Abstract
Consanguineous Pakistani pedigrees segregating deafness have contributed decisively to the discovery of 31 of the 68 genes associated with nonsyndromic autosomal recessive hearing loss (HL) worldwide. In this study, we utilized genome-wide genotyping, Sanger and exome sequencing to identify 163 DNA variants in 41 previously reported HL genes segregating in 321 Pakistani families. Of these, 71 (43.6%) variants identified in 29 genes are novel. As expected from genetic studies of disorders segregating in consanguineous families, the majority of affected individuals (94.4%) are homozygous for HL-associated variants, with the other variants being compound heterozygotes. The five most common HL genes in the Pakistani population are SLC26A4, MYO7A, GJB2, CIB2 and HGF, respectively. Our study provides a profile of the genetic etiology of HL in Pakistani families, which will allow for the development of more efficient genetic diagnostic tools, aid in accurate genetic counseling and guide application of future gene-based therapies. These findings are also valuable in interpreting pathogenicity of variants that are potentially associated with HL in individuals of all ancestries. The Pakistani population, and its infrastructure for studying human genetics, will continue to be valuable to gene discovery for HL and other inherited disorders.
Keywords: Deafness, genetic spectrum, Pakistan, allelic heterogeneity, autosomal recessive hearing loss, DFNB, pathogenic variant
Introduction
Normal hearing ability requires many specialized cell types. In the inner ear there are intricately-structured mechanosensory hair cells overlaid by an acellular tectorial membrane, the stria vascularis, which generates the potassium-rich endolymph, and several supporting cell types, which in concert are necessary for transduction of sound (Burns, Kelly, Hoa, Morell, & Kelley, 2015). A failure of normal development, loss of any one of several essential physiological or biochemical functions or degeneration of one or more of these cell types in the inner ear can cause hearing loss (HL).
Today, ~466 million people have disabling HL (WHO, March 2018), including 1 in 500 newborns (Morton & Nance, 2006), 34 million children and one-half of all individuals over the age of 75 (Yamasoba et al., 2013). HL can be one feature of a more complex clinical syndrome (Gettelfinger & Dahl, 2018). Isolated hearing loss, referred to as nonsyndromic autosomal recessive hearing loss (NSARHL), is also genetically heterogenous. As of June 2018, 105 NSARHL loci have been mapped and 68 identified (http://hereditaryhearingloss.org). Close to half of these genes (31 out of 68) were identified through studies of large consanguineous families from Pakistan. Subsequently, additional mutations in these same genes were recognized in diverse populations across the world.
The genetic heterogeneity of hearing disorders can present a challenge for identification of causative molecular lesions and expeditious molecular diagnosis which is critical for prognosis and informed genetic counseling (Askew et al., 2015; Shibata et al., 2016). The availability of next generation sequencing (NGS), particularly exome sequencing, has improved the success for the identification of possible pathogenic variants for inherited disorders, including HL (Gao & Dai, 2014; Sloan-Heggen & Smith, 2016; Vona, Nanda, Hofrichter, Shehata-Dieler, & Haaf, 2015). Using a combination of experimental strategies from classical homozygosity mapping (Friedman et al., 1995) to exome sequencing, we identified 163 variants (71 of which are novel) in 41 reported hearing-associated genes co-segregating with HL in 321 Pakistani families. Our present and previous genetic profiling provides an improved understanding of the causes of inherited HL in the Pakistani population while revealing genes with high population-specific prevalence rates.
Material and Methods
Human subjects
This study followed the tenets of the Declaration of Helsinki. Approval for this study was obtained from institutional review boards at the University of Maryland, School of Medicine, Baltimore, Maryland, USA (HP-00059851), the Baylor College of Medicine and Affiliated Hospitals (H-17566), the Combined Neurosciences Blue Panel at the National Institutes of Health (OH-93-N-016), the National Centre of Excellence in Molecular Biology at the University of the Punjab, Quaid-i-Azam University, and Liaquat University of Medical and Health Sciences. Written informed consent was obtained from all participating individuals in this study.
Screening and identification of HL variants
Three new cohorts of Pakistani families segregating HL were examined by NGS. Cohorts 1, 2 and 3 have 261, 40, and 20 families, respectively, for a total of 321 families. For cohort 1, DNA samples from two affected individuals were first screened for variants in HGF (Schultz et al., 2009), CIB2 [c.272T>C; p.(Phe91Ser)] (Riazuddin et al., 2012), and GJB2 (Santos, Wajid, Pham, et al., 2005), using Sanger sequencing. HL-associated variants were found in 94 of these families. DNA samples from the family members of these 94 families were also Sanger-sequenced to test co-segregation of variants with HL. For the remaining 167 families that were negative for variants of GJB2, CIB2 or HGF, DNA samples for 129 families were genotyped using microsatellite or single nucleotide polymorphism (SNP) markers. Genotype data were checked for errors using PedCheck (O’Connell and Weeks, 1998) and MERLIN (Abecasis, et al. 2002). Analysis was performed using HomozygosityMapper (Seelow, Schuelke, Hildebrandt, & Nurnberg, 2009) and Allegro for linkage analysis (Gubjartsson, et al., 2005). Out of 129 families with genotype data, 46 families were mapped to a region containing a known HL gene and the putatively pathogenic variant was identified and co-segregation with HL confirmed using Sanger sequencing.
The remaining 121 families from cohort 1 included those who were not mapped to known HL genes and were negative when followed-up with Sanger sequencing. Forty additional families (cohort 2) did not have genome-scan genotype data. A DNA sample from an affected individual from each of these families (total 161 DNA samples) were exome sequenced either at the University of Maryland School of Medicine (UMSOM), University of Washington Center for Mendelian Genomics (UWCMG), or at the National Institute of Deafness and Other Communication Disorders (NIDCD) Genomic and Computational Biology Core.
Aside from the first two cohorts of 301 families, an additional 20 families (cohort 3) were screened using a gene panel that includes all known nonsyndromic and selected syndromic HL genes (n=101) (Zein et al., 2014) and the segregation of pathogenic variants identified from this panel was verified by Sanger sequencing the DNA of all informative family members.
Variants were considered further if (a) they have allele frequency <1% and (b) Combined Annotation Dependent Depletion (CADD; (Kircher et al., 2014)) scaled score was ≥20 and/or (c) at least two of the following algorithms predict the mutation to be deleterious: PROVEAN (Y. Choi & Chan, 2015), SIFT (Kumar, Henikoff, & Ng, 2009), MutationTaster (Schwarz, Rodelsperger, Schuelke, & Seelow, 2010), MutationAssessor (Reva, Antipin, & Sander, 2007), FATHMM (Shihab et al., 2013; Shihab et al., 2015) and PolyPhen2 (Adzhubei et al., 2010). Splice site and splice region variants were evaluated using splice site-altering scoring algorithms such as dbscSNV (Jian, Boerwinkle, & Liu, 2014) and Human Splice Finder (Desmet et al., 2009). Further details of bioinformatics analyses are described in Supp. Tables S1 and S2. All variants were tested for co-segregation with HL in the corresponding family via Sanger sequencing. All novel variants have been submitted to ClinVar public databasis (https://www.ncbi.nlm.nih.gov/clinvar/).
Results
Genetic spectrum
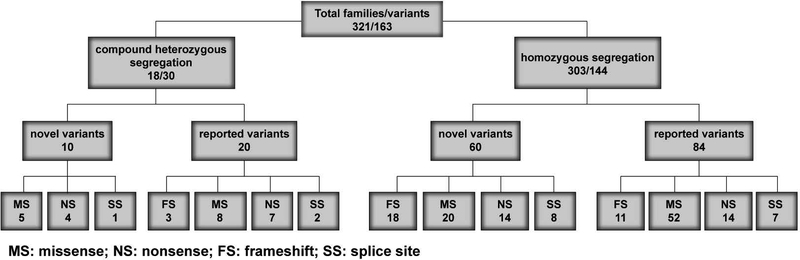
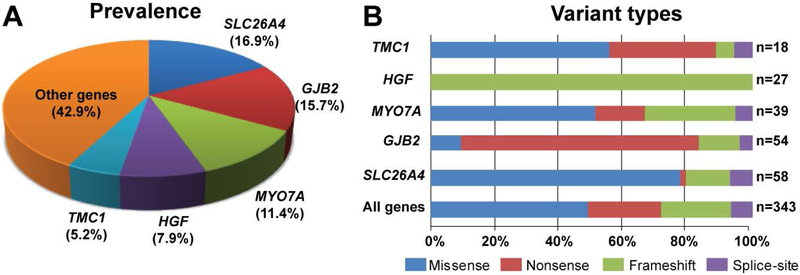
For 41 of the 68 reported NSARHL genes, we identified 163 variants co-segregating with severe-to-profound HL in 321 families (Fig. 1, Table 1). All of these consanguineous families segregate NSARHL associated genes either with homozygous (94.4%; Fig. 2, Table 1) or compound heterozygous (5.6%) variants (Fig. 3, Supp. Table S3). While 92 (56.4%) of the variants have been previously reported to cause HL in humans, we also identified 71 (43.6 %) novel variants associated with HL (Fig. 1). Almost 80% (127 of 163) of the identified variants are present only once in our cohort (Table 1). However, 57.1% of all causative variants identified in this study are found in five genes: SLC26A4, GJB2, MYO7A, HGF, and TMC1 (Fig. 4A).
Figure 1. Overview of genetic studies of 321 Pakistani families segregating recessive hearing loss.
Each box has the number of families over the number of variants found in our study. Of 163 variants, 11 of these variants were identified in multiple families as either homozygous or compound heterozygous. In the bottom two rows, only the number of variants is indicated. MS: missense, NS: nonsense, FS: frameshift, SS: splice site.
Table 1.
Novel and reported variants in this study of genes known to be associated with autosomal recessive hearing loss.
