Abstract
Tumor heterogeneity has been identified at various -omic levels. The tumor genome, transcriptome, proteome, and phenome can vary widely across cells in patient tumors, and are influenced by tumor cell interactions with heterogeneous physical conditions and cellular components of the tumor microenvironment. Here, we explore the concept that while variation exists at multiple -omic levels, changes at each of these levels converge on the same pathways and lead to convergent phenotypes in tumors that can provide common drug targets. These phenotypes include cellular growth and proliferation, sustained oncogenic signaling, and immune avoidance, among others. Tumor heterogeneity complicates treatment of patient cancers as it leads to varied response to therapies. Identification of convergent cellular phenotypes arising in patient cancers and targeted therapies that reverse them has the potential to transform the way clinicians treat these cancers and to improve patient outcome.
Keywords: tumor heterogeneity, convergent phenotypes, tumor microenvironment, cancer therapy
“The demonstration that tumor heterogeneity is a common phenomenon could easily create an air of pessimism among clinical oncologists. After all, it appears that tumors are infinitely adaptable, possessing the ability to metastasize widely prior to clinical detection and rapidly evolve new antigenic properties, hormone receptor levels, and patterns of drug resistance. Tumors, it seems are always one step ahead of the treating physician. The alternative view, however, is that the recognition and understanding of tumor cell heterogeneity may in fact provide the foundation upon which successful new treatment strategies can be developed.” -
Schilsky, Modern Trends in Human Leukemia, Vol. 7, 1987
Introduction
Tumor heterogeneity reflects a state in which cancer cells within the same tumor are fundamentally different from each other. The use of modern sequencing technologies to analyze heterogeneity in patient tumors has confirmed hypotheses formed decades ago regarding different subclonal cell populations within a tumor [1–3]. Further, recent studies have also shown that these subclone populations differ at levels other than merely the genetic, including epigenetic, transcriptomic, proteomic, and, as will be the focus of this review, cellular phenotype [4–8]. Not only have differences at these -omic levels been detected as a cancer progresses through time (temporal heterogeneity), but differences have also been detected across space (spatial heterogeneity) [9–11]. Spatial heterogeneity can be both intratumoral, i.e., within a single primary or metastatic tumor, and intrapatient, i.e, between different tumors within the same person [12, 13]. Thus, as a patient’s cancer progresses, subclones, defined as genetically distinct groups of tumor cells descending from a common ancestor, can be identified, and their evolutionary trajectory can be followed as each subclone responds to or resists treatment.
While cancers have long been considered heterogeneous at the chromosomal level [14, 15], it was not until 1976 that Peter Nowell first theorized that because cancers mostly arise from a single neoplastic cell, they must undergo a stepwise genetic evolution unique to each tumor that gives rise to multiple heterogeneous subclones [1]. This hypothesis formed an early rationale for individualized cancer treatment. This stepwise genetic evolution Nowell attributed to genomic instability inherent to neoplastic cells. Nowell further applied ideas from population biology to tumor cells, positing that some of these acquired mutations would impart selective advantages, allowing certain subclones to expand and proliferate, while other, less fit subclones, would proliferate more slowly and eventually die off due to competition with more dominant subclones. Interestingly, Nowell also posited that as tumors progress, their genetic evolution converges on similar biological characteristics, i.e. phenotypes, a concept we will explore below.
The understanding that genetic variation exists amongst tumor cells engendered Nowell’s notion that these cells may differ in their response to various therapeutic regimens, an idea seconded by Schilsky, who proposed that oncologists adopt treatment strategies capable of eradicating not one homogenous cancer, but various tumor subpopulations [16]. However, little advancement has been made in the clinical realm towards achieving this goal. Currently, the histopathology of tumor biopsies is assessed at the bulk level--biopsies are not dissociated into single cells to identify the subclones before performing laboratory tests or diagnostics. Similarly, clinicians do not routinely re-biopsy tumors or metastases as the disease progresses, steps which could be taken to make changes in treatment based on a tumor’s temporal and spatial heterogeneity and its likely unique evolutionary trajectory. Thus, despite our longstanding awareness of tumor heterogeneity, research efforts in this field have led to little impact on clinical decision making.
In this review, we will discuss the progress that has been made in understanding tumor heterogeneity at various -omic levels, including cellular phenotype. We will also discuss how the field can move closer to incorporating concepts of tumor heterogeneity into the clinic in order to impact patient care.
Tumor Heterogeneity Exists at Multiple -Omic Levels
Tumor -omics vary greatly across cancer cells through time and space. Clinicians use profiling of tumor mutations and gene expression to diagnose, treat, and make prognostic decisions regarding a patient’s cancer [17–19]. Response to targeted therapies requires presence of specific -omic events throughout a patient’s tumor as well as robust methods of detecting these events. Therefore, a full understanding of tumor heterogeneity in primary tumors and metastases and in early versus late stage disease is necessary for determination of efficacious therapeutic regimens and prognostication for patients.
Genomic heterogeneity
Studies of tumor heterogeneity at the genomic level indicate tumor cells are extremely diverse, spatially and temporally. Tumor diversity through space is a phenomenon that has been well-studied by research groups comparing mutations identified in multiple regions sampled from primary tumors or by comparing primary tumor mutations to those identified in metastases [20–23]. For example, whole exome sequencing of 23 regions of a single hepatocellular carcinoma (HC) tumor identified presence of 20 unique subclones, and by extrapolation to the entire volume of the HC tumor, an estimated 100 million somatic coding mutations across all subclones. This finding demonstrates the extreme spatial heterogeneity of a clinically unexceptional tumor, and suggests multiple biopsies may be necessary to capture all clinically-actionable mutations and to accurately determine which mutations are truly clonal and which are actually subclonal [24]. Indeed, in a study of spatial heterogeneity of medulloblastoma, high-grade glioma, and renal cell carcinoma (RCC), Morrissy and colleagues calculated no fewer than 5 biopsies are necessary for an 80% chance of detecting at least 80% of the somatic variants [25]. Similarly, Werner and colleagues calculated that 8 biopsy samples must be taken from clear cell RCC tumors to determine which mutations are truly clonal with a probability of 99% [26]. Other studies have profiled more patients, but fewer regions per tumor to also demonstrate wide spatial heterogeneity throughout a single tumor. Through profiling 4 to 5 regions of primary tumors from 11 patients with HC, Lin and colleagues determined that, on average, 39% of somatic mutations varied across the spatial samples studied from each patient’s tumor [27]. This is similar to the 36% seen across 3 to 4 samplings each from 13 patients with esophageal squamous cell carcinoma (ESCC) [10] and 43% seen in 4 patients with oligodendroglioma [28]. However, this difference in spatial somatic mutation varied widely per patient, ranging from 5–92% in HC, 8–61% in ESCC, and 10–64% in oligodendroglioma, demonstrating the unique evolutionary trajectory inherent to different cancer types and to each individual patient.
Studies of spatial heterogeneity have also shed light on the biology of metastasis. By comparing whole exome sequencing in samples taken from primary FFPE samples and 5–12 metastatic sites during rapid autopsy of 4 patients with metastatic breast cancer, Savas and colleagues demonstrated that metastatic cells are likely capable of cross-seeding sites, and that metastases can be seeded by polyclonal groups of cells [29]. Further, Ng and colleagues found the metastatic breast tumor exome can differ from that of the primary tumor, even in the absence of selection by drug treatment, in patients presenting with untreated metastatic breast cancer [30]. Thus, the tumor genome demonstrates spatial heterogeneity amongst samples from differing regions of the same tumor as well as between primary and metastatic tumor samples.
