summary
Purpose of review
Mucormycosis is an emerging opportunistic fungal infection whose causative agents are found within the Mucorales family. A recent increase in immunocompromised cohorts with solid organ transplants, diabetes mellitus, and other medical conditions have resulted in increased fungal infections including mucormycosis. Our current knowledge about Mucoralean fungi is in its infancy compared to other fungal pathogens, which may be due to lack of robust genetic tools for Mucorales. In this review we summarize recent advances in genetic tools to study the two most prevalent and genetically amenable Mucoralean fungi, Mucor circinelloides and Rhizopus delemar.
Recent findings
There have been advances made in the study of Mucorales family genetics. These findings include the construction of recyclable markers to manipulate the genome, as well as silencing vectors, and the adaptation of the CRISPR/Cas9 gene editing system.
Summary
We present how these genetic methods have been applied to understand basic biology, morphogenesis, pathogenesis, and host-pathogen interactions in the two Mucoralean fungi, M. circinelloides and R. delemar. With these advances in Mucorales the opportunity to further understand the pathogenesis of these organisms is opened.
Keywords: mucormycosis, Mucor, Rhizopus, recyclable marker, CRISPR/Cas9 in fungi
Introduction
Mucormycosis is a fungal infection characterized by rapid progression, and is caused by fungi belonging to the Mucorales family. Mucormycosis has been observed sporadically in the past; however, a continual increase has been observed [1–4]. Mucorales are ubiquitous in nature [5]. The lifestyle of Mucorales can span from being saprobic to being opportunistic pathogens in both humans and animals [5]. A recent increase in fungal infections, including mucormycosis, can be attributed to an influx of susceptible cohorts with solid organ transplants, diabetes mellitus, and other medical conditions that cause impaired immunity [6–8]. Phagocytic cells may play crucial roles to control Mucorales infections because approximately 15% of the patients with severe neutropenia are likely to develop mucormycosis due to defects in phagocytic cell functions [3, 9]. Wüster et al. characterized the response of human mononuclear cells to various Mucorales, in which both inactivated germ tubes and spores significantly stimulated the secretion of proinflammatory cytokines TNF-α and IL1ß [10]. They also observed an upregulation of co-stimulatory molecules on monocyte derived dendritic cells along with T- helper cell activation [10]. Another study found that Toll-like receptor 2 and NF-kB pathway- related genes are induced in human polymorphonuclear neutrophils in response to Rhizopus [11]. The causative agents for mucormycosis include Rhizopus spp., Mucor spp., Rhizomucor spp., Lichtheimia spp., Cunninghamella spp., and others [12, 13, 3]. Mucormycosis presents itself with a mortality rate of ~50% in all cases and over 90% mortality in disseminated cases [7, 14, 15]. Patients with mucormycosis often suffer from permanent disfiguration as a result of surgical debridement of affected tissues followed by antifungal drug treatments [6, 7, 16].
Two pathogenic Mucorales species, Mucor circinelloides and Rhizopus delemar (previously known as R. oryzae, respectively denoted as “Mucor” and “Rhizopus”) are genetically amenable among other human pathogenic Mucorales fungi. These pathogenic Mucorales species typically grow as hyphae, but exhibit dimorphism as they are able to grow as both hyphae and yeast. This dimorphism was found to be regulated by the calcineurin gene [17, 18]. The ability to express dimorphism was also found to have a role in virulence of the Mucorales species [18]. Due to Mucorales naturally growing as moulds, its predominant growth form is that of filamentous hyphae. In a laboratory setting Mucorales can be grown on a variety of media, such as Yeast Peptone Glucose (YPG) or Minimal Media with Casamino acids (MMC) [19]. Mucorales have been observed to grow best at 26°C with plenty of light present [19, 1]. Under these conditions the Mucorales will utilize light to activate carotenoid biosynthesis and asexual sporangiospore production [20]. Mucor and Rhizopus are the most studied Mucorales models today. They are used to study and understand pathogenesis and host-pathogen interactions in mucormycosis. However, the current study of these fungal organisms presents itself with a series of challenges. The genetics of these organisms are far from being extensively understood. A whole-genome duplication occurred early in the Mucormycotina lineage and the duplication of genes may have provided new proteins which expand the sensory and signaling pathways [21]. Mucorales are known to be haploid and present zygotic meiosis when sexually reproducing [22]. Interestingly, Mucorales genomes encode genes that have been known to be metazoan specific, such as GTPases Rab32, the Ras-like GTPase Ral, and their possible positive regulators [23]. The lack of scientific attention this organism receives, and the resulting lack of available genetic tools, could be the key factors as to why it is understudied when compared to other fungal organisms. Nonetheless, the interest in Mucor and Rhizopus as subjects of further study has increased. This review discusses how genetic analysis has been achieved in Mucorales and provides updates on the newly developed genetic systems, which can enhance our understanding of mucormycosis.
