Abstract
The mouth is a first critical interface where most potentially harmful substances or pathogens contact the host environment. Adaptive and innate immune defense mechanisms are established there to inactivate or eliminate pathogenic microbes that traverse the oral environment on the way to their target organs and tissues. Protein and glycoprotein components of saliva play a particularly important role in modulating the oral microbiota and helping with the clearance of pathogens. It has long been acknowledged that glycobiological and glycoimmunological aspects play a pivotal role in oral host-microbe, microbe-host, and microbe-microbe interactions in the mouth. In this review, we aim to delineate how glycan-mediated host defense mechanisms in the oral cavity support human health. We will describe the role of glycans attached to large molecular size salivary glycoproteins which act as a first line of primordial host defense in the human mouth. We will further discuss how glycan recognition contributes to both colonization and clearance of oral microbes.
Keywords: Oral biology, glycobiology, microbiology, host defense, saliva, salivary proteins, mucins, microbial adhesins, glycans, evolution
1. Introduction.
The mouth is the natural port of entry to both the gastrointestinal and respiratory tracts. As such, it is the first critical interface where most potentially harmful substances or pathogens contact the host environment. Adaptive and innate immune defense mechanisms are established in the oral cavity to inactivate or eliminate pathogenic microbes that traverse the oral environment on the way to their target organs and tissues. Starting at birth, the host defense systems of the oral cavity become exposed to microbes. Infants habitually put anything within reach into their mouths. Such early exposure to a wide variety of microbes derived from caretakers and the environment impacts the developing immune system and shapes the developing oral and gut microbiomes. All interactions of microbes with host surfaces in the oral cavity take place in a saliva-soaked environment. Protein and glycoprotein components of saliva play a particularly important role in modulating the oral microbiota. On the one hand they foster and maintain host colonization by a beneficial microflora, and on the other hand they aid in the clearance of pathogens. Over the years, scientists in different disciplines have studied the microbiota of the oral cavity and the oral host molecules that interact with the microbes. It has long been acknowledged that glycobiological and glycoimmunological aspects play a pivotal role in oral host-microbe, microbe-host, and microbe-microbe interactions in the mouth. Historically, some basic, widely applicable insights in glycan-mediated interactions were gained from investigations in the confined area of oral biology, and the ease of retrieving samples from the mouth certainly facilitated that progress.
In this review, we aim to delineate why knowledge of oral glycobiology is of relevance even for immunologists and microbiologists who may not directly be interested in the biology of the oral cavity, but still may want to know how glycan-mediated host defense mechanisms in the oral cavity support human health. We will not cover those immunological and innate host defense mechanisms in which glycans don’t or have not yet been found to play a role. Those interactions, although important for oral health, are in many instances not unique to the oral cavity and are not principally different from host defense mechanisms elsewhere in the body. The immunological host defense mechanisms in the oral cavity have recently been reviewed [1]. We will also not cover the interesting and very relevant ways systemic immune deficiency and salivary gland dysfunction cause dysbiosis in the oral ecosystem and beyond. These topics have also been thoroughly reviewed elsewhere [2–4]. Here, we will discuss the role of glycans attached to mostly large molecular size salivary glycoproteins which act as a first line of primordial host defense in the human mouth. We will describe the various types of proteinglycan interactions between salivary glycoproteins, bacteria, fungi, and viruses that contribute to both colonization and clearance of oral microbes. We will further detail glycan-mediated interactions among different microbes that lead to the formation of multi-species biofilms in the oral cavity. Finally, we discuss the few existing clinical, translational applications of glycans and point to promising future avenues for “glyco-therapy” in the oral cavity.
2. Historical aspects of glycobiology in the oral cavity.
Some of the earliest discoveries in glycoscience were made studying salivary components. Salivary gland-derived mucins were among the first glycoproteins to be chemically analyzed. Eichwald, a physician from Russia working in Scherer’s laboratory in Germany was the first in 1865 to describe the presence of mucins in salivary glands and found that they were composed of protein and sugar [5]. Obolenski in Hoppe-Seyler’s laboratory isolated mucin from bovine submaxillary glands and described it in 1877 as a reducing substance with acidic properties [6]. In 1888, Hammarsten finally demonstrated that carbohydrates were a major component of mucins [7].The German chemist Hoppe-Seyler described mucins isolated from the nests of the Asian swiftlet [8]. These nests are mainly composed of the bird’s dried salivary secretion and are still up-to-today a valuable ingredient in traditional Chinese cuisine for preparing bird’s nest soup.