In contrast to spatial heterogeneity, tracking temporal heterogeneity of the tumor genome has proven to be a greater challenge, as clinical standard of care generally precludes obtaining biopsies throughout the course of patient disease. However, Castellarin and colleagues used cancer cells collected from ascites fluid of patients with high grade serous ovarian carcinoma (HGSOC) to demonstrate that ~90% of mutations found in relapse samples from these patients were detectable in the primary tumor, suggesting temporal evolution in response to drug treatment may be a function of selection of existing cells more than it is a driver of evolution of new mutations [31]. Interestingly, Patch and colleagues also used cells from ascites fluid of patients with HGSOC to confirm increased mutational burden in relapse compared to that seen in primary tumors, and also found that the majority of SNVs and indels identified in relapse samples were identifiable in the primary tumor [32]. This study took findings one step further, however, by identifying recurrent molecular alterations seen in relapse, including reversion mutations in BRCA1 and BRCA2 and translocation of the ABCB1 gene such that it becomes fused to a strong promoter. Interestingly, Aihara and colleagues employed exome sequencing of 12 paired primary and recurrent oligodendrogliomas resected from patients as part of routine clinical care to demonstrate that approximately one-third of mutations from the primary tumor are retained in the recurrent tumor [28]. Thus, despite limited access to sequential samples, several groups identified temporal heterogeneity in various cancers as evidenced by changes in subclonal dynamics and in overall mutational burden over time.
Aside from collection of cancer cells from standard of care methods such as draining of ascites fluid, various studies have employed serial sampling of circulating tumor DNA (ctDNA) to examine temporal heterogeneity. For example, De Mattos-Arruda and colleagues identified all somatic nonsynonymous mutations arising in a primary tumor and liver metastasis of a patient breast cancer over the time course of ipatasertib treatment via massively parallel sequencing of the patient’s ctDNA [33]. Similarly, Frenel and colleagues used identification of somatic mutations in ctDNA to assign patients to clinical trials of therapies targeting these mutations, and tracked response to therapy by measuring changes in variant allele frequency of the mutation over time in ctDNA [34]. Remarkably, clonal hierarchy inferred from SNVs detected in serial ctDNA samples recapitulates the clonal evolution of metastatic lesions and reflects therapy response in a ER+/HER2+ breast cancer patient, confirmed by mutations later identified in samples taken from metastatic sites upon autopsy [35]. The ability to track clonal evolution via serial ctDNA samples has also been demonstrated in non-small cell lung cancer, in which detection of patient-specific somatic variants in post-operative ctDNA samples predates progression detected by more traditional CT imaging by an average of 70 days [36]. Thus, the emergence of technologies capable of capturing and analyzing ctDNA from simple blood draws shows promise in furthering our understanding of temporal heterogeneity in an easily accessible, less invasive manner [37–39].
Recently, technological improvements have allowed for the study of tumor heterogeneity at the single cell level [40]. Thus, rather than disentangling bulk level events by computationally assigning them to specific subclones, co-occurrence of mutations can also be identified in single cells to better map evolutionary and mutational trajectory [41]. An early single-cell study utilized flow sorting of single cell nuclei followed by whole genome amplification to identify copy number differences between single triple negative breast cancer (TNBC) cells [42]. However, these single cell genomes were sequenced at a low depth of 6X and thus recovered only copy number/ploidy data for each cell. Nevertheless, this low-depth data was sensitive enough to identify which subclone from the primary tumor seeded the liver metastasis in the patient studied. Similarly, Wang and colleagues performed whole genome and whole exome sequencing on single cells from one TNBC and one ER+ patient breast tumor, finding that copy number variation was largely similar in all cells studied and thus likely occurs early in cancer initiation, whereas private mutations were more likely to be point mutations than copy number variations [43]. Further, the study found the TNBC cancer had a mutation rate approximately 13 times that of the ER+ cancer. More recently, Li and colleagues demonstrated that nearly twice the number of somatic mutations could be detected by single cell exome sequencing of 20 tumor cells from a patient with RCC than by exome sequencing of bulk tumor tissue, and identified LOC440040 and LOC440563 as potential novel driver genes of RCC stem cells [44]. Thus, study of the tumor genome and/or exome at the single cell level demonstrates utility in identifying and establishing order of mutational events.
Single cell studies have also engendered the confirmation of the bulk sequencing finding that solid tumors are more heterogeneous and possess more subclones than hematologic malignancies [45, 46]. Xu and colleagues examined the exome of 20 tumor and 5 normal cells from a patient with clear cell RCC [47]. Principal components analysis of somatic mutations showed that, instead of being monoclonal, likely many subclones were present in this stage IV patient tumor, as evidenced by detection of multiple somatic mutations unique to single cells. This is to be contrasted with the single cell exome study of 90 cells of a myeloproliferative neoplasm that demonstrated monoclonality of the cancer [48].
Examination of spatial and temporal heterogeneity at the genomic level demonstrates the remarkable diversity of cancer cells and the unique mutational trajectories each cancer type and each individual cancer takes as it evolves through space and over time. Increased spatial sampling of tumors and technological improvements in capturing and analyzing single tumor cells as well as ctDNA will better shape our understanding of spatial and temporal changes in the tumor genome. Thus, a complete understanding of genomic heterogeneity may help guide the use of targeted therapies and improve outcomes in patients.
Transcriptional heterogeneity
Tumor cells demonstrate heterogeneity not only at the genomic level, but also at the transcriptome level. As bulk tumor transcriptome may also reflect signal from normal cells, we discuss here studies of RNA at the single cell level, which have uncovered fascinating findings in the transcriptomes of various cancers [13]. Through the development and application of various computational algorithms, such as spanning-tree progression analysis of density-normalized events (SPADE), data from these single-cell experiments can be used to group cells based on phenotype and to infer hierarchy of subclones [49–51].
Studies of single cell transcriptomics across multiple cancer types support the idea that cellular expression of particular pathways is much more varied than can be concluded by bulk pathway assessment. For example, single cell RNA-sequencing of over 400 primary glioblastoma cells from 5 patients showed these cells vary widely in their expression of cell cycle, hypoxia, and immune/complement gene expression programs [52]. Further, rather than occupying discrete stem and non-stem spaces, these glioblastoma cells are positioned across a stemness gene expression spectrum. Similar variation across the stemness spectrum was identified in a study of over 4,000 cells from 6 patients with oligodendroglioma [53]. In a study of transcriptomic heterogeneity amongst 75 cells from one primary squamous cell carcinoma bladder tumor, Zhang and colleagues identified significant variation in expression of genes of the MAPK signaling pathway in each cell [54]. Further, by identifying genes that were most often co-expressed, the authors identified several “hub” genes coordinating the most common gene expression profiles identified in the cells, including genes such as SCN2A, CENPH, TUBGCP2, LINC00189, and ARHGAP15, encompassing genes both known and previously unknown to have cancer involvement. Chung and colleagues performed single cell RNA-sequencing of cells from 11 patients with various breast cancer subtypes to identify heterogeneity of subtypes even among cells from the same patient tumor and to pinpoint subtype-specific differences in gene expression [55]. Specifically, the group found that TNBC cells have increased expression of epithelial-to-mesenchymal transition (EMT) genes than luminal and HER2+ cells, and that gene expression varies widely in cells of the TNBC subtype. Given the wide variation in expression of pathways discussed above, variable response to therapies, even in tumors within the same patient, is not surprising.
