | E627K: Mammalian host adaptation |
16 |
| HA | 128/138a | S | A | A | A | A | A | A | A | Ab | S138A: Increased virus binding to human-type receptors |
13 |
| 151/160a | A | A | A | A | A | A | A | K | Ab | T160A: Loss of N-glycosylation and increased virus binding to human-type receptors |
15 | |
| 177/186a | G | V | V | V | V | V | V | G | Gb | G186V: Increased virus binding to human-type receptors |
14 | |
| 217/226a | Q | L | L | I | L | L | L | I | Qb | Q226L: Increased virus binding to human-type receptors |
12 | |
| NA | 69–73c | Deletion | Deletion | Deletion | Deletion | Deletion | Deletion | Deletion | No deletion | No deletion | Deletion of amino acids 69–73: Increased virulence in mice |
21 |
| 289/294/292d | K | R | R | R | R | R | R | R | R | R294K: Reduced susceptibility to oseltamivir and zanamivir |
20 | |
| Ml | 30 | D | D | D | D | D | D | D | D/(S) | D | N30D: Increased virulence in mice (most influenza A viruses encode 30D) |
24 |
| 215 | A | A | A | A | A | A | A | A | A | T215A: Increased virulence in mice (most avian influenza A viruses encode 215A) |
24 | |
| M2 | 31 | N | N | N | N | N | N | N | S/N | S/(N) | S31N: Reduced susceptibility to amantadine and rimantadine |
18,19 |
| NS1 | 42 | S | S | S | Nd | S | S | S | S | S/A | P42S: Increased virulence in mice (most avian influenza A viruses encode 42S) |
25 |
| 218–230 | Deletion | Deletion | Deletion | Nd | Deletion | Deletion | Deletion | No deletione | No deletion./ Deletion |
Lack of PDZ domain binding motif: Decreased virulence in mice |
23 |
Substitutions of particular concern are shown in bold.
Nd:no determined.
H7/H3 numbering.
H7 virus.
N9 numbering.
H7N9/avian N9/N2 numbering.
Influenza A(H1N1)pdmo9 virises from the 2009 influenza pandemic have the deletion.
In addition, all seven influenza A(H7N9) viruses possess a T160A substitution (H3 numbering; Table 3) in HA, which is found in recently circulating H7 viruses; this mutation leads to the loss of an N-glycosylation site at position 158 (H3 numbering; position 149 in H7 numbering), which results in increased virus binding to human-type receptors [15].
Lysine at position 627 of the polymerase PB2 protein is essential for the efficient replication of avian influenza viruses in mammals [16] and has been detected in highly pathogenic avian influenza A(H5N1) viruses and in the influenza A(H7N7) virus isolated from the fatal case in the Netherlands in 2003 [17]. PB2–627K is rare among avian H9N2 PB2 proteins (i.e. it has been found in only five of 827 isolates). In keeping with this finding, the avian and environmental influenza A(H7N9) viruses analysed here encode PB2–627E. By contrast, all four human H7N9 viruses analysed here encode PB2–627K (Table 3).
Antiviral compounds are the first line of defense against novel influenza viruses until vaccines become available. All seven novel influenza A(H7N9) viruses sequenced to date encode the S31N substitution in the viral ion channel M2 (encoded by the M segment) (Table 3), which confers resistance to ion channel inhibitors [18,19]. Based on the sequences of their NA proteins, all H7N9 viruses analysed here, with the exception of A/Shanghai/1/2013, should be sensitive to neuraminidase inhibitors (Table 3). However, the R294K mutation in the NA protein of A/Shanghai/1/2013 is known to confer resistance to NA inhibitors in N2 and N9 subtype viruses [20], and is therefore of great concern.
All H7N9 viruses encode a deletion at positions 69–73 of the NA stalk region (Table 3), which is reported to occur upon virus adaptation to terrestrial birds. This finding suggests that the novel H7N9 viruses (or their ancestor) may have circulated in terrestrial birds before infecting humans. Moreover, this deletion is associated with increased virulence in mammals [21].
The influenza A virus PB1-F2 protein (encoded by the PB1 segment) is also associated with virulence. The available sequences indicate that the H7N9 PB1 genes of all of the human viruses encode a full-length PB1-F2 of 90 amino acids, but lack the N66S mutation that is associated with the increased pathogenicity of the 1918 pandemic virus and the highly pathogenic avian influenza A(H5N1) viruses [22]. Interestingly, the pigeon isolate encodes a truncated PB1-F2 of only 25 amino acids; the significance of this truncation is unknown.
The NS1 protein (encoded by the NS segment) is an interferon antagonist with several functions in the viral life cycle. All available H7N9 NS1 sequences lack the C-terminal PDZ domain-binding motif; the lack of the PDZ domain-binding motif may attenuate these viruses in mammals [23].
Other amino acids in the NS1 and matrix (M1; encoded by the M segment) proteins of the novel viruses are also associated with increased virulence (Table 3) [24.25]. However, these amino acids are found in many avian influenza viruses, and therefore, their significance for the biological properties of the novel influenza A(H7N9) viruses is currently unclear.
In conclusion, we here present a biological evaluation of the sequences of the avian influenza A(H7N9) viruses that caused fatal human infections in China. These viruses possess several characteristic features of mammalian influenza viruses, which are likely to contribute to their ability to infect humans and raise concerns regarding their pandemic potential.