Gene Knock-out in Mucor and Rhizopus
Mucoralean fungi are understudied due to the challenges faced by researchers. When compared to other pathogenic fungi, Mucorales research is still in its primordial stages. The two genetically amenable Mucorales, Mucor and Rhizopus, express a greater than usual drug resistance. The idea of traditional genetic manipulation involving drugs as dominant selective markers therefore is not attainable, which significantly limits our ability of genetic manipulation [24]. Auxotrophic markers therefore have been selected to replace a gene in Mucor and Rhizopus. In Mucor, there are two main auxotrophic markers, leuA and pyrG, used for genetic manipulation. The leuA gene is an ortholog of the Saccharomyces cerevisiae LEU1 gene, which encodes an isopropylmalate isomerase that is required for the leucine biosynthesis pathway [25]. The pyrG gene is an ortholog of the S. cerevisiae URA3 gene, which encodes an orotidine- 5’-phosphate (OMP) decarboxylase that participates in de novo biosynthesis of pyrimidines [24, 18]. In Rhizopus, the pyrF gene has been used as a selection marker; pyrF is an ortholog of the URA5 gene that encodes an orotate phosphoribosylatransferase, which is also an enzyme required for the biosynthesis of pyrimidines [26]. These leuA and pyr genes in Mucor and Rhizopus have been used to construct a gene deletion allele.
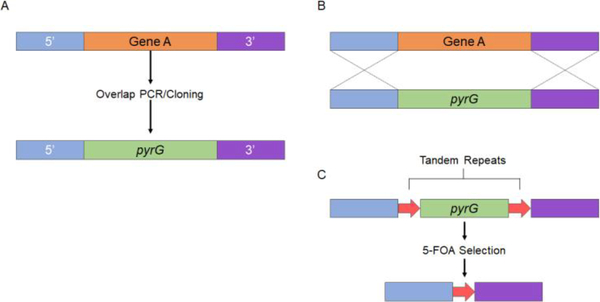
Gene knock in Mucor and Rhizopus has also incorporated conventional gene deletion construction (Fig 1). For example, the approximately 1-kb DNA fragments upstream and downstream of a target gene were put together at the 5’ or 3’ end of the marker gene, in this case pyrG. Overlapping PCR or cloning methods have been used to generate the deletion construct. The deletion construct is then delivered into the fungal cells via protoplasting transformation and/or electroporation, or via biolistic transformations for homologous gene replacement [24, 19]. The juxtaposed ~1-kb fragments of the target gene then enhance the homologous recombination between the gene of interest and the marker.
Figure 1.
Gene deletion in Mucor and Rhizopus. (A) ~1 kb of the 5’ and 3’ end of the target gene is incorporated on their respective sides to the pyrG marker gene. This is accomplished by overlapping PCR or cloning methods. (B) The deletion construct is delivered into the fungal cells via protoplasting transformation and/or electroporation, or via biolistic transformations for homologous gene replacement. (C) To construct a recyclable marker the pyrG gene is flanked on either side with a 237-bp repeat, resulting in the pyrG-dpl237 marker. The tandem repeats of 237-bp around the pyrG gene facilitates the excision of the pyrG marker after target gene deletion.
This conventional gene deletion method advances our understanding of Mucorales. The light sensing mechanisms in Mucor have been elucidated by gene deletion of the related white color genes [27, 28, 24]. These gene knockout methods have also demonstrated the roles of the sexM gene in sexual development, those of rdrp genes, argnaute genes, dicer genes in gene silencing, and those of calcineurin genes in dimorphic transition and virulence [29, 18, 17]. Additionally, the roles of ADP-ribosylation factors and protein kinase A in morphogenesis were elucidated by related gene deletions [30, 31].
In Rhizopus, a similar approach to generate a deletion construct, followed by transformation to generate a deletion allele, is employed [32]. For example, Ibrahim et al. elucidated the function of the high affinity iron permease gene (FRT1) in iron uptake and virulence via this conventional gene knockout approach [33].
Recyclable marker system for genetic manipulation of Mucor
The genomes of Mucor and Rhizopus underwent duplications and have multiple copies of genes in signaling pathways [34, 23]. For example, in Mucor there are three gene copies for the calcineurin A catalytic subunit, two copies for the immunophilin cyclophilin A, and ten copies for the protein kinase A regulatory subunit ([18] and Garcia and Lee unpublished data). Thus, studying a signaling pathway in Mucor is difficult, given the few auxotropic markers available for gene deletion. Garcia et al developed a recyclable marker to achieve serial gene deletions by using the pyrG gene in Mucor (Fig 1). In similar fashion to the URA blast system in Candida albicans, in which a 200-bp tandem repeat was flanked around the URA3 gene [35], the pyrG gene is flanked on either side with a 237-bp repeat, resulting in the pyrG-dpl237 marker [24]. The presence of tandem repeats of 237-bp around the pyrG gene facilitates the excision of the pyrG marker after target gene deletion, thus making it recyclable and possible to achieve a series of gene deletions in a single organism [24].
Selection for mutants that have excised the pyrG marker can be achieved by using the 5-fluoroorotic acid (5-FOA), which is decarboxylated by the pyrG gene product resulting in the production of the toxic compound 5-florouracil (Fig 1) [24]. Thus, mutants that lack the wild-type pyrG gene can survive on the media containing 5-FOA and uracil/uridine. With this method it is possible to selectively grow any mutants that have excised the pyrG marker. With the marker excised, the mutants generated from the first transformation can be used for a second gene deletion.