Proteomic heterogeneity
Some evidence suggests proteomic heterogeneity, while detectable in patient cancer samples, may be less pronounced than that seen at the genetic, epigenetic, and transcriptomic levels. For example, by studying patient colorectal cancer liver metastases, Turtoi and colleagues found that distribution of proteins throughout the metastatic lesions was similar from patient to patient [5]. Peritumoral areas consisting of normal tissue adjacent to the tumor, “rim” tissue consisting of cancerous cells at the outside of the tumor, and inner tumor cells all demonstrated differing proteomes, however, a similar stratification was seen across patients, suggesting the liver microenvironment may impose constraints on cellular phenotype. In terms of temporal heterogeneity, by profiling matched chemo-naive and chemo-resistant cells obtained from ascites fluid of patients with advanced-stage ovarian carcinomas, Ahmed and colleagues identified increases in proteins related to energy metabolism and DNA mismatch repair in chemo-resistant samples [56].
Kim and colleagues demonstrated that proteomic heterogeneity warrants the use of targeted therapies unique to each metastatic organ by identifying site-specific spatial heterogeneity in one pancreatic cancer patient’s lung, liver, and peritoneum metastases [57]. Receptor activity and signal transduction activity were identified as two top classes of proteins most differentially expressed between these metastatic sites. The group further found tyrosine phosphorylation of proteins was highly variable, having greater than 2-fold difference between any of the two metastatic sites, for 84% of the specific phosphorylation sites studied. However, one drawback of this study was that the metastatic cells were cultured prior to proteomic analysis. Exposure to two-dimensional culture may alter gene and protein expression in cancer cells due to adaptation to growth conditions dissimilar to that of the human body [58].
As with the genomic and transcriptomic levels, study of proteomic heterogeneity has also benefitted from single-cell studies. For example, Giesen and colleagues used mass cytometry of single cells laser-ablated from 21 breast cancer FFPE samples to examine proteomic heterogeneity [59]. Interestingly, the group applied SPADE to define subpopulations in these tumors, and found high variability in cytokeratin 8/18, cytokeratin 7, and E-cadherin expression amongst subpopulations of the same tumor. Similarly, Sood and colleagues examined heterogeneity of 27 proteins in single tumor cells from 26 breast cancer FFPE samples via sequential cycles of fluorescence microscopy [60]. Proteins studied, including HER2, ER, PR, PTEN, c-MYC, EGFR, et al., were grouped into 8 coexpression clusters as seen in single patient cells via K-medians clustering. Interestingly, 9 of 26 patient samples expressed a single coexpression cluster in 95% or greater of the cells examined, whereas the remaining patient samples showed multiple coexpression clusters, each present at a lower prevalence. Due to the low number of coexpression clusters detected, the authors of this study concluded that proteomic heterogeneity is less complex than genetic and epigenetic heterogeneity suggests; supporting the idea that tumor cells converge on similar phenotypes.
As evidenced by the above studies, proteomic heterogeneity of cancer cells is a budding field ripe for future discovery. Studies have established that indeed tumors demonstrate proteomic heterogeneity, but the functional links to sources of this heterogeneity require further investigation. Financial and technical challenges of performing whole proteome analysis on single cells have limited progress in this field; a full understanding of proteomic heterogeneity in patients will require both increased patient sample size and increased number of proteins assessed per study.
Despite variation at various -omic levels, tumors converge on recurrent phenotypes
Evidence for evolution of phenotypic convergence
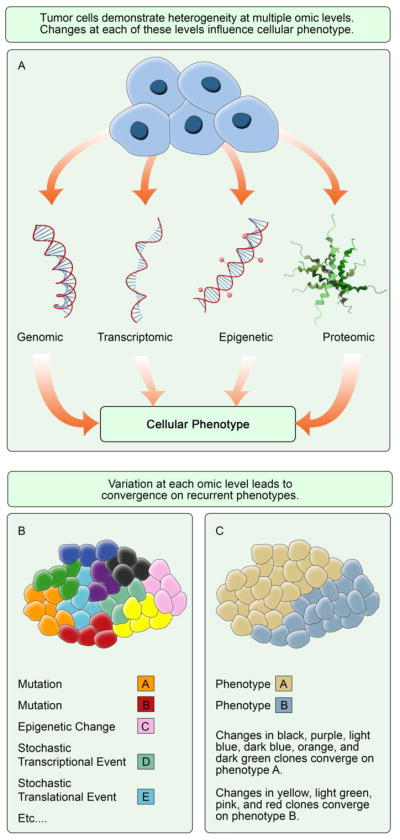
As described above, changes in the tumor genome occur over space and time, and can be monitored at the single cell level. However, it is important to note that while studying heterogeneity allows for the identification of subclones present in a tumor, phenotypic plasticity can occur due to non-genetic factors. Isogenic cancer cells may demonstrate differing phenotypes due to various causes such as stochastic events occurring during transcription and translation or differing interactions with heterogeneous normal cells in the microenvironment (Figure 1) [61]. Despite the pervasive notion that nonsynonymous changes in genotype lead directly to production of a novel phenotype, it has been hypothesized rather that changes in genotype lead to differing frequencies of phenotypes already present in the population [62]. Indeed, the link from tumor cell genotype to output phenotype has yet to be fully elucidated, and microenvironmental impacts on this process have yet to be disentangled. Therefore, the output of heterogeneity at all -omic levels, cellular phenotype, is an important level of data for impacting treatment decision making.
Fig. 1.
Tumor heterogeneity converges on recurrent cellular phenotypes. a Variations at the genomic, transcriptomic, epigenetic, and proteomic levels influence cellular phenotype b Tumor subclones vary widely in terms of mutational status, epigenetic changes, stochastic transcriptional and translational events, etc. c Changes converge on pathways that output recurrent phenotypes A and B
Identification of common pathways instead of uncommon mutations on which individual subclones rely for survival may be an alternate strategy for targeting with drugs. As notably discussed by Hanahan and Weinberg, as cancers progress, they converge on several unifying hallmarks, or phenotypes [63]. These phenotypes include sustained proliferation, invasion and metastasis, and anti-apoptosis signaling, among others, many of which are targetable with drugs currently either in clinical trials or that are FDA-approved. Indeed, targeting common tumor cell phenotypes, rather than the heterogeneous genetic, epigenetic, or tumor microenvironmental (TME) factors underlying them, may prove a more viable strategy in effectively treating cancers.
Several groups have elucidated the specific pathways on which cancers converge to produce hallmark phenotypes. For example, Chen and He found that when measuring gene expression in 3,000 tumors spanning 18 different types of solid cancers, cells evolved towards functional resemblance of embryonic stem cells (ESCs) and away from resemblance of the tissue-of-origin as the cancers progressed [64]. Further, rather than reliance on a specific set of genes, random sampling of 500 genes in the genome used to compare expression profiles of the cancer to ESCs showed equivalent results in prognostic capabilities, suggesting that rather than a set gene expression program, cells converge on a specific ESC functional state or phenotype. This work supports the previous findings by Chen and colleagues that showed that as a cancer evolves, it loses expression of genes required of multicellular organisms and shifts to expressing those of unicellular organisms, thus placing chief importance on self-renewal [65]. One notable limitation of the Chen and colleagues study is that it was performed in only one xenograft tumor--study of more in situ patient tumors is required to further support the notion of devolving from multicellular to unicellular gene expression programs. However, taken together, these two studies indicate that as cancers progress, they converge on gene expression programs of self-renewal that most resembles ESCs rather than tissue of origin, and indicates that therapies reversing the stemness state may lead to lesser self-renewal of advanced cancers and increased sensitivity to chemotherapies.
