Abstract
Since the discovery of HIV as the cause of AIDS, numerous insights have been gained from studies of its natural history and epidemiology. It has become clear that there are substantial interindividual differences in the risk of HIV acquisition and course of disease. Meanwhile, the field of human genetics has undergone a series of rapid transitions that have fundamentally altered the approach to studying HIV host genetics. We aim to describe the field as it has transitioned from the era of candidate-gene studies and the era of genome-wide association studies (GWAS) to its current state in the infancy of comprehensive sequencing. In some ways the field has come full circle, having evolved from being driven almost exclusively by our knowledge of immunology, to a bias-free GWAS approach, to a point where our ability to catalogue human variation far outstrips our ability to biologically interpret it.
Keywords: AIDS, progression, setpoint, GWAS, genome sequencing
INTRODUCTION
Studies of the epidemiology and natural history of HIV/AIDS have found that the risk of infection and clinical course of disease are highly variable across the human population. Over the past three decades, a tremendous amount of research has been directed toward understanding how human genetic variation contributes to the observed differences in acquisition risk and viral control (Figure 1). During this time, the field of human genetics has evolved rapidly as technological breakthroughs have given researchers more powerful tools capable of examining the human genome in increasingly fine detail.
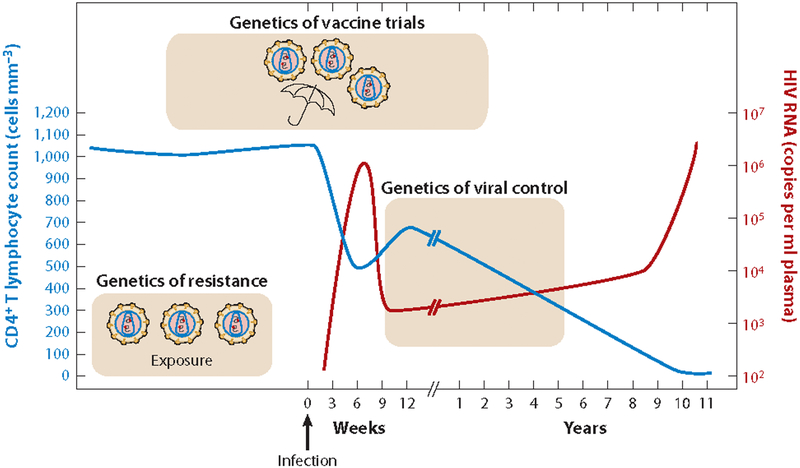
Figure 1.
Graph of the clinical course of a typical HIV infection, highlighting the major areas of HIV host genetics under investigation. Longitudinal measures of HIV viral load in plasma and CD4+ T cell count are shown in red and blue, respectively. Modified from Reference 77 with permission.
HIV HOST GENETICS IN THE CANDIDATE-GENE ERA
Candidate Genes for HIV Acquisition
Prior to the sequencing of the human genome and the development of platforms for genome-wide association studies (GWAS), advances in human genetics were primarily driven by studies of genetic linkage in families or through our understanding of the biology of human disease. In the field of HIV host genetics, discoveries during this candidate-gene era came from insights into the molecular biology and immunology of HIV–host interactions. In particular, identification of host receptor molecules used by HIV and their ligands was one of the most influential forces in the field of HIV host genetics.
CCR5.
Following the discovery of HIV in 1983 (1, 2), it was found that a small fraction of the population appeared to have some form of natural resistance to HIV infection. Study of these resistant individuals revealed that their CD8+ T cells produced increased levels of three β-chemokines: MIP1α, MIP1β, and RANTES (3). In addition, these three chemokines were found to block HIV replication in vitro (4). Subsequently, a CC-chemokine receptor called CCR5 was cloned and found to be potently activated by precisely these three cytokines (5), making it a strong area of interest for candidate-gene studies. CCR5 was found to function as a coreceptor for binding and entry of M-tropic HIV strains into CD4+ T cells (6, 7). Sequencing of CCR5 in individuals thought to be exposed to HIV but not infected (now known as HESN) identified a 32-base-pair deletion common in Caucasians (8–10) but absent in most non-European populations (9, 11, 12). The deletion results in a nonfunctioning receptor protein that fails to localize to the cell surface (13). Individuals with two copies of the CCR5delta32 deletion are highly resistant to M-tropic viruses that use CCR5 as a coreceptor (8–10). However, carriers with a single copy appear to be as susceptible to infection as wildtype homozygotes (8). Genotyping of CCR5delta32 in HESN populations such as hemophiliacs, prostitutes, and intravenous drug users has found that homozygosity for CCR5delta32 is increased in frequency compared to controls (8, 12). Thus, enrichment of CCR5delta32 serves as an important indicator of HIV exposure levels in HESN populations. A number of very rare or population-specific variants have also been found in CCR5, only a fraction of which are known to impact function and modify the risk of HIV acquisition (14–17).
CCR5 Ligands.
The discovery of CCR5 variation as a bona fide resistance factor against HIV infection and the ability of certain β-chemokines to inhibit HIV prompted the investigation of the entire family of CCR5 ligands (RANTES, MIP1α, and MIP1β) as potential candidate genes for HIV resistance. Sequencing of the genes encoding these ligands has identified several polymorphisms that have been associated with HIV acquisition. However, these risk alleles tend to be population specific and are often of low frequency in other populations. Variation in the promoter of the CCL5 gene, encoding RANTES, alters its expression (18) and has been associated with an increased risk of HIV-1 acquisition and disease progression among European-Americans (19,20), but no difference in acquisition risk was seen in Japanese HESNs compared to controls (18). The CCR5 ligands MIP1α and MIP1β are encoded by the CCL3 and CCL4 genes on chromosome 17. A similar population-specific association was reported with CCL3/MIP1α variation and a reduced risk of HIV infection (19, 21). This effect was only seen in populations of African ancestry and was not observed in European-American or Japanese cohorts (19, 21–23). Taken together, these reports suggest that variation in the genes encoding CCR5 ligands may influence acquisition risk in certain populations. However, their contribution to differences in acquisition risk may be minor in other populations due to the low frequency or population-specific nature of risk variants.