| Family | Gene | Variant | Protein change | Reference | Allele frequency | ||
|---|---|---|---|---|---|---|---|
| Pakistan | 1000G | ExAC | |||||
| PKDF1551 | ADGRV1 | c.1055C>T | p.(Pro352Leu) | This study | - | - | 2.5E-05 |
| DEM4304, DEM4384, DEM4558, DEM4150, DEM4576, DEM4592, DEM4593, PKDF970b, PKDF1673, PKSR20 | BSND | c.35T>C | p.(Ile12Thr) | Riazuddin et al., 2009 | - | - | 2E-05 |
| DEM4545 | CABP2 | c.637+1G>T | Splice error | Schrauwen et al., 2012 | - | 0.0008 | 0.001 |
| PKDF1767 | CDH23† | c.2866G>A | p.(Glu956Lys) | Miyagawa, Naito, Nishio, Kamatani, & Usami, 2013 | - | - | - |
| PKDF055 | CDH23† | c.2968G>A | p.(Asp990Asn) | Bork et al., 2001 | - | - | - |
| DEM4556 | CDH23† | c.3481C>T | p.(Arg1161*) | Roux et al., 2006 | - | - | - |
| DEM4504 | CDH23† | c.3880C>T | p.(Gln1294*) | Bork et al., 2001 | - | - | - |
| DEM4324 | CDH23† | c.4892C>T | p.(Ala1631Val) | Shearer et al., 2013 | 0/188 | 0.0010 | 0.0009 |
| DEM4108 | CDH23† | c.5149T>C | p.(Cys1717Arg) | Sloan-Heggen et al., 2015 | 0/188 | - | 8.E-06 |
| PKDF055 | CDH23† | c.6049+1G>A | Splice error | Bork et al., 2001 | - | - | - |
| DEM4363 | CDH23† | c.6050–9G>A | Splice error | von Brederlow et al., 2002 | - | - | - |
| LUAB14 | CDH23† | c.6083A>C | p.(Asp2028Ala) | This study | 0/100 | - | - |
| DEM4112, PKDF1781 | CDH23† | c.6133G>A | p.(Asp2045Asn) | Bork et al., 2001 | - | - | 8E-06 |
| DEM4544 | CDH23† | c.6202A>C | p.(Thr2068Pro) | This study | 0/202 | - | 8E-06 |
| DEM4710 | CDH23† | c.6604G>A | p.(Asp2202Asn) | Bork et al., 2001 | - | - | 8E-06 |
| HL007 | CDH23† | c.7814A>G | p.(Asn2605Ser) | Naz et al., 2017 | 0/125 | - | - |
| DEM4491 | CDH23† | c.7987_7989delTTC | p.(Phe2664del) | Schultz et al., 2011 | 0/188 | - | - |
| DEM4305 | CDH23† | c.8351_8352insCGAT | p.(Leu2785Aspfs*43) | This study | 0/284 | - | - |
| DEM4042, DEM4053, DEM4630, DEM4722, DEM4749, DEM4785, DEM4789, DEM4791, DEM4793, DEM4868, DEM4870, PKDF1093, PKDF1285, PKDF520, PKDF781 | CIB2† | c.272T>C | p.(Phe91Ser) | Riazuddin et al., 2012 | - | - | 5E-05 |
| PKZA13 | CIB2† | c.297C>G | p.(Cys99Trp) | Riazuddin et al., 2012 | - | - | - |
| DEM4312 | CLCNKA | c.1985G>T | p.(Gly662Val) | This study | 0/198 | - | - |
| DEM4101, DEM4210, PKDF958, PKDF1660 | CLDN14† | c.254T>A | p.(Val85Asp) | Lee, Ansar, et al., 2012 | - | - | 8E-06 |
| DEM4005 | EDNRB | c.553G>A | p.(Val185Met) | Puffenberger et al., 1994 | - | - | 0.0001 |
| DEM4134 | EPS8 | c.205–8A>G | Splice error | This study | - | - | - |
| PKDF1550 | ESPN† | c.2496_2496delC | p.(Tyr832*) | This study | - | - | 8.4E-06 |
| DEM4704 | ESRRB† | c.536G>A | p.(Arg179His) | Miyagawa et al., 2013 | - | - | 2E-05 |
| PKDF969, PKDF1442, PKDF1573 | GIPC3† | c.662C>T | p.(Thr221Ile) | Rehman et al., 2011 | - | - | 4E-05 |
| DEM4551B, DEM4582 | GJB2 | c.−23+1G>A | Splice error | Denoyelle et al., 1997 | - | - | - |
| DEM4347, DEM4648, DEM4659, DEM4352A, DEM4871, PKSR8a, PKDF1621 | GJB2 | c.35delG | p.(Gly12Valfs*2) | Zelante et al., 1997 | - | 0.002 | 0.006 |
| DEM4307, DEM4339, DEM4346, DEM4351, DEM4409, DEM4550, DEM4573, DEM4582, DEM4583, DEM4656, DEM4717, DEM4438A, DEM4866, DEM4981, PKDF021, PKDF936, PKDF939, PKDF1368, PKDF1553, PKDF1655, PKDF1760 | GJB2 | c.71G>A | p.(Trp24*) | Kelsell et al., 1997; Santos, Wajid, Pham, et al., 2005 | - | 0.0004 | 0.0006 |
| PKDF1666 | GJB2 | c.223C>T | p.(Arg75Trp) | Richard et al., 1998 | - | - | - |
| DEM4528, DEM4537 | GJB2 | c.224G>A | p.(Arg75Gln) | Uyguner et al., 2002 | - | - | - |
| DEM4015A, DEM4143, DEM4255, DEM4262, DEM4479, DEM4660, DEM4670, DEM4696, DEM4737, DEM4769, DEM4770, DEM4972, PKDF936, PKDF1588, PKDF1633, PKDF1665, PKDF1675, PKDF1790 | GJB2 | c.231G>A | p.(Trp77*) | Santos, Wajid, Pham, et al., 2005 | - | - | 0.0001 |
| DEM4614 | GJB2 | c.355G>T | p.(Glu119*) | This study | 0/454 | - | 7E-05 |
| PKDF1694, PKDF1764 | GJB2 | c.457G>A | p.(Val153Ile) | Marlin et al., 2001 | - | - | - |
| DEM4431, DEM4653 | GPSM2 | c.138C>A | p.(Phe46Leu) | This study | 0/328 | - | 8E-06 |
| PKDF1679 | GRXCR1† | c.229C>T | p.(Gln77*) | Schraders et al., 2010 | - | - | - |
| DEM4153 | GRXCR1† | c.655G>A | p.(Glu219Lys) | This study | 0/114 | - | - |
| DEM4494, DEM4521, DEM4522, DEM4562, DEM4564, DEM4586, DEM4622, DEM4628, DEM4642, DEM4665, DEM4701, DEM4865, DEM4861, DEM4880, DEM4925, DEM4943, PKDF970a, PKDF1017, PKDF1331, PKDF1579, PKDF1604, PKDF1637, PKDF1668, PKDF1754, PKDF1787, PKDF1791 | HGF† | c.482 +1986_88delTGA | Schultz et al., 2009 | - | - | - | |
| PKDF1775 | HGF† | c.482 +1991_2000delGATGATGAAA | Schultz et al., 2009 | - | - | - | |
| PKDF1651 | ILDR1 | c.3G>A | p.(Met1Ile) | Borck et al., 2011 | - | - | - |
| DEM4175 | ILDR1 | c.900delG | p.(Thr301Profs*20) | This study | 0/188 | - | - |
| PKDF1455 | ILDR1 | c.1005_1005delG | p.(Glu335Aspfs*30)) | This study | - | - | - |
| DEM4353 | ILDR1 | c.1029_1030delTT | p.(Trp344Glyfs*17) | This study | 0/282 | - | - |
| PKDF1428 | ILDR1 | c.1384C>T | p.(Arg462*) | This study | - | - | 6E-05 |
| PKDF1311, PKDF1549 | LHFPL5† | c.250delC | p.(Leu84*) | Shabbir et al., 2006 | - | - | - |
| PKDF1178 | LHFPL5† | c.452 G>T | p.(Gly151Val) | This study | - | - | 8.00E-06 |
| PKDF1662 | MARVELD2† | c.1138C>T | p.(Gln380*) | Sloan-Heggen et al., 2015 | - | - | - |
| DEM4204 | MARVELD2† | c.1223_1224insA | p.(Glu408Argfs*4) | This study | 0/280 | - | - |
| DEM4393, DEM4533, PKDF1639, PKDF1641 | MARVELD2† | c.1295+2T>C | Splice error | Riazuddin et al., 2006 | - | - | 4E-05 |
| PKDF1111, PKDF1489, PKDF1606, PKDF1610, PKDF1727 | MSRB3† | c.265T>G | p.(Cys89Gly) | Ahmed et al., 2011 | - | - | 2E-05 |
| PKDF1789 | MSRB3† | c.412–1G>A | Splice error | This study | - | - | 8.2E-06 |
| PKDF019A | MYO7A | c.20G>T | p.(Gly7Val) | This study | 0/170 | - | 1E-05 |
| PKDF173 | MYO7A | c.93C>A | p.(Cys31*) | Weston et al., 1996 | - | 0.017 | 5E-05 |
| DEM4493 | MYO7A | c.247C>A | p.(Arg83Ser) | This study | 0/112 | - | - |
| PKDF312 | MYO7A | c.397dupC | p.(His133Profs*7) | Riazuddin et al., 2008 | - | - | 0.0006 |
| PKDF1465, PKDF1490 | MYO7A | c.496delG | p.(Glu166Argfs*5) | Riazuddin et al., 2008 | - | - | - |
| DEM4141, DEM4299, DEM4365 | MYO7A | c.722G>A | p.(Arg241His) | Cremers et al., 2007 | - | 0.0002 | 2E-05 |
| PKDF370 | MYO7A | c.977T>A | p.(Leu326Gln) | Riazuddin et al., 2008 | - | - | - |
| PKDF537, PKDF1786 | MYO7A | c.1183C>T | p.(Arg395Cys) | Shahzad et al., 2013 | 0/176 | - | - |
| DEM4190, DEM4652 | MYO7A | c.1258A>T | p.(Lys420*) | Le Quesne Stabej et al., 2012 | - | - | 4E-05 |
| DEM4331 | MYO7A | c.1591C>T | p.(Gln531*) | Riazuddin et al., 2008 | - | - | 8E-06 |
| PKDF019A | MYO7A | c.1849T>C | p.(Ser617Pro) | Carss et al., 2017 | 0/160 | - | 5E-05 |
| PKDF721 | MYO7A | c.2339delG | p.(Gly780Valfs*10) | This study | 0/154 | - | - |
| PKDF312 | MYO7A | c.3136dupC | p.(Leu1046Profs*9) | Riazuddin et al., 2008 | - | - | - |
| DEM4561 | MYO7A | c.3364C>A | p.(Leu1122Ile) | This study | - | - | - |
| DEM4281, DEM4692, PKDF628 | MYO7A | c.3502C>T | p.(Arg1168Trp) | Le Guedard-Mereuze et al., 2010 | - | 0.0002 | 0.0001 |
| PKDF1484 | MYO7A | c.3508G>A | p.(Glu1170Lys) | Riazuddin et al., 2008 | - | - | - |
| DEM4052A | MYO7A | c.3590T>C | p.(Leu1197Pro) | This study | 0/282 | - | - |
| PKDF173 | MYO7A | c.3728C>G | p.(Pro1243Arg) | This study | 0/164 | - | - |
| DEM4652 | MYO7A | c.4505A>G | p.(Asp1502Gly) | This study | 0/454 | - | 2E-05 |
| DEM4182, DEM4190, DEM4281, DEM4513, DEM4692, DEM4694 | MYO7A | c.4838delA | p.(Asp1613Valfs*32) | Riazuddin et al., 2008 | - | - | - |
| DEM4433 | MYO7A | c.5326+3A>G | Splice error | This study | 0/118 | - | - |
| DEM4561 | MYO7A | c.5345G>C | p.(Gly1782Ala) | This study | - | - | 4E-05 |
| DEM4206 | MYO7A | c.5434G>A | p.(Glu1812Lys) | Roux et al., 2006 | - | - | - |
| DEM4327 | MYO7A | c.5522C>G | p.(Thr1841Arg) | This study | 0/178 | - | - |
| DEM4725 | MYO7A | c.5660C>T | p.(Pro1887Leu) | Ben-Salem et al., 2014; Bharadwaj, Kasztejna, Huq, Berson, & Dryja, 2000 | - | - | 2E-05 |
| PKDF974 | MYO7A | c.5856+5G>C | Splice error | This study | 0/148 | - | - |