The studies discussed above examined cancer cell phenotypes at primary sites of disease. Other studies have analyzed phenotypes of metastatic cells. To estimate the impact of cellular phenotypes on metastatic success, Cunningham and colleagues employed a computational approach based on evolutionary game theory and the Lotka-Volterra competition model (typically used to model the dynamics between predator and prey populations in ecology), to represent dynamics of cancer cell and normal cell populations at primary and metastatic sites [66]. This model demonstrates that circulating tumor cells attempting to seed a metastatic site whose environment is very different from their tissue of origin cannot compete with normal cells for nutrients and space, and eventually they die. However, cells that seed a metastatic site similar to the tissue type of origin are more likely to express phenotypes that will be successful and expand and proliferate in that environment. Cells that develop successfully into metastatic tumors must adapt to the novel landscape of the metastatic tissue; therefore cells surviving in a specific metastatic site express convergent phenotypes specific to that site, even if their origins are different. Further, circulating tumor cells must strike a balance between employing multiple phenotypic strategies that increase the probability of survival in more metastatic sites and convergence on a mesenchymal phenotype to promote survival in circulation. This in silico model proposed by Cunningham and colleagues requires further testing and validation in vivo, however, initial conclusions from these models indicate that patients should be given targeted therapies specific to the phenotypes employed by cancer cells at each metastatic site. This is often not a reality for patients in the clinic, whose metastases are frequently treated based on characteristics from biopsy of the primary tumor and not the metastatic site.
Convergent phenotypes of metastatic cells have also been identified in patients. In a study of primary and metastatic biopsies from a patient with RCC, Gerlinger and colleagues identified mutation of the same genes involved in histone methylation, SETD2 and KDM5C, in different genomic sites in clones of various lineages, pointing to chromatin modification as an early altered event in this disease [67]. Other studies of metastatic RCC identified convergent evolution towards activation of the mTOR pathway, as evidenced by mutations arising in mTOR pathway members in spatially distinct parts of patient tumors [68, 69]. Additionally, Chen and colleagues identified glucose-independent metabolism pathways, such as gluconeogenesis, as a convergent phenotype in brain metastatic breast cancer cells [70]. Thus, as cancers progress, cells arrive at convergent phenotypes, although these phenotypes may be specific to cancer type.
Further, convergent evolution has been identified in response to therapy. For example, 10 of 14 metastatic lesions from a breast cancer patient harboring an activating mutation in PIK3CA and who was treated with the PI3Kα inhibitor BYL719 showed PTEN loss via mutation or deletion at different sites in each metastatic lesion [71]. Similarly, 5 independent BRCA2 reversion mutations were identified in the metastases of a HGSOC patient with germline BRCA2 mutation treated with platinum-based therapy and the PARP inhibitor olaparib [32]. Interestingly, stochastic transcriptional events have also been linked to establishment of a stable drug-resistance phenotype [72]. Indeed, random fluctuations in gene expression observed in melanoma cell lines demonstrate existence of rare cells in a population that express high levels of resistance markers before ever coming into contact with a drug. Further, after treating these pre-resistant cells with drug, they become stably resistant via chromatin remodeling, leading to differential chromatin accessibility by transcription factors, dedifferentiation, and downstream signaling in new pathways. A similar phenomenon has also been identified in non-small cell lung cancer cell lines, in which initial resistance to the EGFR inhibitor gefitinib is caused by selection of innately resistant clones bearing the EGFRT790M mutation, whereas late-emerging resistance evolves from slow-growing, drug-tolerant clones that initially lack the EGFRT790M mutation, but that evolve this mutation over months of treatment [73]. Therefore, drug treatment shapes evolution of cancer cells towards treatment resistance, either by selection of rare innately resistant cells driven by mutational or stochastic transcriptional changes, or by acquisition of resistant phenotypes.
Research indicates that in the absence of selective pressure, cancers evolve in a neutral fashion [74]. The mutations arising during neutral evolution may produce heterogeneity at various -omic levels within a tumor, but this heterogeneity may not be functionally relevant. Approximately one-third of tumors from 14 different cancer types studied showed evidence of neutral evolution [74]. Therefore, in environments lacking selective pressures, evolution of convergent tumor phenotypes may not occur. Despite evidence supporting Darwinian evolution concepts as applied to tumor ecology and evolution, tumor cell population dynamics between subclones and with normal cells in the microenvironment remain incompletely understood. Eventually, tumor cell populations kill their hosts, and themselves with it, thereby highlighting the difference in modeling cancer versus populations whose end goal is survival of the ‘species’. Further work is required to understand more completely the role of tumor cell interactions between themselves and with the host before tumor heterogeneity and tumor cell behavior can be completely understood.
Mechanisms driving phenotypic convergence
Tumor cells converge on phenotypes promoting tumor growth and survival. Mechanisms driving phenotypic convergence are not completely understood, but are considered to include stochastic evolution and adaptation. Interestingly, stochastic evolution has been shown to lead to convergence. Indeed, convergence driven by random mutation is not a rare event [75]. As discussed above, another mechanism of convergent evolution seen in metastatic tumor cells is adaptation to a novel microenvironment, driving cells away from phenotypes inherent to the primary tumor and towards expression of phenotypes specific to cells native to the metastatic tissue. One way this occurs is through epigenetic remodeling. For example, gene expression profiles of 4 various human cancer cell lines orthotopically transplanted into mouse brain more closely resembled that of mouse brain tissue than the original cell-line specific gene expression profile [76]. Changes in transcriptional profile in these cell lines were attributed to changes in methylation at brain-specific transcription factors. Besides epigenetic changes, direct genetic changes leading to convergent evolution have been observed, as discussed above in RCC subclones developing mutations in genes from the same pathways [67–69], and as evidenced by amplification of similar driver genes occurring in different non-small cell lung cancer clones from the same patients [77].
Treatment of convergent phenotypes
Identification of convergent phenotypes allows for targeting phenotypic dependencies with drugs. While there are effective therapeutic regimens that target specific mutations in tumors, such as EGFR, HER2, and ALK inhibitors, there are few common mutations in progressive resistant cancer, necessitating approaches that are independent of mutations. Therapies targeting the hallmarks of cancer and other more specific phenotypes discussed above are already used in the clinic and could be implemented upon clinical detection of these phenotypes in the patient’s cancer [63]. Therapies targeting convergent phenotypes may prove effective for treatment of various cancer types, given that similar phenotypes are seen across different cancers.
It is necessary to note the limitations of a phenotype-based drug treatment. Even the best therapies targeting convergent phenotypes may not elicit complete response in patients. Zimmer and colleagues demonstrated that drug response, even in clonal cell lines, is subject to stochastic cellular events, whereby the widely differing protein expression in clonal cells leads to populations of both responders and non-responders to drug [78]. Similarly, Sharma and colleagues demonstrated that cancer cells use chromatin remodeling to transiently generate a drug tolerant state [79]. Further, identification of the most appropriate time point to implement candidate therapies may prove challenging. For example, a specific “window of opportunity” may need to be identified in which the tumor has manifested identifiable convergent phenotypes, but before the cancer has metastasized [69]. Further, implementation of phenotype-targeted therapies requires development of a quick and comprehensive method for assaying phenotype. While the cost and high-throughput capabilities of RNA sequencing are improving, this data requires both a good deal of time and expertise to interpret. Other viable options in addition to those discussed may include development of quick phenotypic assays requiring less expertise to interpret, such as high-throughput interrogation of cellular morphology and organization via automated fluorescence microscopy [80].