In 1936 the Swedish scientist Gunnar Blix characterized a particularly acidic carbohydrate as an important component of bovine submaxillary mucin [9]. He named this carbohydrate sialic acid after the Greek word for saliva, τo σιαλov [10]. Sialic acid was obtained in crystalline form from bovine submaxillary mucin in 1954 [11]. In 1958 it was discovered that sialic acid was identical to a sugar isolated from the glycolipids of neural tissue in 1941 by Ernst Klenk [12–14] which he named neuraminic acid after the Greek word νεῦρον for nerve.
Ward Pigman discovered that the sialic acid containing glycans of bovine submaxillary mucin are attached to the protein backbone at serine and threonine residues [15]. The term sialic acid actually describes a family of compounds made of neuraminic acid with various substitutions [16]. The first crystallized form was N-acetylneuraminic acid (Neu5Ac). Another widely distributed form is N-glycolylneuraminic acid (Neu5Gc). Most mammals produce both Neu5Ac and Neu5Gc, but humans have lost the ability to synthesize Neu5Gc [17]. Salivary mucins of different mammalian species carry different ratios of Neu5Ac and Neu5Gc. Mucins were shown to exhibit species specificity, tissue specificity, and interindividual differences. Modifications are introduced by substitutions with O-acetyl groups on various positions of the sialic acid molecule. O-acetylation of salivary mucins also differ between different mammalian species. This was shown in studies analyzing mucins derived from bovine, porcine, ovine, and equine submandibular glands [18–23]. As described further below, the glycans on salivary mucins play important roles in interacting with oral bacteria, fungi, and viruses (Fig. 1). They are believed to be important players in maintaining oral and systemic health. Sialic acids are particularly important for interactions with the oral microbiota as they are prominently exposed as the outermost glycan termini on most mammalian glycans and, as such, are critical determinants of the glycan motifs first encountered by approaching microorganisms. Notably from a historical standpoint, virus binding to sialic acids on salivary mucins was demonstrated as early as 1960 by Gottschalk and coworkers who showed the ability of mucins to inhibit viral hemagglutination [24]. Another important discovery was that blood group antigens are present in salivary secretions [25, 26]. Oral biologists also realized early that sialic acid and other glycans on salivary mucins serve as recognition motifs and binding sites for certain commensal strains of bacteria in the oral cavity [27–29].
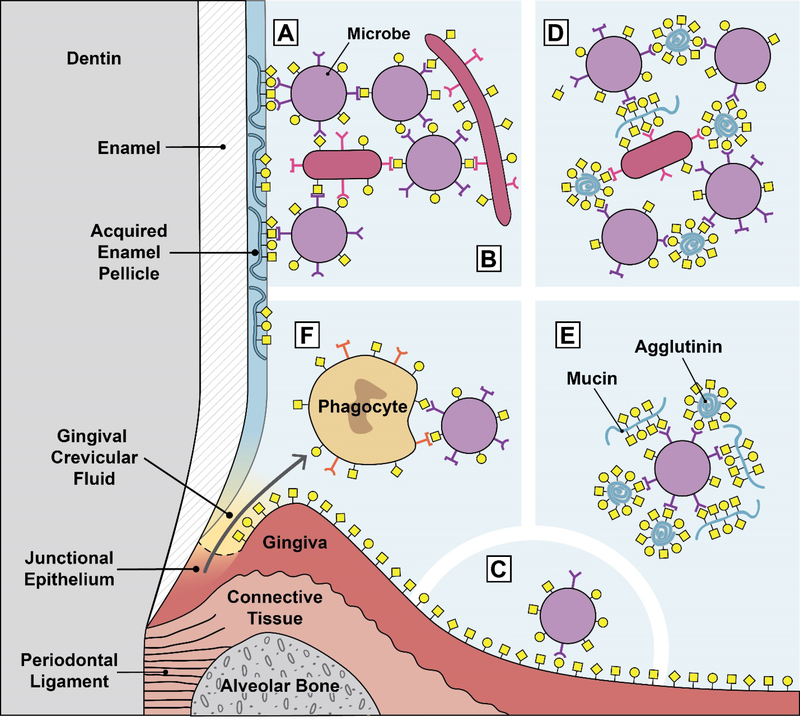
Fig. 1.