| PKDF1382 | MYO15A | c.2456C>A | p.(Ser819*) | Naz et al., 2017 | - | - | - |
| PKDF1657 | MYO15A | c.3843C>T | p.(Arg1169*) | This study | - | - | 8E-06 |
| PKDF1693 | MYO15A | c.4528C>T | p.(Gln1510*) | Rehman, et al., 2016 | - | - | 8E-06 |
| PKDF1693 | MYO15A | c.4570C>T | p.(Gln1524*) | This study | - | - | - |
| PKDF1780 | MYO15A | c.3505C>T | p.(Gln1669*) | This study | - | - | - |
| PKDF016 | MYO15A | c.8224+3A>G | Splice error | Rehman et al., 2016 | - | - | - |
| DEM4725 | OTOA | c.1148C>T | p.(Ala383Val) | EVS variant | - | - | 8E-04 |
| PKDF889 | OTOA | c.1352G>A | p.(Gly451Asp) | Lee et al., 2013 | - | - | 8E-06 |
| DEM4362, DEM4366 | OTOF | c.1550T>C | p.(Leu517Pro) | This study | 0/188 | - | - |
| PKDF1723 | OTOF | c.1904T>A | p.(Val635Asp) | This study | - | - | - |
| PKDF1535 | OTOF | c.2122C>T | p.(Arg708*) | Choi et al., 2009 | - | - | - |
| DEM4078 | OTOF | c.2508C>A | p.(Tyr836*) | Choi et al., 2009 | - | - | 0.0006 |
| DEM4512 | OTOF | c.2896G>A | p.(Glu966Lys) | Choi et al., 2009 | - | - | 8E-06 |
| DEM4548 | OTOF | c.2965_2967delTTC | p.(Phe989del) | Naz et al., 2017 | 0/188 | - | - |
| DEM4756 | OTOF | c.3376dupA | p.(Ile1126Asnfs*5 | This study | - | - | - |
| PKDF1336 | OTOF | c.3514C>T | p.(Arg1172Trp) | This study | - | 0.0002 | 2.7E-05 |
| PKDF1450 | OTOF | c.4799+1G>A | Splice error | This study | - | - | 8.24e-06 |
| PKDF1066a | OTOF | c.5714G>T | p.(Gly1905Val) | This study | 0/168 | - | |
| DEM4372B | OTOG | c.7235delG | p.(Arg2412Hisfs*77) | This study | 1/326 | - | 0.0009 |
| PKDF627 | PCDH15† | deletion of exons 14 and 15 | This study | 0/100 | - | - | |
| PKDF1440 | PCDH15† | c.400C>G | p.(Arg134Gly) | Ahmed et al., 2003 | - | - | - |
| PKDF1499 | PCDH15† | c.788C>A | p.(Pro263Gln) | This study | - | - | - |
| DEM4711 | PCDH15† | c.1737C>A | p.(Tyr584*) | Jaijo et al., 2006 | - | - | - |
| DEM4040 | PJVK | c.158C>G | p.(Ser53*) | This study | 0/284 | - | - |
| DEM4529 | PJVK | c.162_172del | p.(Pro55Glufs*23) | This study | 0/454 | - | - |
| DEM4752 | PJVK | c.406C>T | p.(Arg136*) | This study | - | - | - |
| PKDF880 | PJVK | c.908_910delACA | p.(Asn303del) | This study | 0/168 | - | - |
| PKDF908 | POU3F4 | c.478C>T | p.(Gln160*) | This study | 0/71 | - | - |
| PKDF1595 | POU4F3 | c.374C>T | p.(Pro125Leu) | This study | - | - | - |
| DEM4437 | PTPRQ | c.5158_5159delAT | p.(Ile1720Glnfs*7) | This study | 0/180 | - | - |
| DEM4123 | PTPRQ | c.6739–1G>A | Splice error | This study | 0/172 | - | - |
| DEM4465 | SLC26A4 | c.42delC | p.(Glu15Serfs*51) | This study | 0/454 | - | - |
| DEM4149B, HL005 | SLC26A4 | c.71G>T | p.(Arg24Leu) | Anwar et al., 2009 | - | - | 0.0003 |
| DEM4549 | SLC26A4 | c.154A>T | p.(Lys52*) | This study | 0/454 | - | - |
| DEM4511 | SLC26A4 | c.317C>T | p.(Ala106Val) | This study | 0/454 | - | - |
| DEM4014A | SLC26A4 | c.413T>A | p.(Val138Asp) | Chen, Wang, Sun, & Jiang, 2012 | |||
| DEM4436 | SLC26A4 | c.416G>T | p.(Gly139Val) | Anwar et al., 2009 | - | - | 4E-05 |
| DEM4092, DEM4149B, DEM4152, DEM4220, DEM4241, DEM4271, DEM4290, DEM4292, DEM4293, DEM4295, DEM4296, DEM4429, DEM4482, DEM4483, DEM4523, DEM4575, DEM4726, DEM4766, DEM4192, DEM4291, DEM4481, DEM4982, PKDF1161, PKDF1664 | SLC26A4 | c.716T>A | p.(Val239Asp) | Park et al., 2003 | - | 0.0002 | 0.0003 |
| DEM4186, DEM4454, DEM4455 | SLC26A4 | c.919–2A>G | Splice error | Coucke et al., 1999 | - | - | 0.0003 |
| DEM4336 | SLC26A4 | c.1001G>T | p.(Gly334Val) | Kahrizi et al., 2009 | - | - | 8E-06 |
| DEM4109 | SLC26A4 | c.1198delT | p.(Cys400Valfs*32) | Everett et al., 1997 | - | - | 2.00E-05 |
| PKDF384b | SLC26A4 | c.1226G>A | p.(Arg409His) | Van Hauwe et al., 1998 | - | - | 0.0001 |
| PKDF128, PKDF384b, PKDF910 | SLC26A4 | c.1226G>C | p.(Arg409Pro) | Park et al., 2003 | - | - | - |
| DEM4283, PKDF384b | SLC26A4 | c.1229C>T | p.(Thr410Met) | Coucke et al., 1999 | - | 0.0002 | 0.0002 |
| DEM4355 | SLC26A4 | c.1238A>G | p.(Gln413Arg) | Ji et al., 2009 | - | - | 8E-06 |
| DEM4200 | SLC26A4 | c.1264–3C>G | Splice error | This study | 0/454 | - | - |
| DEM4004, DEM4094A, DEM4220, DEM4276, DEM4574, DEM4695, DEM4702, DEM4966 | SLC26A4 | c.1337A>G | p.(Gln446Arg) | Reardon, CF, Trembath, Jan, & Phelps, 2000 | - | - | |
| DEM4286, DEM4287, DEM4294, DEM4392, DEM4425, DEM4553 | SLC26A4 | c.2106delG | p.(Lys702Asnfs*19) | Yazdanpanahi et al., 2012 | - | - | 8E-06 |
| PKDF1283 | TECTA | c.840_841insT | p.(Val281Cysfs*11) | This study | 0/162 | - | - |
| HL04 | TECTA | c.1247_1248delGG | p.(Gly416Aspfs*24) | This study | 0/100 | - | - |
| DEM4242A | TECTA | c.1774G>A | p.(Val592Met) | This study | 0/132 | - | - |
| PKDF1452 | TECTA | c.2736C>A | p.(Cys912*) | This study | - | - | - |
| PKDF1452 | TECTA | c.6162+5G>A | Splice error | This study | |||
| DEM4367, PKDF1584 | TMC1† | c.100C>T | p.(Arg34*) | Kurima et al., 2002 | - | - | - |
| PKSR39a | TMC1† | c.596A>T | p.(Asn199Ile) | Imtiaz et al., 2016 | 0/148 | - | 8E-06 |
| DEM4451 | TMC1† | c.945G>A | p.(Trp315*) | This study | 0/454 | - | - |
| DEM4396 | TMC1† | c.1114G>A | p.(Val372Met) | Santos, Wajid, Khan, et al., 2005 | - | - | 7E-05 |
| PKSR13b | TMC1† | c.1143C>G | p.(Tyr381*) | This study | 0/166 | - | - |
| DEM4147 | TMC1† | c.1209G>A | p.(Trp403*) | This study | - | - | - |
| DEM4132 | TMC1† | c.1224+2T>C | Splice error | This study | 0/112 | - | - |
| HL006 | TMC1† | c.1259G>A | p.(Cys420Tyr) | This study | 0/125 | - | - |
| PKDF025 | TMC1† | c.1333C>T | p.(Arg445Cys) | Sirmaci et al., 2009 | - | - | - |
| DEM4325 | TMC1† | c.1363T>C | p.(Tyr454His) | NSHI | - | - | - |
| PKDF1449 | TMC1† | c.1543C>T | p.(Cys515Arg) | Kitajiri et al., 2007 | - | - | - |
| DEM4350 | TMC1† | c.1728C>G | p.(Asn576Lys) | This study | 0/454 | - | - |
| PKDF1617 | TMC1† | c. 1753_1754insA | p.(Asn407Lysfs*2) | This study | - | - | - |
| HL006 | TMC1† | c.1788C>A | p.(Ser596Arg) | Imtiaz et al., 2016 | 0/125 | - | - |
| PKSR13b | TMC1† | c.1810C>T | p.(Arg604*) | Sirmaci et al., 2009 | 8E-06 | - | - |
| DEM4370A, DEM4724 | TMC1† | c.2035G>A | p.(Glu679Lys) | Santos, Wajid, Khan, et al., 2005 | - | - | - |
| DEM4486 | TMIE† | c.241C>T | p.(Arg81Cys) | Santos et al., 2006 | - | - | - |
| DEM4687 | TMIE† | c.250C>T | p.(Arg84Trp) | Naz et al., 2002 | - | - | 2E-05 |
| DEM4020, PKDF1053 | TMPRSS3† | c.271C>T | p.(Arg91*) | Bademci et al., 2016 | - | - | 2E-05 |
| DEM4003 | TMPRSS3† | c.323–6G>A | Splice error | Scott et al., 2001 | - | - | 6E-05 |
| PKSR31 | TMPRSS3† | c.581G>T | p.(Cys194Phe) | Ben-Yosef et al., 2001 | - | - | - |
| DEM4106, DEM4124 | TMPRSS3† | c.767C>T | p.(Ala256Val) | Lee, Khan, et al., 2012 | - | - | - |
| PKDF765 | TMPRSS3† | c.783–1G>T | Splice error | This study | 0/130 | - | |
| DEM4046, DEM4202, DEM4372A, DEM4560, LUAB13, HL003, PKDF1057, PKDF1330, PKDF1634 | TMPRSS3† | c.1219T>C | p.(Cys407Arg) | Ben-Yosef et al., 2001 | - | - | 5E-05 |
| HL007, PKDF1756, PKDF1757, PKDF1758 | TPRN† | c.1056G>A | p.(Trp352*) | Rehman et al., 2010 | - | - | - |
| DEM4492 | TRIOBP† | c.3460_3461delCT | p.(Leu1154Alafs*29) | This study | 0/454 | - | - |
| PKDF1430 | TRIOBP† | c.3634_3646delCTGATCCCCCAAG | p.(Leu1212Cysfs*22) | This study | - | - | - |
| DEM4707 | USH1C | c.238dup | p.(Arg80Profs*69) | Bitner-Glindzicz et al., 2000 | - | - | 0.0003 |
| DEM4335B | USH1C | c.463C>T | p.(Arg155*) | Ebermann et al., 2007 | - | - | 8E-06 |
| DEM4675 | USH1G | c.511G>T | p.(Glu171*) | This study | - | 0.0002 | 2E-05 |
| DEM4268 | USH1G | c.812delC | p.(Pro271Argfs*52) | This study | 0/454 | - | - |
| DEM4689 | USH2A | c.3661C>T | p.(Gln1221*) | This study | 0/134 | - | 8E-06 |
| DEM4171 | WHRN | c.2388_2389delCG | p.(Asn796Lysfs*46) | This study | 0/284 | ||
Novel mutations from this study are in bold font.
indicates genes associated with non-syndromic autosomal recessive hearing loss that were discovered through studies in Pakistani pedigrees.
Pak: Pakistani controls, 1000G: 1000 Genomes Project database, ExAc: Exome Aggregation Consortium database.
indicates that the variant is absent from the respective database.