Extrinsic factors impact tumor heterogeneity
Selective pressures on an evolving tumor come not only from the treatment received by the patient, but also from the TME. As discussed above, convergent evolution occurs at metastatic sites as tumor cells adapt to the microenvironmental conditions at that site [66]. Various physical conditions in the TME alter tumor cell phenotype. For example, hypoxia in the microenvironment of the primary tumor leads to heterogeneity of dormancy phenotype expression in tumor cells disseminating from the primary site [81]. Similarly, regional differences in tissue perfusion of non-small cell lung cancers correlated with differences in metabolism--areas of high perfusion used both glucose and alternative fuel sources, whereas areas of lower perfusion used mainly glucose for fuel [82].
Further, heterogeneous normal cell populations resident to the TME impact tumor cell heterogeneity. One way cells of the TME influence cancer cell phenotype is through paracrine signaling. For example, McLean and colleagues found that ovarian carcinoma-associated mesenchymal stem cells (CA-MSCs), a class of non-tumorigenic multipotent cells with a normal genome, promote tumor growth and stemness phenotype and have high BMP2 and BMP4 expression [83]. High BMP4 expression drives production of hedgehog ligands by ovarian tumor cells, which, when released to the stroma, leads to increased BMP4 production by CA-MSCs in a paracrine positive feedback loop [84]. Further, this increased hedgehog signaling activity correlated with chemotherapy resistance, which was reversed by blocking hedgehog signaling with an inhibitor of the hedgehog receptor, smoothened. Similarly, paracrine signaling from cancer associated fibroblast secretion of the ligand IGF-II, which binds to the receptor IGF-1R on cancer stem cells from non-small cell lung cancers, has been shown to induce and maintain stemness phenotype [85]. An analogous event has been identified in colon adenocarcinoma whereby the secretion of hepatocyte growth factor (HGF) by stromal myofibroblasts promotes stemness in colon cancer stem cells and in more differentiated tumor cells alike [86]. Similarly, stromal cell secretion of HGF and subsequent activation of signaling downstream of its receptor, MET, correlates with RAF inhibitor resistance in melanoma [87]. Thus, varying proximity of tumor cells to stromal cells such as fibroblasts and mesenchymal cells leads to phenotypic diversity in drug response and stemness.
Tumor heterogeneity is also shaped by interactions with immune cells in the TME. Immune cells release cytokines which are known to expand cancer stem cell populations [88, 89]. One such cytokine, TGF-β, is secreted by epithelial, immune, and/or tumor cells, and downstream signaling in this pathway can lead to inhibition of cytolytic gene expression and therefore crippling of cytotoxic T cells and immune evasion of tumor cells [90, 91]. Further, differences in tumor cell antigens shape heterogeneity as immune cells kill tumor cells bearing neoantigens, while those cells with lower immunogenicity escape immune surveillance and survive [92]. One mechanism by which this occurs is down-regulation of MHC class I, and therefore decreased antigen presentation, leading to presence of fewer tumor cell antigens on the cell surface [93]. Ability to evade immune surveillance thus selects for presence of low immunogenic cancer cells in patient tumors.
Given the above factors, effective therapeutic strategies targeting tumor heterogeneity must consider not only targeting of the tumor itself, but also of the microenvironment. Indeed, heterogeneity in the microenvironment impacts drug delivery and drug response. For example, Song and colleagues found that heterogeneity of the tumor microenvironment impacted delivery of nanoparticle-bound doxorubicin to tumor cells [94]. Further, physical conditions in the microenvironment impact response to drug and cellular proliferation rates [95]. Changes in glucose, oxygen, and erlotinib concentrations affected proliferation rates of parental HCC827 non-small cell lung cancer cells sensitive to erlotinib and of two different HCC827 erlotinib-resistant lines (one with EGFR mutation, one with MET amplification) in different manners. Compared to erlotinib-resistant cells, erlotinib-sensitive cells have a proliferative advantage in the absence of drug in high glucose and high oxygen environments. However, under nutrient-stressed conditions, resistant cells maintain the growth advantage, even in the absence of drug. While further research is necessary to determine which physical conditions best facilitate drug response for each drug in each cancer type, strategies to target microenvironmental conditions may be a viable option for altering drug response in cancer cells.
Targeting tumor heterogeneity in the clinic
Tumor heterogeneity associates with poor outcome and complicates cancer treatment. Patients from the Cancer Genome Atlas head and neck cancer cohort who had a high mutant-allele tumor heterogeneity (MATH) score, a measure of intratumor heterogeneity based on whole exome sequencing data, had decreased overall survival [96]. Similarly, response to immune checkpoint inhibitors in non-small cell lung cancer correlates with clonality of neoantigens [97]. In the clinic, intrapatient heterogeneity manifests as mixed responses to treatment, where some tumors shrink and others grow when given the same systemic therapy. For example, 21% of patients with non-small cell lung cancer who receive first line therapy with either chemotherapy or tyrosine kinase inhibitors have a mixed response [98]. Because of the presence of growing tumors, oncologists consider these patients to have progressive disease, which often leads to changes in therapy. Indeed, in some settings, mixed response is more common than progression at all sites of cancer in patients with progression [99].
Historically, oncologists would obtain one biopsy of a tumor when a patient presented with metastatic cancer in order to support the diagnosis and to determine the type of cancer. After the initial biopsy was taken, patients were followed clinically with no need to rebiopsy [100]. With the FDA approval of an increased number of targeted treatments, it is becoming more common to biopsy tumor tissue not just at the time of diagnosis but also serially, to guide treatment. Alternatively, “liquid biopsies” of ctDNA are also being used more often [101]. However, without an understanding of intrapatient heterogeneity, we cannot know how useful these biopsies will truly be for guiding treatment. Oncologists now do not know if the biopsy of one site is sufficient; and, if not, how many sites would be needed. Another decision impacted by intrapatient heterogeneity is retreatment. Traditionally, oncologists have thought that once a patient progresses on a treatment, that treatment should not be used again because the cancer cells are now resistant to it. However, if the cancer is made of heterogeneous populations of subclones, sequential cycling between different treatments could be an effective strategy.
Due to the unique evolutionary trajectory of each patient’s cancer, adaptable n-of-1 clinical trials may prove the best model to determine the impact of knowing a patient’s tumor heterogeneity in real time [2, 102, 103]. These trials may employ biopsy of multiple regions to assess spatial heterogeneity and serial liquid biopsies to assess temporal heterogeneity [104, 105]. Unfortunately, lab tests and their interpretation can take a long time and there may be no FDA-approved therapies targeting phenotypes or pathways identified by these tests. However, for those patients with alterations in targetable pathways, identification of intrapatient tumor heterogeneity will promote the use of pathway-targeting drugs to re-sensitize patients to chemotherapy. As targetable mutations are not always clonal, to treat patients effectively, it is likely that therapies must target truncal mutations or pathways shared across subclones [106, 107]. In the absence of these targets, given that each tumor is almost certainly unique, driven by an inimitable combination of genetic alterations spread across a diversity of subclonal populations, it is likely that future clinical regimens will need to account for the distinctive genotypes and phenotypes seen across the genomic profiles of each subclone in a tumor. A full understanding of the causes and the extent of intrapatient heterogeneity may lead to more rational strategies for sequencing of patient biopsies and implementation of a combinatorial treatment approach to target various subclones. Future work will need to assess how to determine which subclones and pathways to target and how to incorporate subclone prevalence into treatment, with targeting of the most dominant subclone(s) a likely starting point. In light of interactions of tumor cells with normal cells in the microenvironment, drugs to target normal cell signaling that promotes cancer cell growth may be a viable option, as seen above with hedgehog signaling in ovarian cancer. Blocking other tumor-stromal and tumor-immune signaling may also prove efficacious.