The region containing CCL3 and CCL4 has been duplicated, and a variable number of copies of these genes are present in the human genome. The duplicated form of CCL3, known as CCL3L1, encodes a protein (MIP1αP) that has even higher affinity for CCR5 than MIP1α has (24). Increased CCL3L1 copy number was found to correlate with higher MIP1αP levels (25) and was associated with a reduced risk of HIV infection in several populations (26). However, this association was not replicated in other independent cohorts (27, 28). Therefore, the role of the CCL3L1 copy-number variant in HIV acquisition remains unclear.
Candidate Genes for HIV Progression
In addition to its role in HIV acquisition, human genetic variation is known to influence the rate at which HIV+ patients progress to AIDS. Genes influencing HIV progression generally fall into three broad categories: those that influence the efficiency of HIV infection, such as the genes for CCR5 and its ligands; those that encode host proteins hijacked by HIV for its own life cycle; and antiviral genes involved in innate and acquired defenses against infection, such as the major histocompatibility complex (MHC) genes.
CCR5 and its ligands.
Variants in CCR5 and its ligands are believed to slow progression by reducing the level of entry receptors present on the cell surface or by increasing chemokine levels so that receptors are either occupied or internalized, thereby limiting the spread of HIV within the body. Although CCR5delta32 does not provide resistance to HIV for carriers with a single copy, these patients do experience a two-year delay, on average, before the onset of AIDS (8). CCR5 promoter variation has also been associated with differences in HIV progression (29, 30). Presumably this occurs through differences in CCR5 expression levels caused by altered transcription factor binding in the CCR5 promoter.
Variants in the RANTES/CCL5 promoter have also been found to be associated with differences in HIV progression. Sequencing of the CCL5 promoter in an HIV+ Japanese cohort found that patients with the –28G allele showed a significantly slower loss of CD4+ T cells compared to wild type (18). This allele was further shown to result in increased expression of RANTES (18), consistent with a model in which increased levels of RANTES occupy more CCR5 binding sites and block HIV entry.
The CCL3L1 copy-number variant has also been associated with HIV progression and set-point viral load level (26). However, this association has not been consistently replicated in independent cohorts (27, 28). Considering that heterozygosity for CCR5delta32 does not appear to influence the risk of HIV acquisition, there is no reason to expect that quantitative variation in CCR5 ligands such as CCL3L1/MIP1αP would influence the risk of HIV acquisition either. In contrast, though, CCR5delta32 heterozygosity has convincingly been associated with lower setpoint viral load and slower AIDS progression, so we might expect that variation in the expression of CCR5 ligands such as MIP1αP may similarly influence progression and viral load. Therefore, it seems likely that sufficiently accurate measurement of CCL3L1 copy-number variation may eventually show some degree of association, even though early reports appear strongly influenced by measuring artifact (27, 28, 31).
Cyclophilin A.
The host cyclophilin A protein is usurped by HIV and is an important part of the viral replication cycle. This role was recognized when it was discovered that cyclophilin A is packaged into the mature viral particle, where it participates in HIV uncoating during infection (32, 33). Sequencing of the PPIA gene revealed five polymorphisms upstream of the coding region (34). A haplotype containing two promoter single-nucleotide polymorphisms (SNPs) was found to be associated with increased PPIA expression and progression to AIDS (34).
APOBEC3s.
The apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 proteins (APOBEC3s) perform diverse mRNA-editing activities. Several members of this family are antiviral proteins that induce hypermutation in extrachromosomal DNA, such as the genomes of viral pathogens. During HIV replication, APOBEC3G is integrated into assembling viral capsids and induces C→U transitions in the HIV genome (35). Sequencing of the APOBEC3G gene has identified multiple variants. One of these, the H186R mutation in exon 4, was associated with more rapid HIV progression in African-Americans (36).
The major histocompatibility complex (MHC) and HIV host genetics.
The human MHC, located on 6p21.3, comprises the richest four-mega-base-pair region of the genome in terms of its associations with human disease as determined by GWAS (http://www.genome.gov/26525384). Human leukocyte antigen (HLA) class I and II genes are housed within the MHC, and they display an extreme degree of polymorphism that is subject to and a result of balancing selection (37), especially in the segments encoding the peptide binding domains (38). HLA is command central of the acquired immune response, dictating the antigens to which we produce antibodies (HLA class II-driven) and cytotoxic T cell responses (primarily HLA class I-driven). The HLA class I molecules have also permeated innate immune activity by serving as ligands for the natural killer (NK) cell receptors known as the killer cell immunoglobulin-like receptors (KIRs) (39). Polymorphism at the HLA class I and KIR loci gives rise to elasticity in the quality of the immune response to a given pathogen, and it is this genetic variation within humans that has shown the greatest influence genome-wide on clinical outcome after HIV infection (40–46).
Human Leukocyte Antigen.