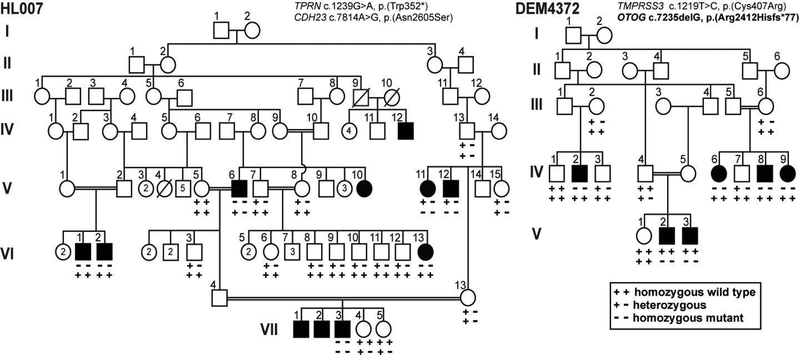
Figure 2. Pakistani families co-segregating autosomal recessive HL due to multiple homozygous variants.
Families HL007 and DEM4372 are examples in which there is intra-familial locus heterogeneity of variants in genes known to cause HL. Circles indicate females, squares indicate males. Black-filled symbols represent affected individuals. Double lines indicate consanguineous marriages. In family HL007, homozygous variants in two different genes TPRN and CDH23 co-segregate with HL. Affected individuals are homozygous for either the TPRN (V:6, VI:1, VI:2, VI:13) or the CDH23 variant (V:11, V:12), except for affected individual VII:3 who is double homozygous. In family DEM4372, variants in two different genes TMPRSS3 and OTOG co-segregate with HL in different branches of this family. Novel mutations from this study are in bold font.
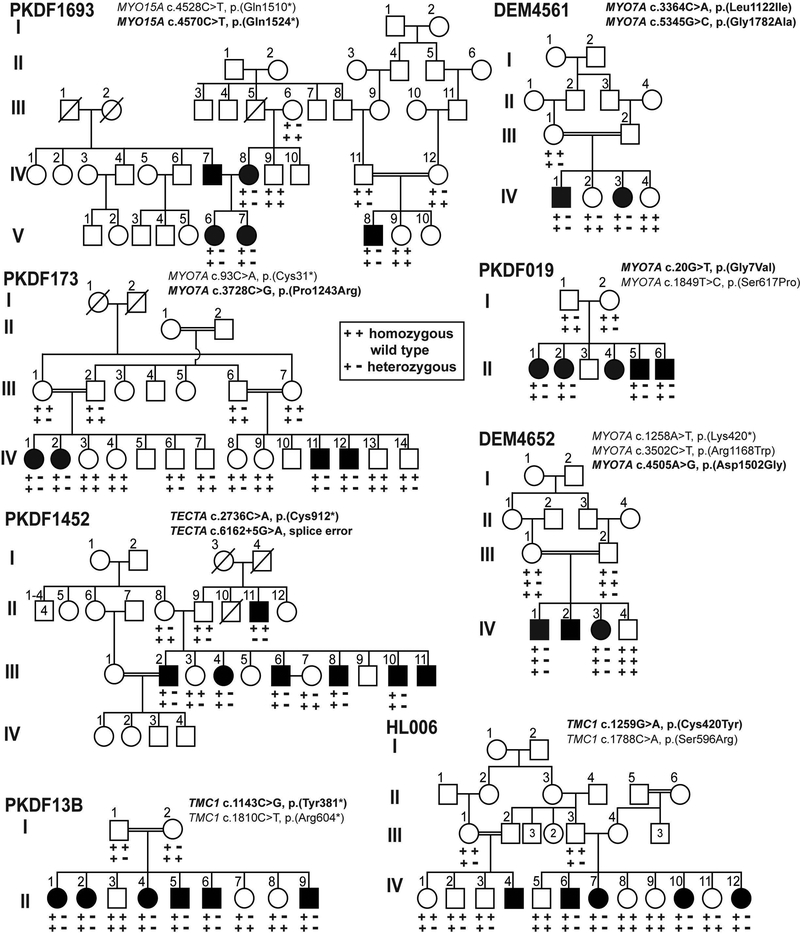
Figure 3. Consanguineous Pakistani families with compound heterozygous variants of reported HL genes.
Eight pedigrees segregating HL due to compound heterozygous variants in MYO7A (PKDF173, DEM4561, DEM4652, PKDF019), TMC1 (HL006, PKDF13B), MYO15A (PKDF1693) and TECTA (PKDF1452) genes. In family DEM4652, the normal hearing father III:2 is heterozygous for two different variants of MYO7A suggesting that the newly identified c.4505A>G variant might be non-pathogenic. Novel mutations from this study are in bold font.
Figure 4. Distribution of variants in the five most prevalent genes associated with nonsyndromic recessively inherited HL in Pakistani families.
(A) Percentage of identified alleles by gene. (B) Percentage of variants by type and occurrence of identified variants. The percentages are based on the total number of variants identified in this study.
Causal variants
Of the causative variants identified in this study 48.7% are missense, 23% are nonsense, 21.6% are small insertion or deletions and 6.7% are predicted to alter splice sites (Fig. 4B). In the five most common HL genes in this study, the distribution of mutation types appears to be gene-specific (Fig. 4B). Pathogenic alleles of HGF are all small deletion variants of an alternative 3’UTR (Schultz et al., 2009), while nonsense variants represent more than 70% of the HL alleles for GJB2 with two nonsense variants [c.71G>A, p.(Trp24*); c.231G>A, p.(Trp77*)] representing 38.9% and 33.3%, respectively, of the GJB2-associated HL mutations (Fig. 4).
A homozygous genomic deletion spanning exons 14 and 15 of PCDH15 was identified in family PKDF627, which causes a shift in the wild type translation reading frame and is predicted to result in a premature truncation of the encoded protein. This deletion has not been previously reported in humans. In mice that are homozygous for the Pcdh15av−5J (Ames Waltzer) allele, skipping of exon 14 due to a splice site variant results in disorganized stereocilia bundles, hearing loss, balance problems (Zheng et al., 2006) and delayed development of retinal sensory function (Haywood-Watson et al., 2006). CNV analyses of other families in our HL cohorts did not reveal any known or potentially HL causal indels. Thus, contrary to previous observations in other cohorts (Shearer et al., 2014), large indels probably rarely cause HL within the Pakistani population.
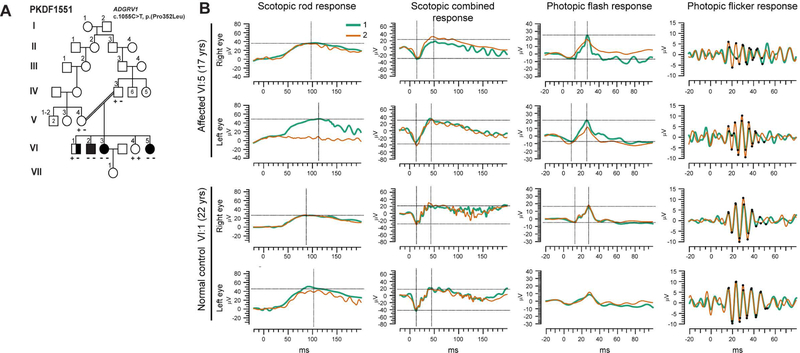
An unreported variant of ADGRV1 (also known as VLGR1, MASS1, KIAA0686 and GPR98 MIM# 602851), c.1055C>T (p.(Pro352Leu)), co-segregates with HL in family PKDF1551 (Fig. 5A). So far, mutations in ADGRV1 have been exclusively associated with Usher syndrome type 2 (USH2C, MIM# 605472) (Besnard et al., 2012; Reddy et al., 2014; Weston, Luijendijk, Humphrey, Moller, & Kimberling, 2004), characterized by congenital mild to severe sensorineural hearing loss, intact vestibular responses and progressive retinitis pigmentosa. In family PKDF1551, affected individuals exhibit mild to severe bilateral sensorineural hearing loss with no vision defects revealed by electroretinogram (Fig. 5B). Usher syndrome type 2 is a progressive disorder and the oldest affected individual in the family was only 17 years old at the time of clinical evaluation. However, USH2C, due to a large deletion in ADGRV1 was previously diagnosed in a 14-year old patient with vision defects (Hilgert et al., 2009).
Figure 5. A new variant of ADGRV1 associated with non-syndromic hearing loss in Pakistani family PKDF1551.
(A) Pedigree of family PKDF1551 segregating HL due to a missense mutation (c.1055C>T, p.(Pro352Leu)) in ADGRV1 (also referred to VLGR1, MIM# 602851, at the USH2C locus). The filled symbols represent affected individuals, and a double horizontal line connecting parents represents a consanguineous marriage. Half-filled symbol represents an individual with potential middle ear infection and/or carrying another variant responsible for his hearing impairment. Exome sequencing was performed on a DNA sample from individual VI:5. (B) Full-field electroretinograms of affected (VI:5, 17 years old) and non-affected individuals (VI:1, 22 years old) revealed no significant difference in a- and b-waves amplitudes among the two subjects, suggesting normal visual function. To show reproducibility, two responses (annotated 1 and 2, green and orange respectively) are superimposed per test and individual. The first crosshair indicates the a-wave while the second crosshair shows the b-wave. No differences in amplitude nor latency for waves a and b were detected in any of the performed tests.
Out of 321 families, a total of 19 families (5.9%) have evidence of intra-familial locus heterogeneity in that some deaf siblings or deaf relatives in other branches of the family are not segregating the same HL-causal variant (data not shown). For instance, two families (HL007 and DEM4372) display locus heterogeneity, within the same or different branches, segregating homozygous variants in two different known NSARHL genes (Fig. 2), which further highlights the unexpected complexity of the genetic diagnosis of HL in consanguineous families. In family HL007, most of the affected individuals are homozygous either for a reported c.1056G>A, p.(Trp352*) variant of TPRN, (Rehman et al., 2010), or another reported c.7814A>G, p.(Asn2605Ser) variant in CDH23 (Naz et al., 2017) a gene known to be involved in either NSARHL or Usher syndrome type 1D (MIM# 601067) (Bork et al., 2001). Affected individual VII:3 is homozygous for both of the likely pathogenic variants of TPRN and CDH23 (Fig. 2). Family DEM4372 is segregating variants for two different known NSARHL genes: OTOG and TMPRSS3 (Fig. 2). Affected individuals V:2 and V:3 are homozygous for a novel OTOG deletion [c.7235delG, p.(Arg2412Hisfs*77)]. Four affected individuals, IV:2, IV:6, IV:8, and IV:9, are homozygous for the recurrent c.1219T>C [p.(Cys407Arg)] variant (Ben-Yosef et al., 2001) in TMPRSS3. These two families (HL007 and DEM4372) exhibit intra-familial locus heterogeneity, which we previously demonstrated in several consanguineous Pakistani families with NSARHL (Rehman, et al., 2015).
Regardless of whether causal variants of one or more HL genes are segregating in a single family, the variants associated with HL are usually but not always homozygous. Affected individuals of family DEM4652 are compound heterozygous for three variants c.1258A>T [p.(Lys420*)], c.3502C>T [p.(Arg1168Trp)] and c.4505A>G [p.(Asp1502Gly)] variants in MYO7A (Fig. 3). While predicted damaging by various algorithms (Mutation Assessor, Mutation Taster, PolyPhen2, SIFT and PROVEAN), we cannot conclude that MYO7A c.4505A>G is pathogenic because normal hearing father III:2 is heterozygous for two different MYO7A variants: the known HL-causing variant c.1258A>T and the newly identified c.4505A>G variant. This family shows that even though multiple rare, predicted-as-damaging variants co-segregate with HL, it is likely that only a subset of these variants are causal, which further highlights the need to evaluate pathogenicity of variants based on multiple lines of evidence even when the variants occur in reported HL genes.