There have been mutants of Mucor developed utilizing this recyclable pyrG marker system. One example would be how Garcia et al generated double deletion mutants with a yeast-locked and albino colony phenotype [24, 18]. Mucor is a dimorphic fungus that can transition between yeast and hyphal forms, and this dimorphism is regulated by the calcineurin pathway [18]. There are three genes encoding calcineurin A catalytic subunits (cnaA, cnaB, and cnaC) and one gene encoding calcineurin B regulatory subunit (cnbR) [18]. The disruption of the cnbR gene in Mucor resulted in mutants that exclusively grow as multi-budded yeast [18]. Mucor produces carotene that results in a brownish colony phenotype, and carotene production in Mucor is governed by car genes. With the recyclable pyrG-dpl237 marker, Garcia et al deleted the carRP gene that encodes for a protein with dual enzyme activities (phytoene synthase and lycopene cyclase), both of which are involved in carotenoid production [27]. The deletion of the carRP gene generated mutants with a white colony phenotype due to a lack of carotene accumulation. The pyrG marker was then excised in the presence of 5-FOA. Eventually the cnbR gene in the albino carRP mutants was replaced with the pyrG marker to generate double mutants that only grow as white yeast. This experiment provides a proof of concept for the use of a recyclable marker system that can be in Mucorales. A recyclable genetic marker is yet to be tested in Rhizopus. It is apparent that development of a recyclable pyrF marker for Rhizopus will further enhance our ability to perform genetic analysis.
Gene Silencing as a genetic tool in Mucor and Rhizopus
Mucor has been found to have a conserved RNAi pathway that allows posttranscriptional gene regulation [36–39]. The endogenous short RNAs (esRNAs) generated by the cleavage of double stranded RNA (dsRNA) regulates expression of endogenous genes by degrading their corresponding mRNAs in Mucor [40]. Transformation of self-replicative plasmids or vectors expressing inverted repeat transgenes resulted in the identification of two different classes of small antisense RNAs linked with silencing [41, 42]. The long antisense RNAs (25nt) are only produced or detected very early in the vegetative growth stage, whereas the short antisense RNAs (21nt) are only detected in the spore RNA (late stage). Two distinct RNA polymerases are involved in the silencing process. The first polymerase (RdRP1) initiates silencing by producing antisense RNA transcripts from the target gene, and the second polymerase (RdRP2) amplifies the silencing signal by producing new dsRNA molecules from target mRNA template [43]. A study in which silencing was triggered by a hairpin RNA shows that a single dicer gene (dcl-2) is essential for gene silencing, and vegetative development [37]. The dcl-2 product cleaves the dsRNA molecules into the two classes of siRNAs which are then loaded onto a single Argonaute gene (ago-1) in order to mediate exogenous and endogenous gene silencing [39]. It is important to note that the RdRP genes are also involved in dicer independent degradation of endogenous mRNA (non-canonical pathway) [44].
Calo et al. have shown that Mucor regulates its epigenetic, RNAi mediated, post- transcriptional silencing of the fkbA gene to confer resistance to FK506 [45]. A mRNA profiling study by Nicolas et al. further confirmed that RNAi machinery controls responses to specific environmental signals [46]. The gene silencing machinery has not only been extensively studied but is also utilized as a molecular tool to perform genetic manipulation in this fungus.
Trieu et al. have developed a silencing vector pMAT1700 to generate a whole-genome silencing library [47]. The vector can also be modified to silence any target gene, and hence this is an important tool for functional genomic screenings. This self-replicative vector comprises of a pyrG selection marker and a silencing cassette. The silencing cassette has two convergent promoters (Pgpdh1 and Pzrt1), between which a multiple cloning site (MCS) is flanked to generate dsRNA that can trigger the gene silencing mechanism in Mucor (Fig 2). The carB gene, which is another car gene responsible for carotene production, is also present between the two promoters to function as a silencing reporter. A vector with target DNA can be transformed into Mucor by electroporation, and successful silencing of the cloned genomic fragment will result in easily detectable white colonies due to the carB gene being silenced simultaneously with the target gene. The authors have used this approach to identify two novel virulence factors - Phospholipase D and Myosin 5. Deletion of these genes resulted in reduced virulence in the two heterologous hosts Galleria mellonella (insect) and Mus musculus (murine). Thus, gene silencing can be used as a tool for screening and identification of unique phenotypes or any other characteristics that are different from the wild-type strain, and then the functional validation can be performed by generating knockdown strains.
Figure 2.