Glycan-mediated interactions in the oral environment. A. Microbial attachment to the tooth surface. Microbial adhesins attach to mostly salivary glycoproteins adsorbed to the tooth surface as part of the acquired enamel pellicle. This is typically the first step in oral biofilm formation. B. Microbial adhesins attach to glycans on other microbes. This example shows microbial coadhesion, the process of attaching to microbes that are part of a biofilm. If this microbe-microbe attachment occurs in suspension, i.e. in the planktonic phase, it is called microbial coaggregation. C. Microbial adhesins attach to glycans on oral epithelia. This interaction normally does not result in long-term colonization because the oral epithelium is a shedding surface. D. Salivary glycoproteins agglutinate microbes. Natural salivary flow causes these aggregates to be swallowed, resulting in clearance of the microbes from the oral cavity. E. Salivary glycoproteins can serve as molecular camouflage. A microbial cells is covered by multiple bound salivary agglutinins and mucins. This might protect the microbe from immune surveillance by masking the underlying microbial cell surface. F. Lectin-mediated phagocytosis. Phagocyte lectins bind to microbial glycans, and microbial adhesins bind to phagocyte glycans. These interactions can both potentially lead to phagocyte activation and phagocytosis of the bacteria.
3. Host-derived innate and immune-related compounds in human saliva.
Saliva plays an important role as a biofluid typical for the oral cavity. Besides its function in preprocessing of food for efficient mastication, and lubrication of the food bolus for swallowing, saliva contributes significantly to protection of the mineralized surface of teeth and the integrity of the epithelial integuments lining the oral cavity. How important saliva is becomes evident when salivary flow is compromised in patients with salivary gland dysfunctions that cause a dry mouth. Inadequate salivary flow causes diffulties masticating food, swallowing, and speaking, interferes with taste perception and digestion, and increases the risk of tooth demineralization, dental caries, oral mucositis, and fungal infections [2, 3, 30]. Many of the beneficial functions of saliva for maintenance of a healthy oral cavity are attributed to the proteins and glycoproteins in saliva. Whole mouth saliva is mainly composed of the secretions of the major and minor salivary glands. Blood plasma tissue exudate mostly derived from gingival crevicular fluid also contributes significantly to whole mouth saliva (Fig. 1F). Salivary gland secretions also contain glycolipids [31]. Several biological and pathological functions have been associated with glycolipids in saliva, but substantial research in this area has not been continued in recent years [32, 33].
Of interest to the immunologist are the numerous antimicrobial, antifungal, and antiviral components of saliva [30, 34–36]. These include most prominently lysozyme, lactoferrin, salivary peroxidase, histatins, cystatins, and defensins. These important components of saliva are reviewed elsewhere [37–40]. The antimicrobial activity of salivary proteins may even extend beyond the oropharynx into the proximal digestive tract, there exerting specific antimicrobial activity against enteric pathogens [41]. Besides antimicrobial components, saliva also contains growth factors and cytokines that likely play a role in wound healing in the oral cavity, oropharynx, and esophagus [42, 43]. All these components will not be covered in this review because glycan-mediated interactions thus far have not been found to play a major role in their function.
The salivary proteome contains more than a thousand proteins and peptides [44]. Yet, only about a dozen of major protein species occur in saliva abundantly enough to be visualized by gel electrophoresis [45, 46]. The majority of salivary protein content is composed of a few major protein families: basic and acidic proline rich proteins, salivary amylase, mucins, salivary agglutinin (gp340/DMBT1), salivary cystatins, histatins, and statherin. By weight, most salivary proteins are glycosylated [47]. Heavily glycosylated salivary proteins include salivary agglutinin (gp340/DMBT1), mucins, proline-rich glycoprotein, and secretory immunoglobulin A. Table 1 provides a more comprehensive list of glycosylated salivary proteins. Other common post-translational modifications of salivary proteins include phosphorylation, sulfation, ubiquitination, and postsecretory proteolytic processing as described in excellent recent reviews [3, 47, 48]. The rich and diverse glycan landscape in the oral environment is shaped by large-molecular-weight, mostly mucous glycoproteins in saliva [49, 50] (Fig. 2). Some of the most abundant and important salivary proteins are listed in Table 1.