Acknowledgements
We are grateful to Dr. Shu Yuelong, Chinese National Influenza Center, Chinese Center for Disease Control and Prevention, Beijing, China, for his rapid publication of the entire gene sequence data of A(H7N9) viruses isolated from human cases in China, and also for his information sharing and advice to this study. We also thank Susan Watson for scientific editing. This work was supported by Grants-in-Aid for Pandemic Influenza Research (TK, SF, HX, and MT) and Grant-in-Aid for Specially Promoted Research (MT) from the Ministry of Health, Labour and Welfare, Japan, by the NIAID-funded Center for Research on Influenza Pathogenesis (CRIP, HHSN266200700010C)(YK), by a Grant-in-Aid for Specially Promoted Research, by the Japan Initiative for Global Research Network on Infectious Diseases from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (YK), and by ERATO, Japan (YK).
Footnotes
Conflict of interest
None declared.
References
- 1.World Health Organization (WHO)/Global Influenza Programme. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2013 Geneva: WHO: 12 March 2013. Available from: http://www.who.int/influenza/human_animal_interface/EN_GIP_20130312CumulativeNumberH5N1cases.pdf [Google Scholar]
- 2.Campbell CH, Webster RG, Breese SS Jr. Fowl plague virus from man. J Infect Dis 1970;122(6):513–6. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC). Update: influenza activity-United States and worldwide, 2003–04 season, and composition of the 2004–05 influenza vaccine. MMWR Morb Mortal Wkly Rep 2004;53(25):547–52. [PubMed] [Google Scholar]
- 4.Editorial team. Avian influenza A/(H7N2) outbreak in the United Kingdom. Euro Surveill 2007;12(22): :pii=3206. Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3206 [PubMed] [Google Scholar]
- 5.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A 2004;101(5):1356–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirst M, Astell CR, Griffith M, Coughlin SM, Moksa M, Zeng T, et al. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg Infect Dis 2004;10(12):2192–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 2004;363(9409):587–93. [DOI] [PubMed] [Google Scholar]
- 8.Kurtz J, Manvell RJ, Banks J. Avian influenza virus isolated from a woman with conjunctivitis. Lancet 1996;348(9031):901–2. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen-Van-Tam JS, Nair P, Acheson P, Baker A, Barker M, Bracebridge S, et al. Outbreak of low pathogenicity H7N3 avian influenza in UK, including associated case of human conjunctivitis. Euro Surveill 2006;11(18): pii=2952. Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=2952 [DOI] [PubMed] [Google Scholar]
- 10.Taylor HR, Turner AJ. A case report of fowl plague keratoconjunctivitis. Br J Ophthalmol 1977;61(2):86–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, et al. Human Illness from Avian Influenza H7N3, British Columbia. Emerg Infect Dis 2004;10(12):2196–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan K, Raman R, Jayaraman A, Viswanathan K, Sasisekharan R. Quantitative description of glycan-receptor binding of influenza A virus H7 hemagglutinin. PLoS One 2013;8(2):e49597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nidom CA, Takano R, Yamada S, Sakai-Tagawa Y, Daulay S, Aswadi D, et al. Influenza A(H5N1) viruses from pigs, Indonesia. Emerg Infect Dis 2010;16(10):1515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog 2010;6(9):e1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Lu B, Zhou H, Suguitan AL Jr, Cheng X, Subbarao K, et al. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J Virol 2010;84(13):6570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 2001;293(5536):1840–2. [DOI] [PubMed] [Google Scholar]
- 17.Munster VJ, de Wit E, van Riel D, Beyer WE, Rimmelzwaan GF, Osterhaus AD, et al. The molecular basis of the pathogenicity of the Dutch highly pathogenic human influenza A H7N7 viruses. J Infect Dis 2007;196(2):258–65. [DOI] [PubMed] [Google Scholar]
- 18.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J 1985;4(11):3021–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell 1992;69(3):517–28. [DOI] [PubMed] [Google Scholar]
- 20.McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, McDonald M, Colman PM, Hart GJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol 1998;72(3):2456–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka Y, Swayne DE, Thomas C, Rameix-Welti MA, Naffakh N, Warnes C, et al. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J Virol 2009;83(9):4704–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 2007;3(10):1414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A 2008;105(11):4381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan S, Deng G, Song J, Tian G, Suo Y, Jiang Y, et al. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology 2009;384(1):28–32. [DOI] [PubMed] [Google Scholar]
- 25.Jiao P, Tian G, Li Y, Deng G, Jiang Y, Liu C, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol 2008;82(3):1146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura M A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16(2):111–20. [DOI] [PubMed] [Google Scholar]
- 27.González-Reiche AS, Morales-Betoulle ME, Alvarez D, Betoulle JL, Müller ML, Sosa SM, et al. Influenza A viruses from wild birds in Guatemala belong to the North American lineage. PLoS One 2012;7(3):e32873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HR, Park CK, Lee YJ, Oem JK, Kang HM, Choi JG, et al. Low pathogenic H7 subtype avian influenza viruses isolated from domestic ducks in South Korea and the close association with isolates of wild birds. J Gen Virol 2012;93(Pt 6):1278–87. [DOI] [PubMed] [Google Scholar]