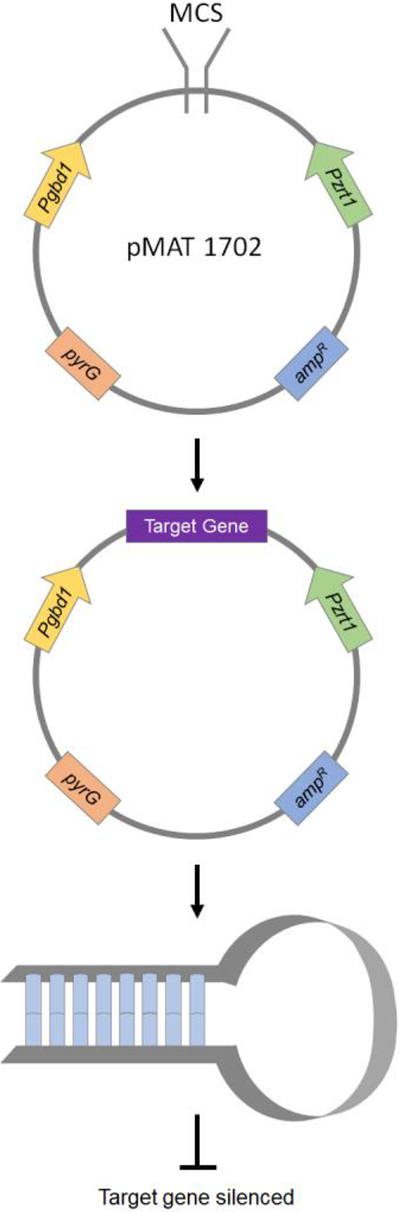
Gene silencing in Mucor and Rhizopus. Two convergent promoters flank a multiple cloning site which can be digested, and a target gene can be placed. The Amp and pyrG act as selectable markers, and the presence of carB gene adjacent to the target gene allows for selection of white colonies post-transformation. The dsRNA resulting from this self-replicative vector can silence the target gene.
A gene silencing system is also used as a molecular tool for genetic manipulation in Rhizopus. A small interfering RNA (siRNA) mediated knockdown of lactate dehydrogenase A resulted in 85.7% reduction in lactic acid but a 15.4% increase in ethanol yield [48]. In patients with chronic kidney failure, Rhizopus utilizes iron from ferrioxamine to boost its growth. Liu et al. have identified two surface proteins/receptors (Fob 1 and Fob 2) in Rhizopus that bind to ferrioxamine and facilitate iron uptake [49]. When RNAi attenuated the expression of Fob 1 and Fob 2, there was a decrease in iron uptake by the fungus.
Recently, it was discovered that a glucose-related protein (GRP78) was acting as an endothelial cell receptor to which Mucorales could bind during host cell invasion [50]. It was then concluded that a spore coating protein homolog (CotH) was the ligand for GRP78. This study resulted in a CotH3, CotH2 Rhizopus mutant (RNAi::CotH2,3) [50]. In a separate study, the RNAi silencing pathway present in Rhizopus was exploited to down-regulate two genes involved in the uptake of iron from ferrioxamine [49]. The exploitation of the RNAi silencing pathway presented by Rhizopus is a solid method for genetic manipulation of this organism. Together the data suggest that RNAi is an important tool for genetic manipulation in Rhizopus.
The main advantage of using silencing over gene knockout is that it allows one to study essential genes in Mucorales. Gene silencing can also be used to study closely related genes with shared sequence similarity such as the CotH family. The gene silencing system compensates for the lack of markers in Mucorales.
CRISPR-Cas9 in Mucor and Rhizopus
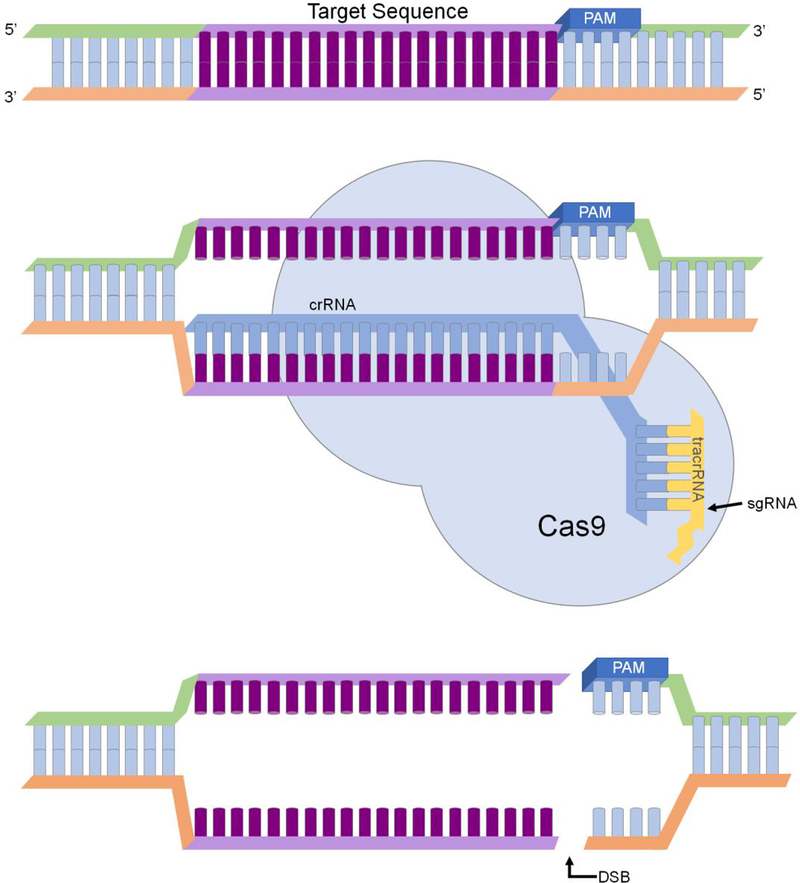
Clustered Regularly Interspaced Short Palindrome Repeats (CRISPR) systems are DNA sequences in bacteria that confer protection against bacteriophages [51]. The Type II CRISPR- Cas system from Streptococcus pyogenes has been extensively studied, and CRISPR- associated protein 9 (Cas9) was determined to be essential for all stages of S. pyogenes immunity [52]. This CRISPR/Cas9 system has been successfully modified to perform gene editing in embryos [47], plants [53], and microbes [54–57]. CRISPR/Cas9 system comprises of two essential components, which are a single guide RNA (sgRNA) and a Cas9 nickase [58]. The sgRNA is obtained by combining CRISPR RNA (crRNA; complementary to a specific part of the target gene) and trans-activating crRNA (tracrRNA). The sgRNA directs the Cas9 nuclease to induce a double-stranded break in the 20bp target DNA with a protospacer adjacent motif (PAM; Fig 3). Studies have further shown that chemical modifications at selected sites of the sgRNA could increase the specificity of the Cas9 enzyme and minimize off-target effects [59].