Many HLA alleles have been associated with various outcomes to HIV infection, and several have shown very consistent effects across studies, including subtypes of B∗57, B∗27, B∗58, and B∗35. HLA class I was known to be associated with HIV outcomes prior to GWAS, but its importance relative to the rest of the genome was not clear until the first GWAS was conducted (41). The protection conferred by B∗57 was strikingly obvious in this study, partly because another SNP on the chip marked the B∗57 allele nearly perfectly, whereas other associated alleles are not well marked by single SNPs on the chip and imputation is needed to determine their contribution to HIV control (44). Although B∗57 has the greatest individual allelic association with HIV control, the basis for this protection is still not clear and probably involves an array of mechanisms (47–56), a scenario that is also likely for all other HLA alleles associating significantly with HIV outcomes. Although B∗57 is highly enriched among the most extreme controllers, the vast majority of B∗57+ patients progress to disease on average as rapidly as those without this allele, an observation that may not be widely recognized. Thus, the influence of HLA on HIV pathogenesis is complex and involves additional genetic, viral, and/or environmental factors that modify the efficacy of specific HLA-restricted responses against the virus.
HLA-C has not been considered a primary contributor to HIV control. Indeed, the functional importance of HLA-C was largely ignored historically because of its limited polymorphism (57) and low cell-surface expression (58, 59), but the discovery of its ligand relationship with KIR revived its status as a mainstay of the innate immune response. One of the most surprising and exciting findings first illuminated through GWAS of HIV cohorts was the strong association between a variant 35 kb upstream of the HLA-C locus, termed −35, and viral control (41). The −35 SNP associates with HLA-C expression in people of European descent (60, 61), due in part to variation in a microRNA binding site of the HLA-C 3′ untranslated region, which also associates strongly with HIV control (62). Varied expression levels of HLA-C may provide the functional explanation for its association with HIV control, although this notion is under debate (63). It appears that higher expression of HLA-C does confer selection pressure on the virus (64, 65), and there is solid justification to probe more fully the impact of HLA-C expression levels on resistance to HIV (66).
Killer cell immunoglobulin-like receptors (KIRs).
The KIR locus mapping to chromosome 19q13.4 encodes receptors that are expressed on NK cells and a subset of CD8+ T cells (39). The KIR genes display allelic and gene content polymorphism and have high sequence similarity, complicating the ability of GWAS to interrogate this region accurately. A growing number of studies have reported associations between HIV outcomes and KIR or functional KIR/HLA combinations (67). The impact of KIR on HIV evolution is beginning to be revealed; the presence of certain KIR genes associates with specific viral mutations on a population level (68). Importantly, naturally occurring mutations in HLA-restricted HIV epitopes have been shown to abrogate (69) or enhance (68) KIR–HLA interactions, thereby altering effector cell response to the virus. The KIR locus not only shows extensive allelic variation but also carries a variety of deletions and duplications. A recent study assessed the copy-number status of the 3DL1/3DS1 locus(70). The analyses showed that individuals with more gene copies had better viral control, but only when the ligand encoded by HLA-B is present (70). Specific counts of 3DL1 and 3DS1 content further revealed that increasing counts of 3DS1 are always beneficial in viral control, but increasing counts of 3DL1 are beneficial only in the presence of at least one copy of 3DS1. These analyses demonstrate a key role for interactions between KIR and HLA in viral control that remain only partially understood at a mechanistic level (70).
GENOME-WIDE ASSOCIATION STUDIES OF HIV ACQUISITION AND CONTROL
The first GWAS of any infectious disease was applied to HIV viral control (41). This marked a conceptual transition in the study of HIV host genetics. Whereas all previous genetic work involved testing candidate genes that had been implicated on the basis of functional work on immunology, the GWAS approach permitted the first truly agnostic approach to gene discovery. Even in the case of CCR5, the identification of CCR5 as a coreceptor for macrophage tropic virus preceded the identification of the deletion polymorphism as responsible for protection. This order is, however, not well appreciated because the key papers were published only a few months apart (7, 8, 10).
The GWAS era therefore brought considerable hope that by moving into the genome as a whole, and not being restricted by what we already know, fundamental new insights about the interaction between HIV and its host would emerge. In the main, this hope unfortunately has not been realized. As described above, the strongest association identified in GWAS points clearly to the already known HLA B∗5701. This is consistent from the first study (41) on viral load to the more recent larger studies on viral load (40) and on comparing progressors to controllers (71). Moreover, a study performed in African-Americans identified the very closely related HLA B∗5703 as the strongest determinant of viral load in that ethnic group (43). As described above, one new pointer did emerge in that HLA-C had not been considered as important in HIV control, but most GWAS show independent association connected to HLA-C (40, 41, 71).
Other GWAS have been performed that are still to be replicated, in particular for acquisition in a case-control setting (72) or discordant-pair design (73) that are reported as negative, and for in vitro permissiveness (74). These studies, however, have been clearly underpowered in comparison with studies of progression and viral control and other human traits. The concern about power is exacerbated by questions about whether there has been effective exposure in the acquisition studies. For example, discordant-pair studies enroll long-term couples in which only one partner is HIV+, and it is unclear which partner is influencing the probability of transmission. In the case-control design, subjects are individuals who report repeated exposure to different sexual partners, and it is often difficult to quantify the actual number of exposures to HIV+ individuals. The cumulative impact of these uncertainties makes the realized power smaller than it would appear from the sample size alone.
In addition to these reports of association, Pereyra et al. have attempted a fine-mapping exercise to identify the functional sites in the HLA-B gene from the fine-scale pattern of association (71). The results are interesting and suggestive, but the very-long-range linkage dis-equilibrium in the HLA region makes identification of causal sites from association data alone a particular challenge, and confirmation that these sites are truly causal as opposed to simply strongly associated will require functional biology.