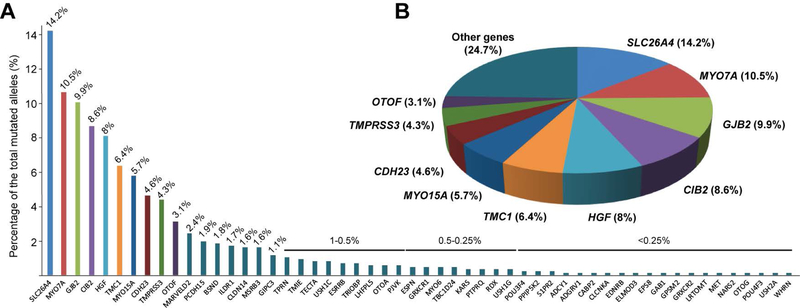
In order to obtain a more precise assessment of the genetic spectrum of HL in this population, we included in our counts not only the data from this specific study but also already published HL variants identified in our previously reported cohorts of Pakistani families (61 publications). Together, our studies have revealed 875 mutant alleles (368 unique variants) in 54 genes (Supp. table S4) and the 10 most prevalent genes are SLC26A4 (14.2%), MYO7A (10.5%), GJB2 (9.9%), CIB2 (8.6%), HGF (8%), TMC1 (6.4%), MYO15A (5.7%), CDH23 (4.6%), TMPRSS3 (4.3%) and OTOF (3.1%). Mutations in these ten genes represent 75.3% of all known causative variants identified in Pakistani families we have studied (Fig. 6, Supp. table S4).
Figure 6. Distribution of variants in the Pakistani population from 62 studies on hearing loss.
(A) Percentage of identified variants by gene. (B) Distribution of the 10 most prevalent genes in our collaborative studies. The percentages are based on the total number of alleles identified in our collaborative studies.
Discussion
Consanguineous families with recessively inherited HL have been a valuable resource for genetic mapping, gene and variant identification and genotype-phenotype correlations. Subsequently, functional studies and knockout animal models that follow gene identification aid in understanding the biology of inner ear development and function. In Pakistan, 45 to 60% of marriages are between relatives (Bittles, 2005; E. M. Scott, 1973), which has increased the coefficient of inbreeding in this population. According to a 2012 World Health Organization report, the prevalence of recessive disorders such as HL is higher in Pakistan (2.4%) compared to a world prevalence of 1.7%. In this study that includes a large cohort of 321 unpublished Pakistani families, we identified 163 unique variants in 41 genes responsible for autosomal recessive hearing loss. As expected, autosomal recessive inheritance due to homozygous variants is demonstrated in the majority of our HL cohort (94.4%), however 5.6% of the variants segregate in a compound heterozygous fashion even in consanguineous families (Fig. 3). Of the 163 identified variants, 71 (43.6%) are novel. In addition, 98.6% of these novel variants are found only once in our cohort. Given the rarity of these private variants, reporting them as damaging based on bioinformatics tools and co-segregation with HL in mostly consanguineous families aids in determining variant pathogenicity (Supp. Table S5) (Richards et al., 2015). The data provided here will also help variant curation in diagnostic laboratories where physician-referred genetic testing is usually performed on singletons.
Despite the heterogeneity of genetic causes of HL, in our study, variants in five known HL genes, SLC26A4 (58), GJB2 (54), MYO7A (39), HGF (27), and TMC1 (18), account for 57.1% (95% CI: 51.7–62.4%) of the identified alleles (Table 1). Variants in SLC26A4 alone account for 16.9% (95% CI: 13.2–21.4%) of variants in our 321 families. SLC26A4 encodes pendrin, a transmembrane anion exchanger for chloride, iodide, bicarbonate and other ions, essential for the development of inner ear and ionic homeostasis (Chattaraj et al., 2017; B. Y. Choi et al., 2011; Everett et al., 2001; Rehman, Friedman, & Griffith, 2017). Consistent with our previous findings (Anwar et al., 2009), c.716T>A [p.(Val239Asp)] is the prevalent variant responsible for SLC26A4-associated HL in Pakistani families. The c.2106delG [p.(Lys702Asnfs*19)] variant, which so far has only been reported in the Iranian population (Yazdanpanahi et al., 2015), is the third most common variant of SLC26A4 in our study, segregating with HL in six Pakistani families from Baluchistan, a province that shares a border with neighboring Iran. Besides these and other common alleles, we also found four novel variants [c.42delC, p.(Glu15Serfs*51); c.154A>T, p.(Lys52*); c.317C>T, p.(Ala106Val); c.1264–3C>G] of SLC26A4. Taken together, our results further confirm the high prevalence of SLC26A4 alleles in Pakistani families with HL (Anwar et al., 2009; Tsukada, Nishio, Hattori, & Usami, 2015).
GJB2 variants are the second most common (15.7%; 95% CI: 12.1–20.1%) cause of HL in our cohort of 321 families. The GJB2 variant spectrum is ancestry-specific and varies significantly around the world. Consistent with previous studies in the Pakistani population, two variants [c.71G>A, p.(Trp24*); c.231G>A, p.(Trp77*)] represent the majority of the GJB2-associated HL (Anjum et al., 2014; Bukhari, Mujtaba, & Naz, 2013; Salman et al., 2015; Santos, Wajid, Pham, et al., 2005; Shafique et al., 2014). We also identified a new nonsense allele [c.355G>T, p.(Glu119*)] segregating with HL in one family (DEM4614), which expands the list of more than 200 variants in the single protein-coding exon of GJB2 (Duman & Tekin, 2012).
Variants of MYO7A account for 11.4% (95% CI: 8.3–15.3%) of HL in our cohort of 321 families, slightly more frequent than in previous reports (Riazuddin et al., 2008; Shahzad et al., 2013). Contrary to SLC26A4 and GJB2, no predominant MYO7A variant in the Pakistani population was uncovered in our study. However, six MYO7A variants (c.496delG, c.722G>A, c.1183C>T, c.1258A>T c.3502C>T, c.4838delA) were present in more than one family (Table 1 and Supp. Table S3). We also identified eight families harboring causative compound heterozygous variants in MYO7A (Supp. Table S3).
Similar to MYO7A, no prevalent allele of TMC1 was revealed in our study and 14 out of 18 identified variants of this gene were found in single families. Seven new likely pathogenic variants (c.945G>A [p.(Trp315*)], c.1143C>G [p.(Tyr381*)], c.1209G>A [p.(Trp403*)], c.1224+2T>C, c.1259G>A [p.(Cys420Tyr)], c.1728C>G [p.(Asn576Lys)] and c. 1753_1754insA [p.( Asn407Lysfs*2)]) were identified in our cohort (Table 1 and Supp. Table S3). In contrast, the c.482 +1986_88delTGA allele of HGF accounts for HL segregating in 26 families (Table 1), which was already identified as a founder allele in 50 other Pakistani families in our collaborative studies (Naz et al., 2017; Rehman et al., 2015; Schultz et al., 2009). Similarly, the c.272T>C allele of CIB2 accounts for the HL phenotype segregating in 15 families (Table 1). Studies in mouse have highlighted the crucial role of CIB2 in the mechanotransduction process and the deleterious effect of this specific missense mutation on hearing (Giese et al., 2017; Michel et al., 2017). Variants of CIB2 have been found in hearing-impaired individuals of Turkish, Caribbean Hispanic, Iranian, Palestinian Arab, European and Dutch origin (Booth et al., 2017; Michel et al., 2017; Patel et al., 2015; Riazuddin et al., 2012; Seco et al., 2016; Shaikh et al., 2017). Together with previously published studies, the CIB2 c.272T>C allele has been found in 85 hearing-impaired Pakistani families (Rehman et al., 2015; Riazuddin et al., 2012; Seco et al., 2016; Shaikh et al., 2017) and is likely to be a founder allele (Riazuddin et al., 2012).
Likely pathogenic variants of other genes associated with hearing loss in our cohort of 321 families include CDH23 (n=16 variants), TMPRSS3 (n=16), OTOF (n=10) and BSND (n=10) (B. Y. Choi et al., 2009; Naz et al., 2017; Rehman et al., 2015; Riazuddin et al., 2012; Shafique et al., 2014). This study highlights the genetic heterogeneity of HL-associated variants as 127 out of 163 (77.1%) identified alleles occur only once in our cohort. The diversity and the growing numbers of identified rare and non-recurrent variants increase the complexity of a molecular genetic diagnosis of HL (Gao & Dai, 2014). These data also predict that the Pakistani population will continue to be valuable for discovering the molecular genetics not only of HL but also of a multitude of other understudied monogenic and complex disorders such as persistent stuttering (Raza, Amjad, Riazuddin, & Drayna, 2012; Raza, Riazuddin, & Drayna, 2010; Riaz et al., 2005).
To better evaluate the genetic spectrum of recessive nonsyndromic HL in the Pakistani population, we combined data from these 321 new families with data from 61 of our previously published collaborative studies in Pakistan since 1999 (Fig. 6, Supp. table S4). Out of 875 identified mutant alleles in 54 genes (368 unique variants), more than 75% (75.3%) of the pathogenic alleles were found in 10 genes: SLC26A4, MYO7A, GJB2, CIB2, HGF, TMC1, MYO15A, CDH23, TMPRSS3 and OTOF. MYO15A variants were recently reviewed (Rehman et al., 2016) and represent 5.7% of the total predicted causative mutations, while alleles in BSND a gene involved in either Bartter syndrome or sensorineural HL with mild renal dysfunction (MIM# 602522), occur in 1.8% of the identified variants. We emphasize that the families in these studies were recruited on the initial assumption that they were segregating non-syndromic HL although a syndromic deafness often becomes evident only after identifying the responsible variants and completing phenotypic analyses and functional studies of the gene in mutant and wild type animal models (Imtiaz et al., 2018).
When the Pakistani population is compared to other populations i.e. Iranian (Ghasemnejad, Shekari Khaniani, Zarei, Farbodnia, & Mansoori Derakhshan, 2017; Sloan-Heggen et al., 2015), Indian (Yan et al., 2015), Chinese (Jiang et al., 2015; Xin et al., 2013; Zhang et al., 2016), African-descent (Rudman et al., 2017), European ancestry (Yan et al., 2017) or multi-ethnic (Sloan-Heggen et al., 2016; Yan et al., 2016), the distribution of genes associated with nonsyndromic HL and the variants reveal differences and commonalities. Some of the most prevalent “HL genes” in all of these populations are SLC26A4, GJB2 and MYO7A.
In these three genes, hundreds of pathogenic variants associated with HL have been identified while only 7 to 50 variants have occurred in the other most prevalent genes in our cohort with the exception of CDH23 (Human Gene Mutation Databases). Increased mutational events might explain the higher incidence of HL-associated variants, independently of the ethnicity of the considered population.
In conclusion, this study expands our understanding and knowledge of the allelic spectrum of HL genes in the Pakistani population, highlighting the extreme locus and variant heterogeneity of this disorder, sometimes even among siblings (e.g. see Fig. 3 in (Rehman et al., 2015)). This implies that even though we identified the genetic etiology of HL in hundreds of families within coding regions of known NSARHL genes, due to intra-familial genetic heterogeneity, we expect to identify at least 1000 additional variants in perhaps hundreds more novel genes in our unsolved families. Given the complexity of inner ear structure and function of the mechano-chemical transduction of sound, it has been speculated that there will be as many as 1000 genes that are involved in HL in mammals (Steel, 2014), which is further supported by yet undiscovered genetic lesions in hundreds of individuals and families with HL in our cohorts. With the rapid evolution of sequencing technologies and bioinformatics tools (He et al., 2017; Ott, Wang, & Leal, 2015), and with whole-genome sequencing more affordable, we anticipate the identification of more rare variants within HL genes. Identifying the molecular lesions will be critical for a more complete understanding of how we hear and for accomplishing personalized therapies (Askew et al., 2015; Chien et al., 2016; Shibata et al., 2016).