Figure 3.
Cas9 induced double-strand break in a target gene. The sgRNA comprises of tracrRNA and crRNA. The crRNA is specific to the target DNA, and the Cas9 enzyme creates a double-strand break upstream of the PAM motif.
Low gene editing frequencies pose a significant problem for genetic manipulation in filamentous fungi. CRISPR-Cas9 has been applied to overcome this problem in several fungi, such as Aspergillus spp.[60, 61, 56], Tichoderma reseei [62], Neurospora crassa [63], and others. Very recently CRISPR-Cas9 started gaining attention in genetic manipulation of mucormycosis causing pathogens.
In Rhizopus, a plasmid-based strategy was implemented to assemble all the required elements into one vector to disrupt a target gene [64]. A biolistic delivery system was used to transform the vector into the M16 strain (pyrF-). The transformation resulted in five stable transformants, and partial deletion of the target gene was confirmed by Southern blotting.
On the contrary, a plasmid-free system without in vitro RNP formation was used to perform gene editing in Mucor [65]. A polyethylene glycol (PEG)-mediated transformation method [66] was adapted to introduce gRNA and Cas9 enzyme with or without donor DNA. This CRISPR-Cas9 system was optimized by first disrupting the carB gene which encodes for a carotenogenic enzyme phytoene dehydrogenase [67]. Interestingly, when non-homologous end joining (NHEJ) mediated mutagenesis was adapted (no donor DNA) to disrupt the carB gene, it resulted in disruption of neighboring the carRP gene as well. However, introduction of donor DNA resulted in the stable integration of the deletion cassette at the target site via homologous recombination. The authors were also able to disrupt HMG-CoA reductase genes using a similar strategy. Together, the data suggest that homology-directed repair combined with CRISPR- Cas9 is more suitable for genetic manipulation in Mucor. This powerful tool can be used to disrupt multiple genes in order to elucidate new mechanisms which contribute to the pathogenicity of this organism.
Conclusions
Fungi belonging to the Mucorales family have proven to play many roles in industry, research, and the medical field. The interest in further study these fungi has increased, but unfortunately the tools to genetically manipulate these fungi are limited. Nonetheless, progress towards overcoming the challenges faced by scientists studying these organisms is being made. Although we are far from fully understanding the Mucorales family, we have presented many methods by which scientists are pioneering new tools for investigating Mucor. By having the ability to genetically manipulate Mucor and Rhizopus, we may be able to find novel methods of treatment for mucormycosis, further manipulate the organism for better biofuel production, and possibly establish a new fungal model organism.