In totality, it is perhaps surprising that genome-wide association has not brought about more progress. This has at least two possible explanations: (a) the variants that are most important to influence viral control are too rare to be well detected by GWAS, and (b) the remaining unexplained variation in viral control is primarily nongenetic. Neither of these possibilities is favored by solid data. Perhaps the only sure way to know the explanation is to complete sequencing-based studies of comparable size and rigor to what is ongoing in GWAS.
NEXT-GENERATION SEQUENCING IN HIV HOST GENETICS
GWAS are designed and have adequate statistical power to interrogate much of the common variation present throughout the genome. Many successes have been described, although few have led to the identification of the causative variant. However, GWAS are not designed to capture rare variation (<1% minor allele frequency) in the human genome. Currently the only way to catalogue the entire spectrum of common and rare variation is to use comprehensive genome or exome sequencing. Over the past few years, the rapid development of next-generation sequencing (NGS) platforms has made this possible to do in an efficient manner. However, the cost of NGS is still too high for most studies to include thousands of individuals. As a result, most sequencing studies have focused on smaller numbers. This reduces the potential power of a genetic study, so study design must be carefully considered.
Study Designs for Next-Generation Sequencing in HIV Host Genetics
With the exception of family studies, which have a limited role in HIV host genetics, three main approaches will likely be used in the design of NGS projects. The first is the case-control study design, where a small number of affected individuals are sequenced and compared to a large number of controls, such as those from the 1000 Genomes Project (http://www.1000genomes.org/). An example of this approach is a study to identify variants responsible for HIV acquisition/protection. Here many HIV+ individuals are sequenced and compared to population controls who are presumed to have low or no HIV exposure. This case-control design should identify variants that are enriched in the HIV+ (or HIV−) group. However, with the limited number of cases that can be included in genome or exome sequencing projects, these studies may be statistically underpowered to detect very rare variants.
One possible solution to this problem is the use of an extreme-trait study design. In this approach, individuals are carefully selected from opposite ends of well-defined phenotypic distribution. An example of this design would be studying individuals who rapidly (within one year) progress to AIDS versus long-term nonprogressors, who show no evidence of AIDS progression after ten years; another example would be comparing HIV+ patients to HESN cohorts. In this latter example, protective alleles would be enriched in the HESN samples and depleted from the HIV+ group. Variants identified in these studies can then be screened in hundreds or thousands of additional cases and controls to increase power.
A third approach is sequencing to evaluate a quantitative trait, such as HIV viral load. This approach is in its infancy for sequencing-based studies because hundreds of samples need to be sequenced to obtain enough power to identify rare variants. Numerous GWAS have been performed to evaluate the role of common genetic variation in the control of HIV viral load. However, their findings can explain only a small fraction of the variation in viral load among individuals (40, 41, 43). Thus, implementing a well-powered, sequencing-based approach to identify rare variants involved with HIV viral load should lead to the identification of additional associated/causative variants. In the next few years, the design of sequencing studies will change greatly as reduced cost and increased sequencing throughput will allow the rapid and cost-effective sequencing of thousands of individuals.
Challenges Associated with Next-Generation Sequencing
Sequencing a human genome or exome has become a relatively straightforward task. However, there are many analytical challenges. Although the use of an extreme-trait study design or larger sample sizes can improve the ability of a study to detect rare causative variation, exceedingly rare or private variants still pose a statistical problem to any study design using single-variant tests of association. One recent solution to this challenge is to collapse each of the variants in a gene down to a single binary variable, thus simply asking whether an individual has variation in that gene or not. In early approaches, all variants were collapsed, but it was soon realized that noise from common, nonpathogenic variants tended to dominate the signal from rare causative alleles. Therefore, more advanced methods were developed, such as the combined multivariate and collapsing (CMC) method, in which rare variants are collapsed and common variants are tested independently (75). New methods are currently being developed that include weighting of variants by frequency, direction of effect, and functional prediction.
A typical genome will have ~3.5 million single-nucleotide variant (SNV) calls and ~700,000 small insertions or deletions (indels). Some fraction of these will be artifacts. Because of the small read-length of NGS output and the repetitive nature of the human genome, it is impossible to reconstruct the genome using de novo assembly methods. Therefore, aligning sequencing reads to a reference genome is necessary. This step can cause misalignments, leading to incorrect variant calling. For the most part, the calling of SNVs is fairly accurate. However, the calling of indels can be greatly overestimated. New methods that involve local realignment of the reads in the region of an indel can increase the quality of alignments/calling, but it is still likely that short-read sequencing technology and the process of aligning reads to a reference genome will always lead to less accurate calling of indels than of SNVs. The solution to many of these issues lies with newer sequencing technologies that are much higher throughput and have much longer read-lengths (1–10 kb), allowing the use of de novo assembly methods.
Another challenge when analyzing sequencing data, as with GWAS, is batch effects. For NGS studies that examine more than a handful of cases and controls, library creation and sequencing require that samples be processed in batches. This can lead to subtle, batch-dependent differences in sequencing results. With NGS technologies evolving extremely rapidly, maintaining reagent and analysis software versions consistently for the duration of a large sequencing project is virtually impossible. Therefore, it is necessary to account for the many variables introduced during the sequencing and analysis phases.
For the field of HIV host genetics, the shift to NGS will allow a detailed examination of the human genome in connection with host traits. The use of efficient study designs and analytical methods will improve the power of these initial sequencing studies. However, the fact that these traits are complex and likely to be genetically heterogeneous will require the sequencing of large numbers of samples in order to provide convincing statistical evidence in light of the large numbers of putatively functional variants present in the genome.