Table 1.
Glycosylated major salivary proteins
| Protein Name | Functions | Glycosylation | Predicted Molecular Mass [kDa] | Apparent Molecular Mass Including Glycosylation [kDa] |
|---|---|---|---|---|
| Mucin-5B (MUC5B, MG1) | Lubrication, tissue protection, microbial agglutination [52] | O-linked >> N- linked [67, 265] | 596 | >1000 [266] |
| IgA secretory chain (polymeric immunoglobulin receptor, PIGR) | Microbial agglutination, antigen binding [88, 267, 268] | N-linked and O- linked [81, 85, 269] | 83 | ~390 (whole SIgA) [266, 270] |
| Salivary Agglutinin, (SAG, parotid agglutinin, gp-340, DMBT1) | Microbial agglutination [79] | N-linked > O- linked [72, 79, 80, 271] | 261 | 340 [80] |
| Mucin-7 (MUC7, MG2) | Microbial adhesion, microbial agglutination, lubrication [52, 216] | O-linked >> N- linked [49, 265, 272] | 39 | 200–250 [266] |
| Salivary peroxidase (SPO, lactoperoxidase, LPO) | Antimicrobial [38, 40] | N-linked | 80 | 70–100 [266, 273] |
| Lactotransferrin (LTF, lactoferrin, LF) | Iron binding, bacteriostatic [274] | N-linked [275] | 78 | 77 [266] |
| Immunoglobulin alpha heavy chain (IGHA1/2, IgA) | Antigen binding, microbial binding [88, 276] | N-linked (IgA1 carries O- glycans) [81–84, 268, 277] | 38 | ~64 [268] |
| Alpha-amylase 1, salivary amylase (AMY1A) | Starch digestion, bacterial adhesion and agglutination [278–280] | N-linked [281] | 58 | 55–60 [266] |
| Carbonic anhydrase 6 (CA6) | Buffering, reversible hydration of carbon dioxide [269, 282, 283] | O-linked and N- linked [266] | 35 | 42–56 [266] |
| Immunoglobulin gamma chain (IGHG1/2, IgG) | Antigen binding [269] | N-linked | 36 | ~51 [284, 285] |
| Zinc-a2-glyco-protein (AZGP1) | Lipid degradation [286] | N-linked | 34 | 41–42 [287] |
| Glandular kallikreins (KLK1, KLK2, etc.) | Serine proteases | O-linked and N- linked | 27–30 | 38–40 [288] |
| Proline-rich glycoprotein (PRG, GI) | Lubrication, microbial agglutination [140] | N-linked [140, 289] | 39 | 3 9 [266] |
| BPI fold-containing family A member 2 (BPIFA2, parotid secretory protein, PSP, SPLUNC2) | Host defense, binds bacterial lipopolysaccharide [290] | N-linked [291, 292] | 27 | 27 [292, 293] |
| Prolactin-inducible protein (PIP) | Aspartic-type proteinase | N-linked | 17 | 17–20 [294, 295] |
| Beta-2-microglobulin | Similar to MHC class 1, bacterial agglutination | N-linked | 14 precursor | 12 [296] |
| Basic salivary proline- rich proteins 1–4 (PRB1/2/3/4) | Dietary tannin precipitation [297] | O-linked and N- linked (some members) [298] | 31–41 proteolytically cleaved into smaller peptides | 5–11 [299–301] |
Fig. 2.