Compliance with Ethics Guidelines
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors
References
Recently published papers of particular interest have been highlighted as:
• Of importance
•• Of major importance
- 1.Lanternier F, Dannaoui E, Morizot G, Elie C, Garcia-Hermoso D, Huerre M et al. A global analysis of mucormycosis in France: the RetroZygo study (2005–2007). Clinical Infectious Diseases. 2012;54(suppl 1):S35–S43. [DOI] [PubMed] [Google Scholar]
- 2.Kauffman CA. Zygomycosis: reemergence of an old pathogen. Clinical Infectious Diseases. 2004;39(4):588–90. [DOI] [PubMed] [Google Scholar]
- 3.Chayakulkeeree M, Ghannoum M, Perfect J. Zygomycosis: the re-emerging fungal infection. European Journal of Clinical Microbiology & Infectious Diseases. 2006;25(4):215–29. [DOI] [PubMed] [Google Scholar]
- 4.Brown J Zygomycosis: An emerging fungal infection. American Journal of Health-System Pharmacy. 2005;62(24):2593–6. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann K, Paw, owska J, Walther G, Wrzosek M, de Hoog GS et al. The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia MolecularPhylogeny and Evolution of Fungi. 2013;30(1):57–76. doi: 10.3767/003158513X666259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spellberg B, Edwards J, Ibrahim A Jr. Novel perspectives on mucormycosis: pathophysiology, presentation, and management. Clinical Microbiology Reviews. 2005; 18(3):556–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarkisova TA, Schaufele RL et al. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clinical Infectious Diseases. 2005;41(5):634–53. [DOI] [PubMed] [Google Scholar]
- 8.Marr KA, Carter RA, Crippa F, Wald A, Corey L. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clinical Infectious Diseases. 2002;34(7):909–17. [DOI] [PubMed] [Google Scholar]
- 9.Ibrahim AS, Spellberg B, Walsh TJ, Kontoyiannis DP. Pathogenesis of mucormycosis. Clinical Infectious Diseases. 2012;54(suppl 1):S16–S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurster S, Thielen V, Weis P, Walther P, Elias J, Waaga-Gasser AM et al. Mucorales spores induce a proinflammatory cytokine response in human mononuclear phagocytes and harbor no rodlet hydrophobins. Virulence. 2017;8(8):1708–18. doi: 10.1080/21505594.2017.1342920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamilos G, Ganguly D, Lande R, Gregorio J, Meller S, Goldman WE et al. Generation of IL-23 Producing Dendritic Cells (DCs) by Airborne Fungi Regulates Fungal Pathogenicity via the Induction of TH-17 Responses. PLOS ONE. 2010;5(9):e12955. doi: 10.1371/journal. pone.0012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neblett Fanfair R, Benedict K, Bos J, Bennett SD, Lo Y-C, Adebanjo T et al. Necrotizing cutaneous mucormycosis after a tornado in Joplin, Missouri, in 2011. New England Journal of Medicine. 2012;367(23):2214–25. doi: 10.1056/NEJMoa1204781. [DOI] [PubMed] [Google Scholar]
- 13.Ibrahim AS, Spellberg B. Zygomycetes as agents of infectious disease in humans In: Heitman J, Filler SG, Edwards JE Jr, Mitchell AP, editors. Molecular Principles of Fungal Pathogenesis. Washington, DC: ASM Press; 2006. p. 429–40. [Google Scholar]
- 14.Ribes JA, Vanover-Sams CL, Baker DJ. Zygomycetes in human disease. Clinical Microbiology Reviews. 2000;13(2):236–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanternier F, Sun H-Y, Ribaud P, Singh N, Kontoyiannis DP, Lortholary O. Mucormycosis in organ and stem cell transplant recipients. Clinical Infectious Diseases. 2012;54(11):1–8. [DOI] [PubMed] [Google Scholar]
- 16.Kontoyiannis DP, Lewis RE. Invasive zygomycosis: update on pathogenesis, clinical manifestations, and management. Infectious Disease Clinics of North America. 2006;20(3):581–607. [DOI] [PubMed] [Google Scholar]
- 17.Lee SC, Li A, Calo S, Inoue M, Tonthat NK, Bain JM et al. Calcineurin orchestrates dimorphic transitions, antifungal drug responses, and host-pathogen interactions of the pathogenic mucoralean fungus Mucor circinelloides. Molecular Microbiology. 2015;97(5):844–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SC, Li A, Calo S, Heitman J. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathogens. 2013;9:e1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vellanki S, Navarro-Mendoza MI, Garcia AE, Murcia L, Perez-Arques C, Garre V, Nicolas FE, & Lee SC. Mucor circinelloides: Growth, maintenance, and genetic manipulation. Current Protocols in Microbiology. 2018(49). doi: 10.1002/cpmc.53.** This paper includes the most current protocols utilized for the growth and maintenance of Mucor. Includes a variety of protocols ranging from simple growth methods of Mucor to genetic manipulation of Mucor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva F, Torres-Martinez S, Garre V. Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Molecular Microbiology. 2006;61(4):1023–37. doi: 10.1111/j.1365-2958.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- 21.Corrochano LM, Kuo A, Marcet-Houben M, Polaino S, Salamov A, Villalobos-Escobedo JM et al. Expansion of signal transduction pathways in fungi by extensive genome duplication. Current biology : CB. 2016;26(12):1577–84. doi: 10.1016/j.cub.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin-Sardin S, Nodet P, Coton E, Jany J-L. Mucor: A Janus-faced fungal genus with human health impact and industrial applications. Fungal Biology Reviews. 2017;31(1):12–32. 10.1016/j.fbr.2016.11.002. [DOI] [Google Scholar]