INTEGRATION OF GENETIC ANALYSIS WITH FUNCTIONAL EVALUATION
NGS technologies provide investigators with a nearly comprehensive ability to catalogue all of the genetic variation present in an individual’s genome. However, one of the most fundamental challenges to these approaches is the problem of how to identify the causal variant or variants among a veritable sea of nonpathogenic background variation. With study designs utilizing trios, this problem becomes more tractable, as cosegregation of variants with the disease phenotype provides a useful filter for reducing the number of variants of interest. However, for studies using case-control or extreme-trait designs, this issue requires alternative ways of prioritizing variants. One of the most promising approaches is to integrate data from functional studies of HIV biology and pathogenesis.
RNAi Screens
RNA interference (RNAi) is a powerful experimental tool that allows silencing of mRNA expression from selected genes. RNAi techniques have been adapted into genome-wide, high-throughput screens that allow a forward genetic analysis of genes that contribute to a particular phenotype of interest. These screens generate a tremendous wealth of functional information that can then be integrated into sequencing studies to prioritize genetic variants for further investigation.
The first genome-wide use of RNAi to screen for host genes involved in HIV infection was reported by Brass et al. (76). In this forward genetic analysis, they examined the host factors needed for primary infection as well as production of infectious virus. Small interfering RNAs (siRNAs) were used to knock down expression of >21,000 host genes in a HeLa-derived cell line prior to infection. Brass et al. identified 272 host factors that could be validated. Their findings implicated several pathways not previously known to participate in the HIV life cycle and identified several new host factors on which HIV was dependent, including vesicular transport proteins Rab6 and Vps53 in viral entry and Transportin 3 in viral integration.
A second genome-wide siRNA study by Konig et al. focused specifically on early infection through the use of a replication-deficient HIV strain (77). The authors infected 293T cells using a virus expressing the VSV-G protein (vesicular stomatitis virus G protein), allowing broad cellular tropism. A protein interaction network was then generated by integrating data from human and HIV-human yeast two-hybrid experiments with the 295 host genes identified in the screen. The resulting network implicated several host pathways, including DNA damage repair, nucleotide binding, ubiquitinylation, and nuclear import. Interestingly, even when similar filtering criteria were applied, only 60 host genes were found to be common with the study by Brass et al. (77).
In a third report, Zhou et al. used a two-stage experimental design virtually identical to that of Brass et al. A HeLa-derived cell line was infected with HIV 24 hours after siRNA transfection. Supernatants were then used to infect fresh cells. The authors identified 232 host genes required for infection that were expressed in activated T cells and macrophages (78). Despite the similarity in experimental design, only 13 of the host factors identified overlapped with those reported by Brass et al.
Although these three studies provide important information about host factors in HIV infection, one potential concern is that the cell types used in these experiments (cervical and kidney epithelial) are not a physiologically relevant model of HIV infection in T cells and macrophages. However, these relevant cell types have low transfection efficiency. To avoid this issue, Yeung et al. used a lentivirus vector system expressing short hairpin RNAs (shRNA) to knock down target mRNAs in Jurkat T cells (79). This system stably expresses shRNAs, allowing examination of the course of infection over a longer time period than would normally be possible when using a transient transfection assay. Again, very little overlap was observed: three genes were common with Konig et al., three separate genes were common with Zhou et al., and no genes were common with Brass et al. (79).
Using an approach opposite to the four previous studies, Liu et al. used a genome-wide siRNA screen to identify host factors that naturally inhibit HIV replication (80). In this screen, host factors were systematically knocked down in a cell line that normally restricts HIV replication in order to identify factors whose loss resulted in increased HIV proliferation. They identified 192 initial candidate genes, 114 of which were able to be replicated. Functional analysis of these genes revealed a diverse set of pathways including vesicle trafficking, mRNA processing, and cross-nuclear membrane transport. These findings provide important insight into the host factors that restrict HIV replication. The limited amount of overlap between the genes identified in this study and those in the previous four studies (two genes in common with Konig et al., two separate genes in common with Yeung et al., and one in common with Brass et al.) is not unexpected given that the authors sought to identify naturally restrictive rather than dependency factors.
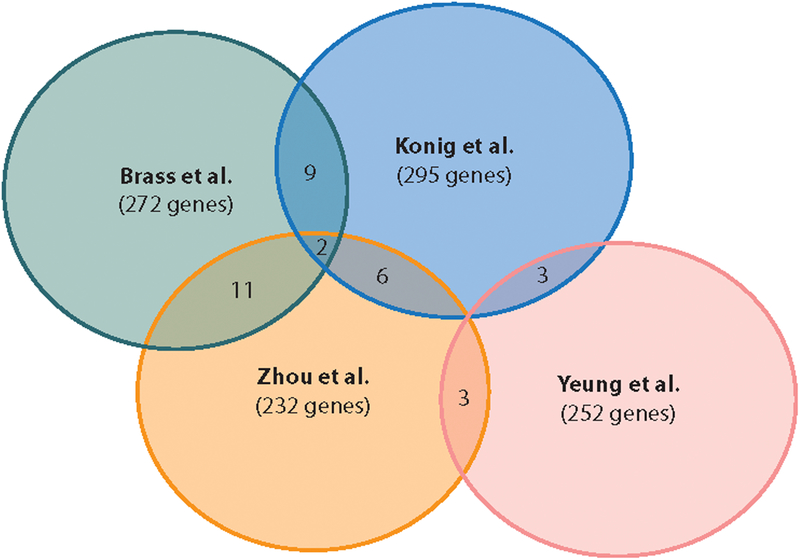
Forward genetic screens of host factors involved in HIV infection and replication are a very powerful resource to help identify causative genetic variants among the long list of statistically significant findings typically generated by NGS of large case-control studies. However, the striking lack of overlap between the findings of the first four studies (Figure 2) does merit concern about their reproducibility. It is important to note that these four studies examined three distinct phenotypes: Konig et al. focused on early-stage HIV infection, the Brass and Zhou groups examined both early- and late-stage infection, and Yeung et al. looked at long-term infection. To some degree the genes identified by Konig et al. should represent a subset of those found in studies of the entire HIV life cycle. However, their use of a VSV-pseudotyped virus could reduce the overlap between findings, as genes involved in CD4-mediated entry would be absent from their dataset. The use of different HIV strains and host cell lines could also lead to the discordance between the results of these studies. Differences in siRNA libraries and their application, the use of different readout assays (p24 staining, reporter expression, or shRNA barcodes), or other technical differences may also have a large impact on reproducibility. Despite the lack of concordance, a number of host factors, such as Transportin 3, Med6, and RelA, were replicated across multiple screens.