Conclusion
Tumor heterogeneity has been identified at various -omic levels. Recurrent tumor cell phenotypes emerge despite spatial and temporal diversity at the genomic, transcriptomic, and proteomic levels. Assessment of intrapatient tumor heterogeneity and cellular phenotypes has the potential to redefine cancer care. As next generation sequencing costs decrease, patients will have increased access to the molecular profiling of their primary tumors, and potentially even of their metastatic tumors and/or ctDNA. The interpretation of data accrued from these sources and how it can best shape implementation of therapies must be explored further through clinical trials. Future clinical trials must follow patients over time and incorporate adaptable measures and outcomes unique to each patient’s cancer. Assessment of cellular phenotypes is crucial to targeting cancer cell weaknesses, and development of routine assays that determine these phenotypes is necessary before phenotypic targeting can be fully implemented in the clinic.
Acknowledgments
This work was supported by funding from the National Institutes of Health (U54CA209978). The authors wish to thank Dr. Samuel W. Brady for manuscript editing.
References
- 1.Nowell PC. The Clonal Evolution of Tumor Cell Populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational Implications of Tumor Heterogeneity. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:1258–1266. doi: 10.1158/1078-0432.CCR-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turtoi A, Blomme A, Debois D, Somja J, Delvaux D, Patsos G, Di Valentin E, Peulen O, Mutijima EN, De Pauw E, et al. Organized proteomic heterogeneity in colorectal cancer liver metastases and implications for therapies. Hepatology. 2014;59:924–934. doi: 10.1002/hep.26608. [DOI] [PubMed] [Google Scholar]
- 6.Brocks D, Assenov Y, Minner S, Bogatyrova O, Simon R, Koop C, Oakes C, Zucknick M, Lipka Daniel B, Weischenfeldt J, et al. Intratumor DNA Methylation Heterogeneity Reflects Clonal Evolution in Aggressive Prostate Cancer. Cell Reports. 2014;8:798–806. doi: 10.1016/j.celrep.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 7.Gorges TM, Kuske A, Röck K, Mauermann O, Müller V, Peine S, Verpoort K, Novosadova V, Kubista M, Riethdorf S, et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clinical Chemistry. 2016;62:1504. doi: 10.1373/clinchem.2016.260299. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, Patel J, Dillon R, Vijay P, Brown AL, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nature medicine. 2016;22:792–799. doi: 10.1038/nm.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz RF, Ng CKY, Cooke SL, Newman S, Temple J, Piskorz AM, Gale D, Sayal K, Murtaza M, Baldwin PJ, et al. Spatial and Temporal Heterogeneity in High-Grade Serous Ovarian Cancer: A Phylogenetic Analysis. PLOS Medicine. 2015;12:e1001789. doi: 10.1371/journal.pmed.1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao J-J, Lin D-C, Dinh HQ, Mayakonda A, Jiang Y-Y, Chang C, Jiang Y, Lu C-C, Shi Z-Z, Xu X, et al. Spatial intratumor heterogeneity of genetic, epigenetic alterations and temporal clonal evolution in esophageal squamous cell carcinoma. Nature genetics. 2016;48:1500–1507. doi: 10.1038/ng.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanton C. Intratumour Heterogeneity: Evolution through Space and Time. Cancer research. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sveen A, Løes IM, Alagaratnam S, Nilsen G, Høland M, Lingjærde OC, Sorbye H, Berg KCG, Horn A, Angelsen J-H, et al. Intra-patient Inter-metastatic Genetic Heterogeneity in Colorectal Cancer as a Key Determinant of Survival after Curative Liver Resection. PLOS Genetics. 2016;12:e1006225. doi: 10.1371/journal.pgen.1006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tellez-Gabriel M, Ory B, Lamoureux F, Heymann M-F, Heymann D. Tumour Heterogeneity: The Key Advantages of Single-Cell Analysis. International Journal of Molecular Sciences. 2016;17:2142. doi: 10.3390/ijms17122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winge Ö. Zytologische Untersuchungen über die Natur maligner Tumoren. Zeitschrift für Zellforschung und Mikroskopische Anatomie. 1930;10:683–735. [Google Scholar]
- 15.Levan A. CHROMOSOMES IN CANCER TISSUE. Annals of the New York Academy of Sciences. 1956;63:774–792. doi: 10.1111/j.1749-6632.1956.tb50892.x. [DOI] [PubMed] [Google Scholar]
- 16.Schilsky RL. Clinical Implications of Tumor Heterogeneity. In: Neth R, Gallo RC, Greaves MF, Kabisch H, editors. Modern Trends in Human Leukemia VII: New Results in Clinical and Biological Research Including Pediatric Oncology. Springer Berlin Heidelberg; Berlin, Heidelberg: 1987. pp. 278–282. [Google Scholar]
- 17.Trainer AH, Lewis CR, Tucker K, Meiser B, Friedlander M, Ward RL. The role of BRCA mutation testing in determining breast cancer therapy. Nat Rev Clin Oncol. 2010;7:708–717. doi: 10.1038/nrclinonc.2010.175. [DOI] [PubMed] [Google Scholar]
- 18.Cagle PT, Allen TC. Lung Cancer Genotype-Based Therapy and Predictive Biomarkers: Present and Future. Archives of Pathology & Laboratory Medicine. 2012;136:1482–1491. doi: 10.5858/arpa.2012-0508-RA. [DOI] [PubMed] [Google Scholar]
- 19.Jekunen A. Clinicians’ Expectations for Gene-Driven Cancer Therapy. Clinical Medicine Insights Oncology. 2014;8:159–164. doi: 10.4137/CMO.S20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, Follows GA, Green AR, Futreal PA, Stratton MR. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 22.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 23.Hardiman KM, Ulintz PJ, Kuick RD, Hovelson DH, Gates CM, Bhasi A, Rodrigues Grant A, Liu J, Cani AK, Greenson JK, et al. Intra-tumor genetic heterogeneity in rectal cancer. Lab Invest. 2016;96:4–15. doi: 10.1038/labinvest.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling S, Hu Z, Yang Z, Yang F, Li Y, Lin P, Chen K, Dong L, Cao L, Tao Y, et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E6496–E6505. doi: 10.1073/pnas.1519556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrissy AS, Cavalli FMG, Remke M, Ramaswamy V, Shih DJH, Holgado BL, Farooq H, Donovan LK, Garzia L, Agnihotri S, et al. Spatial heterogeneity in medulloblastoma. Nat Genet. 2017;49:780–788. doi: 10.1038/ng.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werner B, Traulsen A, Sottoriva A, Dingli D. Detecting truly clonal alterations from multi-region profiling of tumours. Scientific Reports. 2017;7:44991. doi: 10.1038/srep44991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin D-C, Mayakonda A, Dinh HQ, Huang P, Lin L, Liu X, Ding L-w, Wang J, Berman BP, Song E-W, et al. Genomic and Epigenomic Heterogeneity of Hepatocellular Carcinoma. Cancer Research. 2017;77:2255–2265. doi: 10.1158/0008-5472.CAN-16-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aihara K, Mukasa A, Nagae G, Nomura M, Yamamoto S, Ueda H, Tatsuno K, Shibahara J, Takahashi M, Momose T, et al. Genetic and epigenetic stability of oligodendrogliomas at recurrence. Acta Neuropathologica Communications. 2017;5:18. doi: 10.1186/s40478-017-0422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savas P, Teo ZL, Lefevre C, Flensburg C, Caramia F, Alsop K, Mansour M, Francis PA, Thorne HA, Silva MJ, et al. The Subclonal Architecture of Metastatic Breast Cancer: Results from a Prospective Community-Based Rapid Autopsy Program “CASCADE”. PLoS Medicine. 2016;13:e1002204. doi: 10.1371/journal.pmed.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng CKY, Bidard F-C, Piscuoglio S, Geyer FC, Lim RS, de Bruijn I, Shen R, Pareja F, Berman SH, Wang L, et al. Genetic Heterogeneity in Therapy-Naïve Synchronous Primary Breast Cancers and Their Metastases. Clinical Cancer Research. 2017 doi: 10.1158/1078-0432.ccr-16-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castellarin M, Milne K, Zeng T, Tse K, Mayo M, Zhao Y, Webb JR, Watson PH, Nelson BH, Holt RA. Clonal evolution of high-grade serous ovarian carcinoma from primary to recurrent disease. The Journal of Pathology. 2013;229:515–524. doi: 10.1002/path.4105. [DOI] [PubMed] [Google Scholar]