- 23.Ma L-J, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M et al. Genomic Analysis of the Basal Lineage Fungus Rhizopus oryzae Reveals a Whole-Genome Duplication. PLOS Genetics. 2009;5(7):e1000549. doi: 10.1371/journal.pgen.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia A, Adedoyin G, Heitman J, Lee SC. Construction of a recyclable genetic marker and serial gene deletions in the human pathogenic mucorales Mucor circinelloides. G3: Genes|Genomes|Genetics. 2017;7(7):2047–54. doi: 10.1534/g3.117.041095. ** Development of the first recyclable marker system for Mucor. This tool opens the opportunity to achieve serial gene deletion, which was not previously possible. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roncero MIG, Jepsen LP, Strøman P, van Heeswijck R. Characterization of a leuA gene and an ARS element from Mucor circinelloides. Gene. 1989;84(2):335–43. [DOI] [PubMed] [Google Scholar]
- 26.Skory CD, Ibrahim AS. Native and modified lactate dehydrogenase expression in a fumaric acid producing isolate Rhizopus oryzae 99–880. Current Genetics. 2007;52(1):23–33. doi: 10.1007/s00294-007-0135-0. [DOI] [PubMed] [Google Scholar]
- 27.Velayos A, Lôpez-Matas MaA, Ruiz-Hidalgo MaJ, Eslava AP. Complementation analysis of carotenogenic mutants of Mucor circinelloides. Fungal Genetics and Biology. 1997;22(1):19–27. doi: 10.1006/fgbi.1997.0998. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Frômeta RA, Gutiérrez A, Torres-Martinez S, Garre V. Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl Microbiol Biotechnol. 2013;97(7):3063–72. doi: 10.1007/s00253-012-4432-2. [DOI] [PubMed] [Google Scholar]
- 29.Lee SC, Heitman J. Sex in the Mucoralean Fungi. Mycoses. 2014;57(s3):18–24. doi: 10.1111/myc.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patino-Medina JA, Maldonado-Herrera G, Perez-Arques C, Alejandre-Castaneda V, Reyes- Mares NY, Valle-Maldonado MI et al. Control of morphology and virulence by ADP-ribosylation factors (Arf) in Mucor circinelloides. Curr Genet. 2017. doi: 10.1007/s00294-017-0798-0. [DOI] [PubMed] [Google Scholar]
- 31.Perina D, Mikoč A, Ahel J, Ćetković H, Žaja R, Ahel I. Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Repair. 2014;23:4–16. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skory C Homologous recombination and double-strand break repair in the transformation of Rhizopus oryzae. Molecular Genetics and Genomics. 2002;268(3):397–406. doi: 10.1007/s00438-002-0760-8. [DOI] [PubMed] [Google Scholar]
- 33.Ibrahim AS, Gebremariam T, Lin L, Luo G, Husseiny MI, Skory CD et al. The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Molecular Microbiology. 2010;77(3):587–604. doi:doi: 10.1111/j.1365-2958.2010.07234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corrochano Luis M, Kuo A, Marcet-Houben M, Polaino S, Salamov A, Villalobos-Escobedo José M et al. Expansion of Signal Transduction Pathways in Fungi by Extensive Genome Duplication. Current Biology.26(12): 1577–84. doi: 10.1016/j.cub.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson BR, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product- directed gene disruptions. Yeast. 2000;16(1):65–70. doi:. [DOI] [PubMed] [Google Scholar]
- 36.Nicolás FE, Santiago TM, M. RVR. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO Journal. 2003;22(15):3983–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Haro JP, Calo S, Cervantes M, Nicolás FE, Torres-Martínez S, Ruiz-Vázquez RM. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryotic Cell. 2009;8(10):1486–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Billmyre RB, Calo S, Feretzaki M, Wang X, Heitman J. RNAi function, diversity, and loss in the fungal kingdom. Chromosome Res. 2013;21(6–7):561–72. doi: 10.1007/s10577-013-9388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cervantes M, Vila A, Nicolas FE, Moxon S, de Haro JP, Dalmay T et al. A single argonaute gene participates in exogenous and endogenous RNAi and controls cellular functions in the basal fungus Mucor circinelloides. PLoS ONE. 2013;8(7):e69283. doi: 10.1371/journal.pone.0069283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicolas FE, Moxon S, de Haro JP, Calo S, Grigoriev IV, Torres-Martinez S et al. Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic acids research. 2010;38(16):5535–41. doi: 10.1093/nar/gkq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolás FE, Torres‐Martínez S, Ruiz‐Vázquez RM. Two classes of small antisense RNAs in fungal RNA silencing triggered by non‐integrative transgenes. EMBO Journal. 2003;22(15):3983–91. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicolas F, Ruiz-Vâzquez R. Functional diversity of RNAi-associated sRNAs in fungi. International Journal of Molecular Sciences. 2013;14(8):15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calo S, Nicolâs FE, Vila A, Torres-Martinez S, Ruiz-Vâzquez RM. Two distinct RNA- dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides. Molecular Microbiology. 2012;83(2):379–94. doi: 10.1111/j.1365-2958.2011.07939.x. [DOI] [PubMed] [Google Scholar]