Figure 2.
Venn diagram showing the overlap in HIV host dependency factors identified by the four siRNA screens (76–79).
Protein–Protein Interaction Experiments
Substantial experimental data are also available on interactions between HIV and host proteins. An initial study catalogued the host proteins that physically interact with the HIV transcriptional activator, Tat (81). Subsequently, a more global HIV–host interactome was reported, in which all 18 HIV proteins and polyproteins were used to identify the entire landscape of HIV–host protein–protein interactions (82). In addition, the National Institute of Allergy and Infectious Diseases maintains a database of all direct and indirect HIV–host protein interactions reported in the literature (http://www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions/).
Integrating Functional Information into Genetic Studies
The most basic method of integrating functional data into genetic studies involves a simple cross-reference in which a list of genes found to have variants with significant p-values is compared to lists of genes identified by candidate-gene studies or RNAi/PPI screens. The selection of resources for inclusion also demands careful consideration. Some contain large numbers of reported host factors, so limited power of discrimination is gained by their inclusion.
Recently, more advanced and statistically rigorous methods of incorporating gene-set enrichment data have been developed. These methods can incorporate disparate functional information such as gene ontologies, RNAi screens, and PPI databases in the form of prior probabilities in a Bayesian framework.
Future Types of Screens and Databases
As sequencing costs continue to decrease, the ability to sequence large numbers of samples will increase demand for new tools to prioritize genetic variants. Current methods will mature to fill some of this demand, but more effective methods of identifying causal variants will be required as well. These may include medium-throughput assays capable of testing cDNAs in cell systems or higher-throughput methods of creating isogenic mutants to directly test the phenotypic impacts of individual variants. Improvement of computational techniques will also be required. Currently, computational methods exist for assessing the functional consequences of coding variants, but there are few options for prioritizing intergenic variants even though they compose the majority of variable sites in the genome. Integration of DNase-hypersensitivity information and improved computational methods for identifying gene regulatory elements will alleviate this immensely.
DISCUSSION
Although NGS is better suited to capturing the full spectrum of genetic variation, it is clear that genome-wide association is not quite dead yet. New chips with denser coverage and rare variants, as well as the ability to impute genotypes, will continue to extend its life until sequencing costs decrease sufficiently that tens or hundreds of thousands of samples can be included. However, GWAS have taught us an important lesson: common genetic variation beyond CCR5 and HLA is unlikely to make a major contribution to the observed variability in HIV acquisition and control. Whether the answer lies in rare variation remains to be seen, but it is certain that significant hurdles will need to be overcome in order to fully determine how much host genetics can teach us about HIV–host interactions. In particular, it is clear that identifying convincing associations between genetic variants and individual-level responses to HIV will require large-scale sequencing of phenotypically well-characterized samples. Perhaps even more challenging, however, will be developing the functional tools necessary to understand how implicated variants influence viral responses.
ACKNOWLEDGMENTS
We thank Jacques Fellay for his assistance in designing and preparing figures.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review. This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, Frederick National Laboratory for Cancer Research.