Supplementary Material
Acknowledgments
We would like to thank all the families for participating in the study. We thank AM Waryah, R. Yousaf and S. Yousaf for their technical help. We also would like to thank the University of Washington Center for Mendelian Genomics.
Contract grant sponsors: NIDCD/NIH extramural research grants (R56 DC011803 to SR, R01 DC011651 to SML, R01 DC003594 to SML, R01 DC012564 to ZMA); and in part from Intramural Research Program of the National Institute on Deafness and Other Communication Disorders at the NIH (DC000039–20 to T.B.F. and DC000086 to R.J.M.); and Higher Education Commission of Pakistan (HEC). Genotyping and exome sequencing performed at the UWCMG was funded by the National Human Genome Research Institute and the National Heart, Lung and Blood Institute grant HG006493 (to D.A.N., M.J.B. and S.M.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work utilized computational resources of the NIH HPC Biowulf cluster.
Footnotes
Disclosure statement: All authors declare no conflicts of interest.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, … Sunyaev SR. (2010). A method and server for predicting damaging missense mutations. Nat Methods, 7(4), 248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Ahmad J, Bernstein SL, Guo Y, Sabar MF, … Wilcox ER. (2003). PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet, 12(24), 3215–3223. doi: 10.1093/hmg/ddg358 [DOI] [PubMed] [Google Scholar]
- Ahmed ZM, Yousaf R, Lee BC, Khan SN, Lee S, Lee K, … Riazuddin S. (2011). Functional null mutations of MSRB3 encoding methionine sulfoxide reductase are associated with human deafness DFNB74. Am J Hum Genet, 88(1), 19–29. doi: 10.1016/j.ajhg.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum S, Azhar A, Tariq M, Baig S, HJ. B, Qayyum M, … Raja G. (2014). GJB2 Gene Mutations Causing Hearing Loss in Consanguineous Pakistani Families. Pak J Life Soc Sci(12), 126–131. [Google Scholar]
- Anwar S, Riazuddin S, Ahmed ZM, Tasneem S, Ateeq ul J, Khan SY, … Riazuddin S. (2009). SLC26A4 mutation spectrum associated with DFNB4 deafness and Pendred’s syndrome in Pakistanis. J Hum Genet, 54(5), 266–270. doi: 10.1038/jhg.2009.21 [DOI] [PubMed] [Google Scholar]
- Askew C, Rochat C, Pan B, Asai Y, Ahmed H, Child E, … Holt JR. (2015). Tmc gene therapy restores auditory function in deaf mice. Sci Transl Med, 7(295), 295ra108. doi: 10.1126/scitranslmed.aab1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bademci G, Foster J 2nd, Mahdieh N, Bonyadi M, Duman D, Cengiz FB, … Tekin M. (2016). Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med, 18(4), 364–371. doi: 10.1038/gim.2015.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Salem S, Rehm HL, Willems PJ, Tamimi ZA, Ayadi H, Ali BR, & Al-Gazali L (2014). Analysis of two Arab families reveals additional support for a DFNB2 nonsyndromic phenotype of MYO7A. Mol Biol Rep, 41(1), 193–200. doi: 10.1007/s11033-013-2851-5 [DOI] [PubMed] [Google Scholar]
- Ben-Yosef T, Wattenhofer M, Riazuddin S, Ahmed ZM, Scott HS, Kudoh J, … Morell RJ. (2001). Novel mutations of TMPRSS3 in four DFNB8/B10 families segregating congenital autosomal recessive deafness. J Med Genet, 38(6), 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard T, Vache C, Baux D, Larrieu L, Abadie C, Blanchet C, … Roux AF. (2012). Non-USH2A mutations in USH2 patients. Hum Mutat, 33(3), 504–510. doi: 10.1002/humu.22004 [DOI] [PubMed] [Google Scholar]
- Bharadwaj AK, Kasztejna JP, Huq S, Berson EL, & Dryja TP (2000). Evaluation of the myosin VIIA gene and visual function in patients with Usher syndrome type I. Exp Eye Res, 71(2), 173–181. doi: 10.1006/exer.2000.0863 [DOI] [PubMed] [Google Scholar]
- Bitner-Glindzicz M, Lindley KJ, Rutland P, Blaydon D, Smith VV, Milla PJ, … Glaser B. (2000). A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet, 26(1), 56–60. doi: 10.1038/79178 [DOI] [PubMed] [Google Scholar]
- Bittles AH (2005). Population stratification and genetic association studies in South Asia. J Mol Genet Med, 1(2), 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth KT, Kahrizi K, Babanejad M, Daghagh H, Bademci G, Arzhangi S, … Smith RJ. (2017). Variants in CIB2 cause DFNB48 and not USH1J. Clin Genet. doi: 10.1111/cge.13170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck G, Ur Rehman A, Lee K, Pogoda HM, Kakar N, von Ameln S, … Kubisch C. (2011). Loss-of-function mutations of ILDR1 cause autosomal-recessive hearing impairment DFNB42. Am J Hum Genet, 88(2), 127–137. doi: 10.1016/j.ajhg.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, … Morell RJ. (2001). Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet, 68(1), 26–37. doi: 10.1086/316954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari I, Mujtaba G, & Naz S (2013). Contribution of GJB2 mutations to hearing loss in the Hazara Division of Pakistan. Biochem Genet, 51(7–8), 524–529. doi: 10.1007/s10528-013-9583-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Kelly MC, Hoa M, Morell RJ, & Kelley MW (2015). Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat Commun, 6, 8557. doi: 10.1038/ncomms9557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carss KJ, Arno G, Erwood M, Stephens J, Sanchis-Juan A, Hull S, … Raymond FL. (2017). Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am J Hum Genet, 100(1), 75–90. doi: 10.1016/j.ajhg.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattaraj P, Munjal T, Honda K, Rendtorff ND, Ratay JS, Muskett JA, … Griffith AJ. (2017). A common SLC26A4-linked haplotype underlying non-syndromic hearing loss with enlargement of the vestibular aqueduct. J Med Genet, 54(10), 665–673. doi: 10.1136/jmedgenet-2017-104721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wang X, Sun L, & Jiang H (2012). Screening of SLC26A4, FOXI1, KCNJ10, and GJB2 in bilateral deafness patients with inner ear malformation. Otolaryngol Head Neck Surg, 146(6), 972–978. doi: 10.1177/0194599812439670 [DOI] [PubMed] [Google Scholar]
- Chien WW, Isgrig K, Roy S, Belyantseva IA, Drummond MC, May LA, … Cunningham LL. (2016). Gene Therapy Restores Hair Cell Stereocilia Morphology in Inner Ears of Deaf Whirler Mice. Mol Ther, 24(1), 17–25. doi: 10.1038/mt.2015.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BY, Ahmed ZM, Riazuddin S, Bhinder MA, Shahzad M, Husnain T, … Friedman TB. (2009). Identities and frequencies of mutations of the otoferlin gene (OTOF) causing DFNB9 deafness in Pakistan. Clin Genet, 75(3), 237–243. doi: 10.1111/j.1399-0004.2008.01128.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BY, Kim HM, Ito T, Lee KY, Li X, Monahan K, … Griffith AJ. (2011). Mouse model of enlarged vestibular aqueducts defines temporal requirement of Slc26a4 expression for hearing acquisition. J Clin Invest, 121(11), 4516–4525. doi: 10.1172/JCI59353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, & Chan AP (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics, 31(16), 2745–2747. doi: 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucke PJ, Van Hauwe P, Everett LA, Demirhan O, Kabakkaya Y, Dietrich NL, … Van Camp G. (1999). Identification of two different mutations in the PDS gene in an inbred family with Pendred syndrome. J Med Genet, 36(6), 475–477. [PMC free article] [PubMed] [Google Scholar]
- Cremers FP, Kimberling WJ, Kulm M, de Brouwer AP, van Wijk E, te Brinke H, … Kremer H. (2007). Development of a genotyping microarray for Usher syndrome. J Med Genet, 44(2), 153–160. doi: 10.1136/jmg.2006.044784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoyelle F, Weil D, Maw MA, Wilcox SA, Lench NJ, Allen-Powell DR, … et al. (1997). Prelingual deafness: high prevalence of a 30delG mutation in the connexin 26 gene. Hum Mol Genet, 6(12), 2173–2177. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, & Beroud C (2009). Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res, 37(9), e67. doi: 10.1093/nar/gkp215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman D, & Tekin M (2012). Autosomal recessive nonsyndromic deafness genes: a review. Front Biosci (Landmark Ed), 17, 2213–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebermann I, Lopez I, Bitner-Glindzicz M, Brown C, Koenekoop RK, & Bolz HJ (2007). Deafblindness in French Canadians from Quebec: a predominant founder mutation in the USH1C gene provides the first genetic link with the Acadian population. Genome Biol, 8(4), R47. doi: 10.1186/gb-2007-8-4-r47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett LA, Belyantseva IA, Noben-Trauth K, Cantos R, Chen A, Thakkar SI, … Green ED. (2001). Targeted disruption of mouse Pds provides insight about the inner-ear defects encountered in Pendred syndrome. Hum Mol Genet, 10(2), 153–161. [DOI] [PubMed] [Google Scholar]
- Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, … Green ED. (1997). Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat Genet, 17(4), 411–422. doi: 10.1038/ng1297-411 [DOI] [PubMed] [Google Scholar]
- Friedman TB, Liang Y, Weber JL, Hinnant JT, Barber TD, Winata S, … Asher JH Jr. (1995). A gene for congenital, recessive deafness DFNB3 maps to the pericentromeric region of chromosome 17. Nat Genet, 9(1), 86–91. doi: 10.1038/ng0195-86 [DOI] [PubMed] [Google Scholar]
- Gao X, & Dai P (2014). Impact of next-generation sequencing on molecular diagnosis of inherited non-syndromic hearing loss. Journal of Otology, 9, 122–125. [Google Scholar]
- Gettelfinger JD, & Dahl JP (2018). Syndromic Hearing Loss: A Brief Review of Common Presentations and Genetics. J Pediatr Genet, 7(1), 1–8. doi: 10.1055/s-0037-1617454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemnejad T, Shekari Khaniani M, Zarei F, Farbodnia M, & Mansoori Derakhshan S (2017). An update of common autosomal recessive non-syndromic hearing loss genes in Iranian population. Int J Pediatr Otorhinolaryngol, 97, 113–126. doi: 10.1016/j.ijporl.2017.04.007 [DOI] [PubMed] [Google Scholar]
- Giese APJ, Tang YQ, Sinha GP, Bowl MR, Goldring AC, Parker A, … Ahmed ZM. (2017). CIB2 interacts with TMC1 and TMC2 and is essential for mechanotransduction in auditory hair cells. Nat Commun, 8(1), 43. doi: 10.1038/s41467-017-00061-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood-Watson RJ 2nd, Ahmed ZM, Kjellstrom S, Bush RA, Takada Y, Hampton LL, … Friedman TB. (2006). Ames Waltzer deaf mice have reduced electroretinogram amplitudes and complex alternative splicing of Pcdh15 transcripts. Invest Ophthalmol Vis Sci, 47(7), 3074–3084. doi: 10.1167/iovs.06-0108 [DOI] [PubMed] [Google Scholar]
- He Z, Zhang D, Renton AE, Li B, Zhao L, Wang GT, … Leal SM. (2017). The Rare-Variant Generalized Disequilibrium Test for Association Analysis of Nuclear and Extended Pedigrees with Application to Alzheimer Disease WGS Data. Am J Hum Genet. doi: 10.1016/j.ajhg.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgert N, Kahrizi K, Dieltjens N, Bazazzadegan N, Najmabadi H, Smith RJ, & Van Camp G (2009). A large deletion in GPR98 causes type IIC Usher syndrome in male and female members of an Iranian family. J Med Genet, 46(4), 272–276. doi: 10.1136/jmg.2008.060947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiaz A, Belyantseva IA, Beirl AJ, Fenollar-Ferrer C, Bashir R, Bukhari I, … Friedman TB. (2018). CDC14A phosphatase is essential for hearing and male fertility in mouse and human. Hum Mol Genet, 27(5), 780–798. doi: 10.1093/hmg/ddx440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiaz A, Maqsood A, Rehman AU, Morell RJ, Holt JR, Friedman TB, & Naz S (2016). Recessive mutations of TMC1 associated with moderate to severe hearing loss. Neurogenetics, 17(2), 115–123. doi: 10.1007/s10048-016-0477-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaijo T, Aller E, Oltra S, Beneyto M, Najera C, Ayuso C, … Millan JM. (2006). Mutation profile of the MYO7A gene in Spanish patients with Usher syndrome type I. Hum Mutat, 27(3), 290–291. doi: 10.1002/humu.9404 [DOI] [PubMed] [Google Scholar]