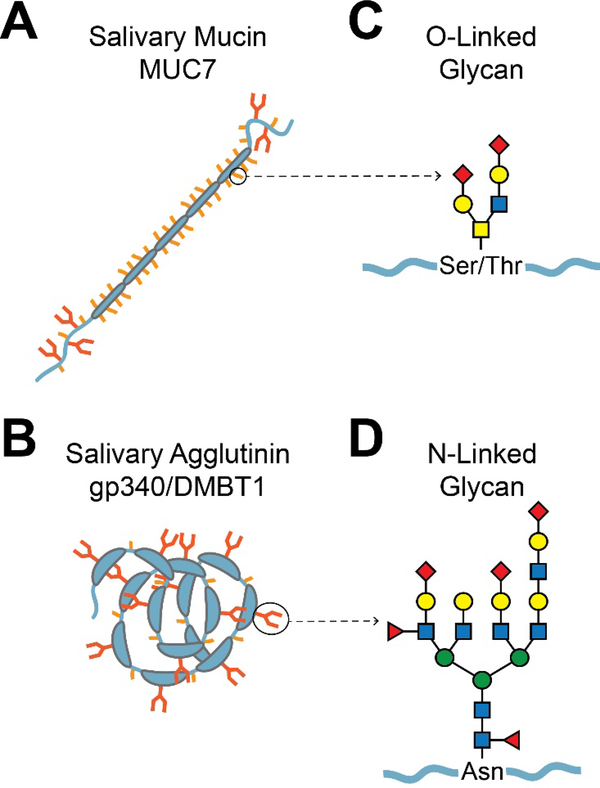
Typical salivary glycoproteins and their glycans. A. Mucins. Shown is a schematic diagram of the salivary mucin MUC7. The mucin protein backbone (apomucin) contains tandem repeats of proline, serine, and threonine-rich (PTS) domains, which are densely O-glycosylated. Non-repeat regions at the N- and C-terminals can be both O- and N-glycosylated. Dense O-glycosylation is believed to prevent typical protein folding, resulting in a bottle-brush-like shape. The larger gel-forming mucins such as MUC5B (not shown) are composed of several covalently linked mucin subunits. B. Agglutinins. Shown is a schematic diagram of the salivary agglutinin gp340/DMBT1. This protein contains several scavenger receptor cysteine-rich (SRCR) domains, which constitute the majority of the protein [263]. The SRCR domains often mediate ligand binding or protein-protein interactions with microbes. N-glycosylation occurs within the SRCR domains, and O-glycosylation occurs between the SRCR domains in serine-threonine-rich motifs that are approximately 20 amino acids in length [264]. C. Representation of a typical O-glycan chain. The base of an O-glycan is typically comprised of GalNAc α-linked to the hydroxyl group of serine or threonine. Other specific saccharide subunits and their attachments vary, but there are often two major branches with a small number of subunits. D. Representation of a typical Nglycan. N-glycans are attached to asparagine. All of the characterized eukaryotic N-glycans begin with a GlcNAcβ1-Asn linkage, but other saccharide linkages have been observed in prokaryotes. N-glycans are generally larger than O-glycans, containing more saccharide subunits and two to four major branches.
3.1. Salivary mucins.
Salivary mucins are the most densely glycosylated proteins in saliva and play an important role in oral host defense [51, 52] (Fig. 2). They are mostly decorated with O-glycans linked to serine and threonine residues in the protein backbone, but also contain some N-glycans linked to asparagine. The dense O-glycosylation confers a bottle-brush like structure to the molecule [53, 54] (Fig. 2A). Salivary mucins from different mammalian species vary widely in the types of sialoglycans attached to the mucin backbone. Variations include different types of terminal sialic acids, their modifications by O-acetylation, and the linkage to their subterminal glycan chains [22, 55]. Bacteria, fungi, and viruses have evolved many types of lectin-like adhesins that facilitate attachment to the many types of glycan moieties on salivary mucins. These interactions are described in more detail further below. The predominant mucins in human saliva are the high-molecular-weight mucin MUC5B and the lower molecular-weight mucin MUC7 [54]. MUC5B is a gel-forming mucin [56, 57] and much larger than the soluble and evolutionary younger mucin MUC7 [58, 59]. Recently, the presence of MUC19, another largemolecular-weight mucin in human saliva has been described [60]. The glycans attached to MUC5B and MUC7 have been identified, and their glycosylation patterns were found to be different [49, 50].