Glossary
- GWAS
genome-wide association study (or studies); a genetic investigation examining hundreds of thousands or millions of single-nucleotide polymorphisms across the genome for correlation with a disease or trait
- HESN
HIV-exposed seronegative; describes an individual with a previous high-risk exposure to HIV who remains serologically negative for evidence of HIV infection
- Haplotype
a combination of more than one genetic variant positioned together along the chromosome
- NGS
next-generation sequencing
- PPI
protein–protein interaction
LITERATURE CITED
- 1.Barre-Sinoussi F, Chermann JC, Rey F, et al. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–71 [DOI] [PubMed] [Google Scholar]
- 2.Gallo RC, Sarin PS, Gelmann EP, et al. 1983. Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS). Science 220:865–67 [DOI] [PubMed] [Google Scholar]
- 3.Paxton WA, Martin SR, Tse D, et al. 1996. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat. Med 2:412–17 [DOI] [PubMed] [Google Scholar]
- 4.Cocchi F, DeVico AL, Garzino-Demo A, et al. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811–15 [DOI] [PubMed] [Google Scholar]
- 5.Samson M, Labbe O, Mollereau C, et al. 1996. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry 35:3362–67 [DOI] [PubMed] [Google Scholar]
- 6.Dragic T, Litwin V, Allaway GP, et al. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–73 [DOI] [PubMed] [Google Scholar]
- 7.Deng H, Liu R, Ellmeier W, et al. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–66 [DOI] [PubMed] [Google Scholar]
- 8.Dean M, Carrington M, Winkler C, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856–62 [DOI] [PubMed] [Google Scholar]
- 9.Samson M, Libert F, Doranz BJ, et al. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722–25 [DOI] [PubMed] [Google Scholar]
- 10.Liu R, Paxton WA, Choe S, et al. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–77 [DOI] [PubMed] [Google Scholar]
- 11.Martinson JJ, Chapman NH, Rees DC, et al. 1997. Global distribution of the CCR5 gene 32-basepair deletion. Nat. Genet 16:100–3 [DOI] [PubMed] [Google Scholar]
- 12.Zimmerman PA, Buckler-White A, Alkhatib G, et al. 1997. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol. Med 3:23–36 [PMC free article] [PubMed] [Google Scholar]
- 13.Rana S, Besson G, Cook DG, et al. 1997. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the delta ccr5 mutation. J. Virol 71:3219–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrington M, Kissner T, Gerrard B, et al. 1997. Novel alleles of the chemokine-receptor gene CCR5. Am. J. Hum. Genet 61:1261–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrington M, Dean M, Martin MP, et al. 1999. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum. Mol. Genet 8:1939–45 [DOI] [PubMed] [Google Scholar]
- 16.Quillent C, Oberlin E, Braun J, et al. 1998. HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet 351:14–18 [DOI] [PubMed] [Google Scholar]
- 17.Blanpain C, Lee B, Tackoen M, et al. 2000. Multiple nonfunctional alleles of CCR5 are frequent in various human populations. Blood 96:1638–45 [PubMed] [Google Scholar]
- 18.Liu H, Chao D, Nakayama EE, et al. 1999. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc. Natl. Acad. Sci. USA 96:4581–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez E, Dhanda R, Bamshad M, et al. 2001. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc. Natl. Acad. Sci. USA 98:5199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDermott DH, Beecroft MJ, Kleeberger CA, et al. 2000. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. AIDS 14:2671–78 [DOI] [PubMed] [Google Scholar]
- 21.Modi WS, Lautenberger J, An P, et al. 2006. Genetic variation in the CCL18-CCL3-CCL4 chemokine gene cluster influences HIV Type 1 transmission and AIDS disease progression. Am. J. Hum. Genet 79:120–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu L, Song W, Brill I, et al. 2012. Genetic variations and heterosexual HIV-1 infection: analysis of clustered genes encoding CC-motif chemokine ligands. Genes Immun. 13:202–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xin X, Nakamura K, Liu H, et al. 2001. Novel polymorphisms in human macrophage inflammatory protein-1 alpha (MIP-1alpha) gene. Genes Immun. 2:156–58 [DOI] [PubMed] [Google Scholar]
- 24.Proost P, Menten P, Struyf S, et al. 2000. Cleavage by CD26/dipeptidyl peptidase IV converts the chemokine LD78beta into a most efficient monocyte attractant and CCR1 agonist. Blood 96:1674–80 [PubMed] [Google Scholar]
- 25.Townson JR, Barcellos LF, Nibbs RJ. 2002. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur. J. Immunol 32:3016–26 [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez E, Kulkarni H, Bolivar H, et al. 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307:1434–40 [DOI] [PubMed] [Google Scholar]
- 27.Urban TJ, Weintrob AC, Fellay J, et al. 2009. CCL3L1 and HIV/AIDS susceptibility. Nat. Med 15:1110–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya T, Stanton J, Kim EY, et al. 2009. CCL3L1 and HIV/AIDS susceptibility. Nat. Med 15: 1112–15 [DOI] [PubMed] [Google Scholar]
- 29.Martin MP, Dean M, Smith MW, et al. 1998. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282:1907–11 [DOI] [PubMed] [Google Scholar]
- 30.McDermott DH, Zimmerman PA, Guignard F, et al. 1998. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS). Lancet 352:866–70 [DOI] [PubMed] [Google Scholar]
- 31.Field SF, Howson JM, Maier LM, et al. 2009. Experimental aspects of copy number variant assays at CCL3L1. Nat. Med 15: 1115–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franke EK, Yuan HE, Luban J. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359–62 [DOI] [PubMed] [Google Scholar]
- 33.Thali M, Bukovsky A, Kondo E, et al. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363–65 [DOI] [PubMed] [Google Scholar]
- 34.An P, Wang LH, Hutcheson-Dilks H, et al. 2007. Regulatory polymorphisms in the cyclophilin A gene, PPIA, accelerate progression to AIDS. PLoS Pathog. 3:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albin JS, Harris RS. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med 12:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An P, Bleiber G, Duggal P, et al. 2004. APOBEC3G genetic variants and their influence on the progression to AIDS. J. Virol 78:11070–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein J, Satta Y, O’hUigin C, et al. 1993. The molecular descent of the major histocompatibility complex. Annu. Rev. Immunol 11:269–95 [DOI] [PubMed] [Google Scholar]
- 38.Hughes AL, Nei M. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–70 [DOI] [PubMed] [Google Scholar]
- 39.Parham P, Norman PJ, Abi-Rached L, et al. 2012. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci 367:800–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fellay J, Ge D, Shianna KV, et al. 2009. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 5:e1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fellay J, Shianna KV, Ge D, et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Limou S, Le Clerc S, Coulonges C, et al. 2009. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J. Infect. Dis 199:419–26 [DOI] [PubMed] [Google Scholar]