- 32.Patch A-M, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, Nones K, Cowin P, Alsop K, Bailey PJ, et al. Whole–genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 33.De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CKY, Nuciforo P, Bidard FC, Aura C, Saura C, Peg V, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Annals of Oncology. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, Riisnaes R, Miranda S, Figueiredo I, NavaRodrigues D, et al. Serial Next Generation Sequencing of Circulating Cell Free DNA Evaluating Tumour Clone Response To Molecularly Targeted Drug Administration. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:4586–4596. doi: 10.1158/1078-0432.CCR-15-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murtaza M, Dawson S-J, Pogrebniak K, Rueda OM, Provenzano E, Grant J, Chin S-F, Tsui DWY, Marass F, Gale D, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. 2015;6:8760. doi: 10.1038/ncomms9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA, Veeriah S, Rosenthal R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulfoni M, Turetta M, Del Ben F, Di Loreto C, Beltrami AP, Cesselli D. Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack. International Journal of Molecular Sciences. 2016;17:1775. doi: 10.3390/ijms17101775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, Kurtz DM, Stehr H, Scherer F, Karlovich CA, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han X, Wang J, Sun Y. Circulating Tumor DNA as Biomarkers for Cancer Detection. Genomics, Proteomics & Bioinformatics. 2017;15:59–72. doi: 10.1016/j.gpb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Research. 2015;25:1499–1507. doi: 10.1101/gr.191098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross EM, Markowetz F. OncoNEM: inferring tumor evolution from single-cell sequencing data. Genome Biology. 2016;17:69. doi: 10.1186/s13059-016-0929-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J. Tumour evolution inferred by single-cell sequencing. Nature. 2011:472. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014:512. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Wu S, Yang Z, Zhang X, Zheng Q, Lin L, Niu Z, Li R, Cai Z, Li L. Single-cell exome sequencing identifies mutations in KCP, LOC440040, and LOC440563 as drivers in renal cell carcinoma stem cells. Cell Res. 2017;27:590–593. doi: 10.1038/cr.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in Acute Myeloid Leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nature reviews Genetics. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012:148. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012:148. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 49.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotech. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anchang B, Hart TDP, Bendall SC, Qiu P, Bjornson Z, Linderman M, Nolan GP, Plevritis SK. Visualization and cellular hierarchy inference of single-cell data using SPADE. Nat Protocols. 2016;11:1264–1279. doi: 10.1038/nprot.2016.066. [DOI] [PubMed] [Google Scholar]
- 51.Saadatpour A, Lai S, Guo G, Yuan G-C. Single-cell analysis in cancer genomics. Trends in genetics : TIG. 2015;31:576–586. doi: 10.1016/j.tig.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Zhang M, Hou Y, Xu L, Li W, Zou Z, Liu C, Xu A, Wu S. Single-cell analyses of transcriptional heterogeneity in squamous cell carcinoma of urinary bladder. Oncotarget. 2016;7:66069–66076. doi: 10.18632/oncotarget.11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chung W, Eum HH, Lee H-O, Lee K-M, Lee H-B, Kim K-T, Ryu HS, Kim S, Lee JE, Park YH, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nature Communications. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed N, Greening D, Samardzija C, Escalona RM, Chen M, Findlay JK, Kannourakis G. Unique proteome signature of post-chemotherapy ovarian cancer ascites-derived tumor cells. 2016;6:30061. doi: 10.1038/srep30061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim M-S, Zhong Y, Yachida S, Rajeshkumar NV, Abel ML, Marimuthu A, Mudgal K, Hruban RH, Poling JS, Tyner JW, et al. Heterogeneity of Pancreatic Cancer Metastases in a Single Patient Revealed by Quantitative Proteomics. Molecular & Cellular Proteomics : MCP. 2014;13:2803–2811. doi: 10.1074/mcp.M114.038547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay and Drug Development Technologies. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Meth. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 60.Sood A, Miller AM, Brogi E, Sui Y, Armenia J, McDonough E, Santamaria-Pang A, Carlin S, Stamper A, Campos C, et al. Multiplexed immunofluorescence delineates proteomic cancer cell states associated with metabolism. JCI Insight. 2016;1:e87030. doi: 10.1172/jci.insight.87030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta Piyush B, Fillmore Christine M, Jiang G, Shapira Sagi D, Tao K, Kuperwasser C, Lander Eric S. Stochastic State Transitions Give Rise to Phenotypic Equilibrium in Populations of Cancer Cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 62.Nichol D, Robertson-Tessi M, Jeavons P, Anderson ARA. Stochasticity in the Genotype-Phenotype Map: Implications for the Robustness and Persistence of Bet-Hedging. Genetics. 2016;204:1523–1539. doi: 10.1534/genetics.116.193474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 64.Chen H, He X. The Convergent Cancer Evolution toward a Single Cellular Destination. Molecular Biology and Evolution. 2016;33:4–12. doi: 10.1093/molbev/msv212. [DOI] [PubMed] [Google Scholar]
- 65.Chen H, Lin F, Xing K, He X. The reverse evolution from multicellularity to unicellularity during carcinogenesis. 2015;6:6367. doi: 10.1038/ncomms7367. [DOI] [PubMed] [Google Scholar]
- 66.Cunningham JJ, Brown JS, Vincent TL, Gatenby RA. Divergent and convergent evolution in metastases suggest treatment strategies based on specific metastatic sites. Evolution, Medicine, and Public Health. 2015;2015:76–87. doi: 10.1093/emph/eov006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. New England Journal of Medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voss MH, Hakimi AA, Pham CG, Brannon AR, Chen Y-B, Cunha LF, Akin O, Liu H, Takeda S, Scott SN, et al. Tumor Genetic Analyses of Patients with Metastatic Renal Cell Carcinoma and Extended Benefit from mTOR Inhibitor Therapy. Clinical Cancer Research. 2014;20:1955–1964. doi: 10.1158/1078-0432.CCR-13-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei EY, Hsieh JJ. A river model to map convergent cancer evolution and guide therapy in RCC. Nat Rev Urol. 2015;12:706–712. doi: 10.1038/nrurol.2015.260. [DOI] [PubMed] [Google Scholar]