The glycans attached to the mucin backbone, particularly the negatively charged terminal sialic acid residues, confer viscoelastic properties to saliva. This aids in food bolus formation and lubrication thereby facilitating the swallowing of food [54]. Salivary mucins, along with other salivary proteins, adsorb to the tooth enamel, there forming a thin proteinaceous film, called the acquired enamel pellicle [61, 62] (Fig. 1). This film protects the mineralized surface of teeth from erosion by acids in the diet or acids produced by oral bacteria, the latter being a first step in the initiation of dental caries [63–65]. The lubricating effect of glycosylated mucins also protects the integrity of the tooth occlusal surfaces from attrition through antagonistic teeth and excessive wear through abrasive food components. As described further below, many oral bacteria and systemic pathogens possess adhesins that bind to sialic acids on salivary mucins and other sialoglycoproteins in saliva.
Both MUC5B and MUC7 are the major carriers of blood group glycan motifs in saliva [49, 66–68]. Blood group glycans on salivary mucins likely evolved to capture microbial pathogens, and thus play a role in host defense [57]. It has been shown for instance that the gastric pathogen Helicobacter pylori binds to salivary mucin MUC5B and salivary agglutinin through recognition of Lewis b glycan motifs [69–73], but the biological outcome of that interaction on host defense is open to debate. Agglutination of microbes in the mouth normally leads to clearance and destruction of these organisms by the stomach environment. However, in the case of H. pylori, “clearance” of this pathogen from the oral cavity deposits H. pylori in its preferred environment, the stomach. Helicobacter has also been detected in dental plaque, but in most studies oral colonization did not correlate with stomach colonization or gastric inflammation [71, 74]. One of the main receptors for H. pylori on the gastric epithelium is the mucin MUC5AC, which carries Lewis b glycan motifs [75, 76] just like the salivary mucin MUC5B [69, 73]. If salivary MUC5B was bound by H. pylori in the mouth and still remained bound once in the stomach, it could possibly prevent binding to MUC5AC and, thus, inhibit stomach colonization. Salivary mucin MUC5B bound to the surface of Helicobacter may also serve the pathogen as a molecular camouflage to prevent its detection by the immune system [70] (Fig. 1E).
3.2. Salivary agglutinins.
Particular components of saliva, called agglutinins which include the above-mentioned mucins, can clump bacteria, viruses, and fungi as long as these organisms are suspended in the fluid, i.e. in planktonic phase, leading to their clearance from the oral environment through swallowing [77]. A number of such agglutinins have been identified by in vitro studies. Among them are the salivary mucins MUC5B [50] and MUC7 [49, 59, 78] (Fig. 2A), salivary agglutinin gp340/DMBT1 [72, 79, 80] (Fig. 2B), secretory immunoglobulin A [81–84], and free secretory component [85]. Salivary secretory IgA not only plays an important role in mucosal immune defense in the mouth [86, 87], but is also recognized by glycan-binding adhesins of oral actinomyces and streptococci [88]. Only a few and sometimes contradictory studies have been performed in the past to investigate the in-vivo significance of salivary agglutinins for oral and systemic health. There is evidence that salivary agglutination of S. gordonii enhances its phagocytosis by neutrophils [89]. It remains still a matter of dispute whether and how salivary agglutinins participate in clearance of oral bacteria causing dental caries, such as Streptococcus mutans and S. sobrinus [90–92], or in agglutination of systemic pathogens such as Staphylococcus aureus [93] and Helicobacter pylori [70] (Fig. 1D). Conversely, agglutinins have been described to facilitate bacterial attachment to the tooth surface thereby promoting colonization [72, 94–96]. Agglutinins bound to bacteria may potentially also lead to interspecies attachment during the establishment of oral biofilms [77]. The role of glycoprotein agglutinins in biofilm formation is discussed in more detail in section 8.