- 43.Pelak K, Goldstein DB, Walley NM, et al. 2010. Host determinants of HIV-1 control in African Americans. J. Infect. Dis 201:1141–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereyra F, Jia X, McLaren PJ, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guergnon J, Dalmasso C, Broet P, et al. 2012. Single-nucleotide polymorphism-defined class I and class III major histocompatibility complex genetic subregions contribute to natural long-term nonprogression in HIV infection. J. Infect. Dis 205:718–24 [DOI] [PubMed] [Google Scholar]
- 46.Dalmasso C, Carpentier W, Meyer L, et al. 2008. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PLoS One 3:e3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiepiela P, Ngumbela K, Thobakgale C, et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med 13:46–53 [DOI] [PubMed] [Google Scholar]
- 48.Altfeld M, Kalife ET, Qi Y, et al. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8+ T cell response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Picado J, Prado JG, Fry EE, et al. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol 80:3617–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crawford H, Lumm W, Leslie A, et al. 2009. Evolution of HLA-B∗5703 HIV-1 escape mutations in HLA-B∗5703-positive individuals and their transmission recipients. J. Exp. Med 206:909–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kloverpris HN, Stryhn A, Harndahl M, et al. 2012. HLA-B∗57 micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control. J. Virol 86:919–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu XG, Lichterfeld M, Chetty S, et al. 2007. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J. Virol 81:1619–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kosmrlj A, Read EL, Qi Y, et al. 2010. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 465:350–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin MP, Qi Y, Gao X, et al. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet 39:733–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elahi S, Dinges WL, Lejarcegui N, et al. 2011. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat. Med 17:989–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendoza D, Royce C, Ruff LE, et al. 2012. HLA B∗5701-positive long-term nonprogressors/elite controllers are not distinguished from progressors by the clonal composition of HIV-specific CD8+ T cells.J. Virol 86:4014–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zemmour J, Parham P. 1992. Distinctive polymorphism at the HLA-C locus: implications for the expression of HLA-C. J. Exp. Med 176:937–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCutcheon JA, Gumperz J, Smith KD, et al. 1995. Low HLA-C expression at cell surfaces correlates with increased turnover of heavy chain mRNA. J. Exp. Med 181:2085–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snary D, Barnstable CJ, Bodmer WF, et al. 1977. Molecular structure of human histocompatibility antigens: the HLA-C series. Eur. J. Immunol 7:580–85 [DOI] [PubMed] [Google Scholar]
- 60.Thomas R, Apps R, Qi Y, et al. 2009. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet 41:1290–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stranger BE, Forrest MS, Clark AG, et al. 2005. Genome-wide associations of gene expression variation in humans. PLoS Genet. 1:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kulkarni S, Savan R, Qi Y, et al. 2011. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature 472:495–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Corrah TW, Goonetilleke N, Kopycinski J, et al. 2011. Reappraisal of the relationship between the HIV-1-protective single-nucleotide polymorphism 35 kilobases upstream of the HLA-C gene and surface HLA-C expression. J. Virol 85:3367–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Specht A, Telenti A, Martinez R, et al. 2010. Counteraction of HLA-C-mediated immune control of HIV-1 by Nef. J. Virol 84:7300–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blais ME, Zhang Y, Rostron T, et al. 2012. High frequency of HIV mutations associated with HLA-C suggests enhanced HLA-C-restricted CTL selective pressure associated with an AIDS-protective polymorphism. J. Immunol 188:4663–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kulpa DA, Collins KL. 2011. The emerging role of HLA-C in HIV-1 infection. Immunology 134:116–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bashirova AA, Thomas R, Carrington M. 2011. HLA/KIR restraint of HIV: surviving the fittest. Annu. Rev. Immunol 29:295–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alter G, Heckerman D, Schneidewind A, et al. 2011. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 476:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brackenridge S, Evans EJ, Toebes M, et al. 2011. An early HIV mutation within an HLA-B∗57-restricted T cell epitope abrogates binding to the killer inhibitory receptor 3DL1. J. Virol 85:5415–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pelak K, Need AC, Fellay J, et al. 2011. Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 9:e1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pereyra F, Jia X, McLaren PJ, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petrovski S, Fellay J, Shianna KV, et al. 2011. Common human genetic variants and HIV-1 susceptibility: a genome-wide survey in a homogeneous African population. AIDS 25:513–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lingappa JR, Petrovski S, Kahle E, et al. 2011. Genomewide association study for determinants of HIV-1 acquisition and viral set point in HIV-1 serodiscordant couples with quantified virus exposure. PLoS One 6:e28632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loeuillet C, Deutsch S, Ciuffi A, et al. 2008. In vitro whole-genome analysis identifies a susceptibility locus for HIV-1. PLoS Biol. 6:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li B, Leal SM. 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet 83:311–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brass AL, Dykxhoorn DM, Benita Y, et al. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–26 [DOI] [PubMed] [Google Scholar]
- 77.Konig R, Zhou Y, Elleder D, et al. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou H, Xu M, Huang Q, et al. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504 [DOI] [PubMed] [Google Scholar]
- 79.Yeung ML, Houzet L, Yedavalli VS, et al. 2009. A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem 284:19463–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu L, Oliveira NM, Cheney KM, et al. 2011. A whole genome screen for HIV restriction factors. Retrovirology 8:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gautier VW, Gu L, O’Donoghue N, et al. 2009. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology 6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jager S, Cimermancic P, Gulbahce N, et al. 2012. Global landscape of HIV–human protein complexes. Nature 481:365–70 [DOI] [PMC free article] [PubMed] [Google Scholar]