- 70.Chen J, Lee H-J, Wu X, Huo L, Kim S-J, Xu L, Wang Y, He J, Bollu LR, Gao G, et al. Gain of glucose-independent growth upon metastasis of breast cancer cells to the brain. Cancer research. 2015;75:554–565. doi: 10.1158/0008-5472.CAN-14-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)K[agr] inhibitor. Nature. 2015;518:240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, Beqiri M, Sproesser K, Brafford PA, Xiao M, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431–435. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang H-eC, Krishnamurthy Radhakrishna V, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stayton CT. Is convergence surprising? An examination of the frequency of convergence in simulated datasets. Journal of Theoretical Biology. 2008;252:1–14. doi: 10.1016/j.jtbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 76.Park ES, Kim SJ, Kim SW, Yoon S-L, Leem S-H, Kim S-B, Kim SM, Park Y-Y, Cheong J-H, Woo HG, et al. Cross-species hybridization of microarrays for studying tumor transcriptome of brain metastasis. Proceedings of the National Academy of Sciences. 2011;108:17456–17461. doi: 10.1073/pnas.1114210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. New England Journal of Medicine. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 78.Zimmer A, Amar-Farkash S, Danon T, Alon U. Dynamic proteomics reveals bimodal protein dynamics of cancer cells in response to HSP90 inhibitor. BMC Systems Biology. 2017;11:33. doi: 10.1186/s12918-017-0410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu P-H, Phillip JM, Khatau SB, Chen W-C, Stirman J, Rosseel S, Tschudi K, Van Patten J, Wong M, Gupta S, et al. Evolution of cellular morpho-phenotypes in cancer metastasis. Scientific Reports. 2015;5:18437. doi: 10.1038/srep18437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fluegen G, Avivar-Valderas A, Wang Y, Padgen MR, Williams JK, Nobre AR, Calvo V, Cheung JF, Bravo-Cordero JJ, Entenberg D, et al. Phenotypic heterogeneity of disseminated tumour cells is preset by primary tumour hypoxic microenvironments. Nat Cell Biol. 2017;19:120–132. doi: 10.1038/ncb3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McLean K, Gong Y, Choi Y, Deng N, Yang K, Bai S, Cabrera L, Keller E, McCauley L, Cho KR, et al. Human ovarian carcinoma-associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered BMP production. J Clin Invest. 2011;121:3206–3219. doi: 10.1172/JCI45273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coffman LG, Choi Y-J, McLean K, Allen BL, di Magliano MP, Buckanovich RJ. Human carcinoma-associated mesenchymal stem cells promote ovarian cancer chemotherapy resistance via a BMP4/HH signaling loop. Oncotarget. 2016:3. doi: 10.18632/oncotarget.6870. Advance Online Publications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen W-J, Ho C-C, Chang Y-L, Chen H-Y, Lin C-A, Ling T-Y, Yu S-L, Yuan S-S, Louisa Chen Y-J, Lin C-Y, et al. Cancer-associated fibroblasts regulate the plasticity of lung cancer stemness via paracrine signalling. 2014;5:3472. doi: 10.1038/ncomms4472. [DOI] [PubMed] [Google Scholar]
- 86.Vermeulen L, De Sousa E, Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 87.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. The Journal of Clinical Investigation. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F, Korkaya H, Heath A, Dutcher J, Kleer CG, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71:614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thomas DA, Massagué J. TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 8:369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 91.Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, Lichtor T, Decker WK, Whelan RL, Kumara HMCS, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Seminars in Cancer Biology. 2015;35:S185–S198. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 92.de Charette M, Marabelle A, Houot R. Turning tumour cells into antigen presenting cells: The next step to improve cancer immunotherapy? European Journal of Cancer. 2016;68:134–147. doi: 10.1016/j.ejca.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 93.Haworth KB, Leddon JL, Chen C-Y, Horwitz EM, Mackall CL, Cripe TP. Going Back to Class I: MHC and Immunotherapies for Childhood Cancer. Pediatric blood & cancer. 2015;62:571–576. doi: 10.1002/pbc.25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Song G, Darr DB, Santos CM, Ross M, Valdivia A, Jordan JL, Midkiff BR, Cohen S, Feinberg NN, Miller CR, et al. Effects of Tumor Microenvironment Heterogeneity on Nanoparticle Disposition and Efficacy in Breast Cancer Tumor Models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:6083–6095. doi: 10.1158/1078-0432.CCR-14-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mumenthaler SM, Foo J, Choi NC, Heise N, Leder K, Agus DB, Pao W, Michor F, Mallick P. The Impact of Microenvironmental Heterogeneity on the Evolution of Drug Resistance in Cancer Cells. Cancer Informatics. 2015;14:19–31. doi: 10.4137/CIN.S19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mroz EA, Tward AM, Hammon RJ, Ren Y, Rocco JW. Intra-tumor Genetic Heterogeneity and Mortality in Head and Neck Cancer: Analysis of Data from The Cancer Genome Atlas. PLOS Medicine. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016 doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dong ZY, Zhai HR, Hou QY, Su J, Liu SY, Yan HH, Li YS, Chen ZY, Zhong WZ, Wu YL. Mixed Responses to Systemic Therapy Revealed Potential Genetic Heterogeneity and Poor Survival in Patients with Non-Small Cell Lung Cancer. The Oncologist. 2017;22:61–69. doi: 10.1634/theoncologist.2016-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee Y, Kim HY, Lee S-H, Lim KY, Lee GK, Yun T, Han J-Y, Kim HT, Lee JS. Clinical Significance of Heterogeneity in Response to Retreatment With Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Patients With Lung Cancer Acquiring Secondary Resistance to the Drug. Clinical Lung Cancer. 2014;15:145–151. doi: 10.1016/j.cllc.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 100.Connolly JLSS, Wang HH, Longtine JA, Dvorak A, Dvorak HF. Role of the Surgical Pathologist in the Diagnosis and Management of the Cancer Patient. In: Kufe DWPR, Weichselbaum RR, et al., editors. Holland-Frei Cancer Medicine. BC Decker; Hamilton (ON): 2003. [Google Scholar]
- 101.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 102.Lillie EO, Patay B, Diamant J, Issell B, Topol EJ, Schork NJ. The n-of-1 clinical trial: the ultimate strategy for individualizing medicine? Personalized medicine. 2011;8:161–173. doi: 10.2217/pme.11.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Catenacci DVT. Next-generation clinical trials: Novel strategies to address the challenge of tumor molecular heterogeneity. Molecular Oncology. 2015;9:967–996. doi: 10.1016/j.molonc.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Joung J-G, Bae JS, Kim SC, Jung H, Park W-Y, Song S-Y. Genomic Characterization and Comparison of Multi-Regional and Pooled Tumor Biopsy Specimens. PLOS ONE. 2016;11:e0152574. doi: 10.1371/journal.pone.0152574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lennon NJ, Adalsteinsson VA, Gabriel SB. Technological considerations for genome-guided diagnosis and management of cancer. Genome Medicine. 2016;8:112. doi: 10.1186/s13073-016-0370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lohr Jens G, Stojanov P, Carter Scott L, Cruz-Gordillo P, Lawrence Michael S, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, et al. Widespread Genetic Heterogeneity in Multiple Myeloma: Implications for Targeted Therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Science translational medicine. 2015;7:283ra254–283ra254. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]