- Ji YB, Han DY, Wang DY, Zhou Y, Zhao C, Wang H, … Wang QJ. (2009). [Evaluation of deaf-mute patients with sensitive deafness gene screening in Shandong province]. Zhonghua Yi Xue Za Zhi, 89(36), 2531–2535. [PubMed] [Google Scholar]
- Jian X, Boerwinkle E, & Liu X (2014). In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res, 42(22), 13534–13544. doi: 10.1093/nar/gku1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Huang S, Deng T, Wu L, Chen J, Kang D, … Dai P. (2015). Mutation Spectrum of Common Deafness-Causing Genes in Patients with Non-Syndromic Deafness in the Xiamen Area, China. PLoS One, 10(8), e0135088. doi: 10.1371/journal.pone.0135088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahrizi K, Mohseni M, Nishimura C, Bazazzadegan N, Fischer SM, Dehghani A, … Najmabadi H. (2009). Identification of SLC26A4 gene mutations in Iranian families with hereditary hearing impairment. Eur J Pediatr, 168(6), 651–653. doi: 10.1007/s00431-008-0809-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, … Leigh IM. (1997). Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature, 387(6628), 80–83. doi: 10.1038/387080a0 [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, & Shendure J (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet, 46(3), 310–315. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajiri SI, McNamara R, Makishima T, Husnain T, Zafar AU, Kittles RA, … Griffith AJ. (2007). Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin Genet, 72(6), 546–550. doi: 10.1111/j.1399-0004.2007.00895.x [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, & Ng PC (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc, 4(7), 1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, … Griffith AJ. (2002). Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet, 30(3), 277–284. doi: 10.1038/ng842 [DOI] [PubMed] [Google Scholar]
- Le Guedard-Mereuze S, Vache C, Baux D, Faugere V, Larrieu L, Abadie C, … Tuffery-Giraud S. (2010). Ex vivo splicing assays of mutations at noncanonical positions of splice sites in USHER genes. Hum Mutat, 31(3), 347–355. doi: 10.1002/humu.21193 [DOI] [PubMed] [Google Scholar]
- Le Quesne Stabej P, Saihan Z, Rangesh N, Steele-Stallard HB, Ambrose J, Coffey A, … Bitner-Glindzicz M. (2012). Comprehensive sequence analysis of nine Usher syndrome genes in the UK National Collaborative Usher Study. J Med Genet, 49(1), 27–36. doi: 10.1136/jmedgenet-2011-100468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Ansar M, Andrade PB, Khan B, Santos-Cortez RL, Ahmad W, & Leal SM (2012). Novel CLDN14 mutations in Pakistani families with autosomal recessive non-syndromic hearing loss. Am J Med Genet A, 158A(2), 315–321. doi: 10.1002/ajmg.a.34407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Chiu I, Santos-Cortez RL, Basit S, Khan S, Azeem Z, … Leal SM. (2013). Novel OTOA mutations cause autosomal recessive non-syndromic hearing impairment in Pakistani families. Clin Genet, 84(3), 294–296. doi: 10.1111/cge.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Khan S, Islam A, Ansar M, Andrade PB, Kim S, … Leal SM. (2012). Novel TMPRSS3 variants in Pakistani families with autosomal recessive non-syndromic hearing impairment. Clin Genet, 82(1), 56–63. doi: 10.1111/j.1399-0004.2011.01695.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlin S, Garabedian EN, Roger G, Moatti L, Matha N, Lewin P, … Denoyelle F. (2001). Connexin 26 gene mutations in congenitally deaf children: pitfalls for genetic counseling. Arch Otolaryngol Head Neck Surg, 127(8), 927–933. [DOI] [PubMed] [Google Scholar]
- Michel V, Booth KT, Patni P, Cortese M, Azaiez H, Bahloul A, … El-Amraoui A. (2017). CIB2, defective in isolated deafness, is key for auditory hair cell mechanotransduction and survival. EMBO Mol Med. doi: 10.15252/emmm.201708087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa M, Naito T, Nishio SY, Kamatani N, & Usami S (2013). Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of Japanese deafness patients. PLoS One, 8(8), e71381. doi: 10.1371/journal.pone.0071381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CC, & Nance WE (2006). Newborn hearing screening--a silent revolution. N Engl J Med, 354(20), 2151–2164. doi: 10.1056/NEJMra050700 [DOI] [PubMed] [Google Scholar]
- Naz S, Giguere CM, Kohrman DC, Mitchem KL, Riazuddin S, Morell RJ, … Wilcox ER. (2002). Mutations in a novel gene, TMIE, are associated with hearing loss linked to the DFNB6 locus. Am J Hum Genet, 71(3), 632–636. doi: 10.1086/342193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz S, Imtiaz A, Mujtaba G, Maqsood A, Bashir R, Bukhari I, … Friedman TB. (2017). Genetic causes of moderate to severe hearing loss point to modifiers. Clin Genet, 91(4), 589–598. doi: 10.1111/cge.12856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J, Wang J, & Leal SM (2015). Genetic linkage analysis in the age of whole-genome sequencing. Nat Rev Genet, 16(5), 275–284. doi: 10.1038/nrg3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, … Griffith AJ. (2003). Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet, 40(4), 242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K, Giese AP, Grossheim JM, Hegde RS, Delio M, Samanich J, … Morrow BE. (2015). A Novel C-Terminal CIB2 (Calcium and Integrin Binding Protein 2) Mutation Associated with Non-Syndromic Hearing Loss in a Hispanic Family. PLoS One, 10(10), e0133082. doi: 10.1371/journal.pone.0133082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffenberger EG, Hosoda K, Washington SS, Nakao K, deWit D, Yanagisawa M, & Chakravart A (1994). A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung’s disease. Cell, 79(7), 1257–1266. [DOI] [PubMed] [Google Scholar]
- Raza MH, Amjad R, Riazuddin S, & Drayna D (2012). Studies in a consanguineous family reveal a novel locus for stuttering on chromosome 16q. Hum Genet, 131(2), 311–313. doi: 10.1007/s00439-011-1134-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza MH, Riazuddin S, & Drayna D (2010). Identification of an autosomal recessive stuttering locus on chromosome 3q13.2–3q13.33. Hum Genet, 128(4), 461–463. doi: 10.1007/s00439-010-0871-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon W, CF OM, Trembath R, Jan H, & Phelps PD (2000). Enlarged vestibular aqueduct: a radiological marker of pendred syndrome, and mutation of the PDS gene. QJM, 93(2), 99–104. [DOI] [PubMed] [Google Scholar]
- Reddy R, Fahiminiya S, El Zir E, Mansour A, Megarbane A, Majewski J, & Slim R (2014). Molecular genetics of the Usher syndrome in Lebanon: identification of 11 novel protein truncating mutations by whole exome sequencing. PLoS One, 9(9), e107326. doi: 10.1371/journal.pone.0107326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Bird JE, Faridi R, Shahzad M, Shah S, Lee K, … Friedman TB. (2016). Mutational Spectrum of MYO15A and the Molecular Mechanisms of DFNB3 Human Deafness. Hum Mutat, 37(10), 991–1003. doi: 10.1002/humu.23042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Friedman TB, & Griffith AJ (2017). Unresolved questions regarding human hereditary deafness. Oral Dis, 23(5), 551–558. doi: 10.1111/odi.12516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Gul K, Morell RJ, Lee K, Ahmed ZM, Riazuddin S, … Friedman TB. (2011). Mutations of GIPC3 cause nonsyndromic hearing loss DFNB72 but not DFNB81 that also maps to chromosome 19p. Hum Genet, 130(6), 759–765. doi: 10.1007/s00439-011-1018-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Morell RJ, Belyantseva IA, Khan SY, Boger ET, Shahzad M, … Friedman TB. (2010). Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am J Hum Genet, 86(3), 378–388. doi: 10.1016/j.ajhg.2010.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Santos-Cortez RL, Drummond MC, Shahzad M, Lee K, Morell RJ, … Leal SM. (2015). Challenges and solutions for gene identification in the presence of familial locus heterogeneity. Eur J Hum Genet, 23(9), 1207–1215. doi: 10.1038/ejhg.2014.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, & Sander C (2007). Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol, 8(11), R232. doi: 10.1186/gb-2007-8-11-r232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz N, Steinberg S, Ahmad J, Pluzhnikov A, Riazuddin S, Cox NJ, & Drayna D (2005). Genomewide significant linkage to stuttering on chromosome 12. Am J Hum Genet, 76(4), 647–651. doi: 10.1086/429226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Ahmed ZM, Fanning AS, Lagziel A, Kitajiri S, Ramzan K, … Friedman TB. (2006). Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet, 79(6), 1040–1051. doi: 10.1086/510022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Anwar S, Fischer M, Ahmed ZM, Khan SY, Janssen AG, … Fahlke C. (2009). Molecular basis of DFNB73: mutations of BSND can cause nonsyndromic deafness or Bartter syndrome. Am J Hum Genet, 85(2), 273–280. doi: 10.1016/j.ajhg.2009.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Belyantseva IA, Giese AP, Lee K, Indzhykulian AA, Nandamuri SP, … Ahmed ZM. (2012). Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat Genet, 44(11), 1265–1271. doi: 10.1038/ng.2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, Nazli S, Ahmed ZM, Yang Y, Zulfiqar F, Shaikh RS, … Friedman TB. (2008). Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum Mutat, 29(4), 502–511. doi: 10.1002/humu.20677 [DOI] [PubMed] [Google Scholar]