- 44.Trieu TA, Calo S, Nicolás FE, Vila A, Moxon S, Dalmay T et al. A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi. PLoS Genetics. 2015; 11 (4). doi: 10.1371/journal. pgen. 1005168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calo S, Shertz-Wall C, Lee SC, Bastidas RJ, Nicolás FE, Granek JA et al. Antifungal drug resistance evokedvia RNAi-dependent epimutations. Nature. 2014;513(7519):555–8. doi: 10.1038/nature13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicolâs FE, Vila A, Moxon S, Cascales MD, Torres-Martinez S, Ruiz-Vâzquez RM et al. The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides. BMC Genomics. 2015;16(1):237. doi: 10.1186/s12864-015-1443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trieu TA, Navarro-Mendoza MI, Pérez-Arques C, Sanchis M, Capilla J, Navarro-Rodriguez P et al. RNAi-based functional genomics identifies new virulence determinants in mucormycosis. PLOS Pathogens. 2017;13(1):e1006150. doi: 10.1371/journal.ppat.1006150. * This work shows the development of an RNAi based genomic platform to study virulence in Mucor. It led to the identification of two genes that present themselves as promising targets for future antifungals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gheinani AH, Jahromi NH, Feuk-Lagerstedt E, Taherzadeh MJ. RNA silencing of lactate dehydrogenase gene in Rhizopus oryzae. Journal of RNAi and Gene Silencing : An International Journal of RNA and Gene Targeting Research. 2011;7:443–8. [PMC free article] [PubMed] [Google Scholar]
- 49.Liu M, Lin L, Gebremariam T, Luo G, Skory CD, French SW et al. Fob1 and Fob2 proteins are virulence determinants of Rhizopus oryzae via facilitating iron uptake from ferrioxamine. PLoS Pathogens. 2015;11(5):e1004842. doi: 10.1371/journal.ppat.1004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gebremariam T, Liu M, Luo G, Bruno V, Phan QT, Waring AJ et al. CotH3 mediates fungal invasion of host cells during mucormycosis. The Journal of Clinical Investigation. 2014;124(1):237–50. doi: 10.1172/JCI71349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR-Cas immune system: biology, Mechanisms and applications. Biochimie. 2015;117:119–28. doi:. [DOI] [PubMed] [Google Scholar]
- 52.Marraffini LA. The CRISPR-Cas system of Streptococcus pyogenes: function and applications In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes : Basic Biology to Clinical Manifestations. Oklahoma City (OK): University of Oklahoma Health Sciences Center.; 2016. [PubMed] [Google Scholar]
- 53.Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnology Advances. 2015;33(1):41–52. doi:. [DOI] [PubMed] [Google Scholar]
- 54.Zheng YM, Lin FL, Gao H, Zou G, Zhang JW, Wang GQ et al. Development of a versatile and conventional technique for gene disruption in filamentous fungi based on CRISPR-Cas9 technology. Scientific reports. 2017;7(1):9250. doi: 10.1038/s41598-017-10052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin H, Xiao H, Zou G, Zhou Z, Zhong J-J. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species. Process Biochemistry. 2017;56:57–61. doi:. [DOI] [Google Scholar]
- 56.Fuller KK, Chen S, Loros JJ, Dunlap JC. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryotic Cell. 2015;14(11):1073–80. doi: 10.1128/EC.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi T-Q, Liu G-N, Ji R-Y, Shi K, Song P, Ren L-J et al. CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl Microbiol Biotechnol. 2017;101(20):7435–43. doi: 10.1007/s00253-017-8497-9. [DOI] [PubMed] [Google Scholar]
- 58.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8(11):2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryan DE, Taussig D, Steinfeld I, Phadnis SM, Lunstad BD, Singh M et al. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic acids research. 2018;46(2):792–803. doi: 10.1093/nar/gkx1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katayama T, Tanaka Y, Okabe T, Nakamura H, Fujii W, Kitamoto K et al. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnology letters. 2016;38(4):637–42. doi: 10.1007/s10529-015-2015-x. [DOI] [PubMed] [Google Scholar]
- 61.Kuivanen J, Wang Y-MJ, Richard P. Engineering Aspergillus niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microbial Cell Factories. 2016;15(1):210. doi: 10.1186/s12934-016-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu R, Chen L, Jiang Y, Zhou Z, Zou G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discovery. 2015;1:15007. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsu-ura T, Baek M, Kwon J, Hong C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biology and Biotechnology. 2015;2(1):4. doi: 10.1186/s40694-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baldin C, Soliman S, Jeon H, Skory C, Edwards J, Ibrahim A. Optimization of the CRISPR/Cas9 system to manipulate gene function in Rhizopus delemar. Open Forum Infectious Diseases. 2017;4(Suppl 1): S116–S. doi: 10.1093/ofid/ofx163.136. [DOI] [Google Scholar]
- 65.Nagy G, Szebenyi C, Csernetics A, Vaz AG, Toth EJ, Vagvolgyi C et al. Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Scientific reports. 2017;7(1):16800. doi: 10.1038/s41598-017-17118-2. * Depicts the first implementation of the CRISPR-Cas9 system for genetic manipulation of Mucor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Heeswijck R, Roncero MIG. High frequency transformation of Mucor with recombinant plasmid DNA. Carlsberg Research Communications. 1984;49(7):691. doi: 10.1007/bf02907500. [DOI] [Google Scholar]
- 67.Velayos-Baeza A,L Blasco J,I Alvarez M, Iturriaga E,P Eslava A. Blue-light regulation of phytoene dehydrogenase (carB) gene expression in Mucor circinelloides. 2000. [DOI] [PubMed]