- Richard G, White TW, Smith LE, Bailey RA, Compton JG, Paul DL, & Bale SJ (1998). Functional defects of Cx26 resulting from a heterozygous missense mutation in a family with dominant deaf-mutism and palmoplantar keratoderma. Hum Genet, 103(4), 393–399. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, … Committee, A. L. Q. A. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 17(5), 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux AF, Faugere V, Le Guedard S, Pallares-Ruiz N, Vielle A, Chambert S, … French Usher Syndrome, C. (2006). Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90%. J Med Genet, 43(9), 763–768. doi: 10.1136/jmg.2006.041954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudman JR, Kabahuma RI, Bressler SE, Feng Y, Blanton SH, Yan D, & Liu XZ (2017). The genetic basis of deafness in populations of African descent. J Genet Genomics, 44(6), 285–294. doi: 10.1016/j.jgg.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Salman M, Bashir R, Imtiaz A, Maqsood A, Mujtaba G, Iqbal M, & Naz S (2015). Mutations of GJB2 encoding connexin 26 contribute to non-syndromic moderate and severe hearing loss in Pakistan. Eur Arch Otorhinolaryngol, 272(8), 2071–2075. doi: 10.1007/s00405-015-3523-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RL, El-Shanti H, Sikandar S, Lee K, Bhatti A, Yan K, … Leal SM. (2006). Novel sequence variants in the TMIE gene in families with autosomal recessive nonsyndromic hearing impairment. J Mol Med (Berl), 84(3), 226–231. doi: 10.1007/s00109-005-0015-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RL, Wajid M, Khan MN, McArthur N, Pham TL, Bhatti A, … Leal SM. (2005). Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum Mutat, 26(4), 396. doi: 10.1002/humu.9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RL, Wajid M, Pham TL, Hussan J, Ali G, Ahmad W, & Leal SM (2005). Low prevalence of Connexin 26 (GJB2) variants in Pakistani families with autosomal recessive non-syndromic hearing impairment. Clin Genet, 67(1), 61–68. doi: 10.1111/j.1399-0004.2005.00379.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraders M, Lee K, Oostrik J, Huygen PL, Ali G, Hoefsloot LH, … Kremer H. (2010). Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment. Am J Hum Genet, 86(2), 138–147. doi: 10.1016/j.ajhg.2009.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Helfmann S, Inagaki A, Predoehl F, Tabatabaiefar MA, Picher MM, … Van Camp G. (2012). A mutation in CABP2, expressed in cochlear hair cells, causes autosomal-recessive hearing impairment. Am J Hum Genet, 91(4), 636–645. doi: 10.1016/j.ajhg.2012.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz JM, Bhatti R, Madeo AC, Turriff A, Muskett JA, Zalewski CK, … Friedman TB. (2011). Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J Med Genet, 48(11), 767–775. doi: 10.1136/jmedgenet-2011-100262 [DOI] [PubMed] [Google Scholar]
- Schultz JM, Khan SN, Ahmed ZM, Riazuddin S, Waryah AM, Chhatre D, … Morell RJ. (2009). Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am J Hum Genet, 85(1), 25–39. doi: 10.1016/j.ajhg.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, & Seelow D (2010). MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods, 7(8), 575–576. doi: 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- Scott EM (1973). Genetic disorders in isolated populations. Arch Environ Health, 26(1), 32–35. [DOI] [PubMed] [Google Scholar]
- Scott HS, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, … Antonarakis SE. (2001). Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet, 27(1), 59–63. doi: 10.1038/83768 [DOI] [PubMed] [Google Scholar]
- Seco CZ, Giese AP, Shafique S, Schraders M, Oonk AM, Grossheim M, … Kremer H. (2016). Novel and recurrent CIB2 variants, associated with nonsyndromic deafness, do not affect calcium buffering and localization in hair cells. Eur J Hum Genet, 24(4), 542–549. doi: 10.1038/ejhg.2015.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelow D, Schuelke M, Hildebrandt F, & Nurnberg P (2009). HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res, 37(Web Server issue), W593–599. doi: 10.1093/nar/gkp369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabbir MI, Ahmed ZM, Khan SY, Riazuddin S, Waryah AM, Khan SN, … Riazuddin S. (2006). Mutations of human TMHS cause recessively inherited non-syndromic hearing loss. J Med Genet, 43(8), 634–640. doi: 10.1136/jmg.2005.039834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafique S, Siddiqi S, Schraders M, Oostrik J, Ayub H, Bilal A, … Qamar R. (2014). Genetic spectrum of autosomal recessive non-syndromic hearing loss in Pakistani families. PLoS One, 9(6), e100146. doi: 10.1371/journal.pone.0100146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad M, Sivakumaran TA, Qaiser TA, Schultz JM, Hussain Z, Flanagan M, … Ahmed ZM. (2013). Genetic analysis through OtoSeq of Pakistani families segregating prelingual hearing loss. Otolaryngol Head Neck Surg, 149(3), 478–487. doi: 10.1177/0194599813493075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh H, Waryah AM, Narsani AK, Iqbal M, Shahzad M, Waryah YM, … Mahmood A. (2017). Genetic Testing of Non-familial Deaf Patients for CIB2 and GJB2 Mutations: Phenotype and Genetic Counselling. Biochem Genet, 55(5–6), 410–420. doi: 10.1007/s10528-017-9828-3 [DOI] [PubMed] [Google Scholar]
- Shearer AE, Black-Ziegelbein EA, Hildebrand MS, Eppsteiner RW, Ravi H, Joshi S, … Smith RJ. (2013). Advancing genetic testing for deafness with genomic technology. J Med Genet, 50(9), 627–634. doi: 10.1136/jmedgenet-2013-101749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Kolbe DL, Azaiez H, Sloan CM, Frees KL, Weaver AE, … Smith RJ. (2014). Copy number variants are a common cause of non-syndromic hearing loss. Genome Med, 6(5), 37. doi: 10.1186/gm554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata SB, Ranum PT, Moteki H, Pan B, Goodwin AT, Goodman SS, … Smith RJ. (2016). RNA Interference Prevents Autosomal-Dominant Hearing Loss. Am J Hum Genet, 98(6), 1101–1113. doi: 10.1016/j.ajhg.2016.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, … Gaunt TR. (2013). Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat, 34(1), 57–65. doi: 10.1002/humu.22225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day IN, … Campbell C. (2015). An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics, 31(10), 1536–1543. doi: 10.1093/bioinformatics/btv009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirmaci A, Duman D, Ozturkmen-Akay H, Erbek S, Incesulu A, Ozturk-Hismi B, … Tekin M. (2009). Mutations in TMC1 contribute significantly to nonsyndromic autosomal recessive sensorineural hearing loss: a report of five novel mutations. Int J Pediatr Otorhinolaryngol, 73(5), 699–705. doi: 10.1016/j.ijporl.2009.01.005 [DOI] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Babanejad M, Beheshtian M, Simpson AC, Booth KT, Ardalani F, … Najmabadi H. (2015). Characterising the spectrum of autosomal recessive hereditary hearing loss in Iran. J Med Genet, 52(12), 823–829. doi: 10.1136/jmedgenet-2015-103389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, … Smith RJ. (2016). Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet, 135(4), 441–450. doi: 10.1007/s00439-016-1648-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, & Smith RJ (2016). Navigating genetic diagnostics in patients with hearing loss. Curr Opin Pediatr, 28(6), 705–712. doi: 10.1097/MOP.0000000000000410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel KP (2014). What’s the Use of Genetics? In Popper AN & Fay RR (Eds.), Perspectives on Auditory Research (pp. 569–584). New York: Springer. [Google Scholar]
- Tsukada K, Nishio SY, Hattori M, & Usami S (2015). Ethnic-specific spectrum of GJB2 and SLC26A4 mutations: their origin and a literature review. Ann Otol Rhinol Laryngol, 124 Suppl 1, 61S–76S. doi: 10.1177/0003489415575060 [DOI] [PubMed] [Google Scholar]
- Uyguner O, Tukel T, Baykal C, Eris H, Emiroglu M, Hafiz G, … Wollnik B. (2002). The novel R75Q mutation in the GJB2 gene causes autosomal dominant hearing loss and palmoplantar keratoderma in a Turkish family. Clin Genet, 62(4), 306–309. [DOI] [PubMed] [Google Scholar]
- Van Hauwe P, Everett LA, Coucke P, Scott DA, Kraft ML, Ris-Stalpers C, … Van Camp G. (1998). Two frequent missense mutations in Pendred syndrome. Hum Mol Genet, 7(7), 1099–1104. [DOI] [PubMed] [Google Scholar]
- von Brederlow B, Bolz H, Janecke A, La OCA, Rudolph G, Lorenz B, … Gal A. (2002). Identification and in vitro expression of novel CDH23 mutations of patients with Usher syndrome type 1D. Hum Mutat, 19(3), 268–273. doi: 10.1002/humu.10049 [DOI] [PubMed] [Google Scholar]
- Vona B, Nanda I, Hofrichter MA, Shehata-Dieler W, & Haaf T (2015). Non-syndromic hearing loss gene identification: A brief history and glimpse into the future. Mol Cell Probes, 29(5), 260–270. doi: 10.1016/j.mcp.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Weston MD, Kelley PM, Overbeck LD, Wagenaar M, Orten DJ, Hasson T, … Kimberling WJ. (1996). Myosin VIIA mutation screening in 189 Usher syndrome type 1 patients. Am J Hum Genet, 59(5), 1074–1083. [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Luijendijk MW, Humphrey KD, Moller C, & Kimberling WJ (2004). Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am J Hum Genet, 74(2), 357–366. doi: 10.1086/381685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin F, Yuan Y, Deng X, Han M, Wang G, Zhao J, … Dai P. (2013). Genetic mutations in nonsyndromic deafness patients of Chinese minority and Han ethnicities in Yunnan, China. J Transl Med, 11, 312. doi: 10.1186/1479-5876-11-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba T, Lin FR, Someya S, Kashio A, Sakamoto T, & Kondo K (2013). Current concepts in age-related hearing loss: epidemiology and mechanistic pathways. Hear Res, 303, 30–38. doi: 10.1016/j.heares.2013.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Kannan-Sundhari A, Vishwanath S, Qing J, Mittal R, Kameswaran M, & Liu XZ (2015). The Genetic Basis of Nonsyndromic Hearing Loss in Indian and Pakistani Populations. Genet Test Mol Biomarkers, 19(9), 512–527. doi: 10.1089/gtmb.2015.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Tekin D, Bademci G, Foster J 2nd, Cengiz FB, Kannan-Sundhari A, … Tekin M. (2016). Spectrum of DNA variants for non-syndromic deafness in a large cohort from multiple continents. Hum Genet, 135(8), 953–961. doi: 10.1007/s00439-016-1697-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Xiang G, Chai X, Qing J, Shang H, Zou B, … Liu XZ. (2017). Screening of deafness-causing DNA variants that are common in patients of European ancestry using a microarray-based approach. PLoS One, 12(3), e0169219. doi: 10.1371/journal.pone.0169219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanpanahi N, Chaleshtori MH, Tabatabaiefar MA, Noormohammadi Z, Farrokhi E, Najmabadi H, … Hosseinipour A. (2012). Two novel SLC26A4 mutations in Iranian families with autosomal recessive hearing loss. Int J Pediatr Otorhinolaryngol, 76(6), 845–850. doi: 10.1016/j.ijporl.2012.02.056 [DOI] [PubMed] [Google Scholar]
- Yazdanpanahi N, Tabatabaiefar MA, Bagheri N, Azadegan Dehkordi F, Farrokhi E, & Hashemzadeh Chaleshtori M (2015). The role and spectrum of SLC26A4 mutations in Iranian patients with autosomal recessive hereditary deafness. Int J Audiol, 54(2), 124–130. doi: 10.3109/14992027.2014.944276 [DOI] [PubMed] [Google Scholar]
- Zein WM, Falsini B, Tsilou ET, Turriff AE, Schultz JM, Friedman TB, … Sieving PA. (2014). Cone responses in Usher syndrome types 1 and 2 by microvolt electroretinography. Invest Ophthalmol Vis Sci, 56(1), 107–114. doi: 10.1167/iovs.14-15355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante L, Gasparini P, Estivill X, Melchionda S, D’Agruma L, Govea N, … Fortina P. (1997). Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet, 6(9), 1605–1609. [DOI] [PubMed] [Google Scholar]
- Zhang F, Xiao Y, Xu L, Zhang X, Zhang G, Li J, … Wang H. (2016). Mutation Analysis of the Common Deafness Genes in Patients with Nonsyndromic Hearing Loss in Linyi by SNPscan Assay. Biomed Res Int, 2016, 1302914. doi: 10.1155/2016/1302914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QY, Yu H, Washington JL 3rd, Kisley LB, Kikkawa YS, Pawlowski KS, … Alagramam KN. (2006). A new spontaneous mutation in the mouse protocadherin 15 gene. Hear Res, 219(1–2), 110–120. doi: 10.1016/j.heares.2006.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.