4. Evolution & coevolution of host glycans and microbes in the oral cavity.
The wide variety of mutual interactions between host glycoproteins and oral microorganisms have been shaped by coevolution of the microorganisms with their host [97]. This coevolution has led to an ecosystem that generally benefits both the host and its indigenous microbiota. In the oral cavity, glycoproteins in saliva can serve as both substrates for attachment and as agents of bacterial clearance. Salivary glycoproteins on the tooth surface or the oral epithelium serve as anchors for bacterial adhesins, allowing the bacteria to take a foothold in the oral cavity and multiply. This can eventually lead to the formation of dental and oral biofilms, a necessary adaptation to avoid being flushed away by the constant salivary flow in the oral environment [30]. Conversely, salivary agglutinins are believed to encourage clearance of bacteria by facilitating the swallowing of bacteria along with saliva [77]. Colonization by beneficial bacteria is tolerated because it creates an environment that likely prevents colonization and infection by more pathogenic microbes. Human and bacterial coevolution has thus developed mechanisms to encourage attachment and colonization of commensal bacteria while discouraging the growth of pathogens. Oral bacterial adhesins have presumably gone through millions of years of rapid evolution to maximize binding to specific aspects of the glycome or sialoglycome in their ecological niche or host species [97]. It is also likely that bacterial adhesins have evolved the means to recognize glycans produced by other microbes. These may include common nonulosonic acids and “exotic” non-mammalian glycans. This bacteria-bacteria binding may occur between members of the same species or with other members of the biofilm community.
The fact that salivary glycoproteins, in particular the salivary mucins, are decorated with an incredible variety of different glycans can be explained by the so-called Red Queen effect. Microbes evolved to express novel glycan-binding adhesins to colonize host surfaces, and the host in turn evolved novel glycan motifs to which pathogenic microbes cannot bind. According to the Red Queen effect, this race continues by microbes again evolving novel or modified adhesins that are able to recognize those new host glycan motifs, and so on [98]. One important and recently evolved glycan motif in humans is sialic acid. The rich variety of structural subtypes of sialic acids on salivary mucins and their differences in different mammalian species are likely the result of microbial host coevolution [99, 100]. Also, the loss of Neu5Gc in the human lineage about 2 million years ago could have been the result of selective pressure by an unknown pathogen on our hominin ancestors [101]. Microbial-host coevolution may not only affect the structure of glycans, but also their numbers, densities, and sterical presentations on their protein or lipid backbone, and perhaps in a larger sense on any cell or biological surface [102]. An example for that effect may be the observed variations in the number of densly O-glycosylated PTS repeat domains in salivary agglutinins [103] and mucins [104, 105] which are known to interact with microbes through glycan-mediated binding (sections 3.1 and 3.2). For example, variations in subexonic PTS copy number repeats in salivary mucin MUC7 underwent rapid evolution in the primate lineage and among other mammalian species [59], and similar mechanisms involving possible pathogenic pressure may explain the observable variations in MUC7 among geographically distinct human populations [58].
5. Oral microbial glycan-binding molecules.
Attachment to glycans on host tissues is a common first step toward colonization, and specific glycan structures sometimes contribute to the characteristic tropism to particular sites of infection observed in many microbes (Fig. 1A and C). Many oral bacteria have evolved an astonishing tropism for the oral cavity and even for specific sites within that space, e.g. the tooth surface, the interdental approximal space, the periodontal pocket, the surface of the tongue, or the oral mucosal epithelium. Mutans streptococci in particular are extremely specialized for their ecological habitat on the surface of the tooth, and are rarely found in the oral cavity before tooth eruption [106–108].
Initial attraction of oral bacteria to surfaces in the oral cavity is mediated by physicochemical forces that lead to a primary association of the bacteria with the surface [109]. For more permanent adhesion to take place, bacteria express highly specialized proteins, called adhesins, which bind to specific cognate motifs on host molecules. These specific binding interactions are mediated by stereochemical and electrostatic forces, and can be based on proteinprotein or protein-glycan binding. While the recognition of amino acid motifs by oral bacterial adhesins certainly plays an important role for binding to a variety of biomolecules [110–113], this review focuses on the recognition of glycan motifs mediated by glycan-binding adhesins expressed by oral bacteria. Glycan-binding proteins are often highly specific, but have low affinity [114–116]. Low affinity interactions can become biologically relevant because microbial cells often express many copies of an adhesin protein on their surface, and glycoproteins, particularly mucins, often carry clustered glycan receptors. The effect is not simply additive, because the initial interaction increases the chances of further interactions, thus increasing avidity of microbial binding. In most of the examples presented below, the bacterial adhesins do not function as independent points of attachment between bacteria and host, but instead behave in a concerted fashion. Multiple weak adhesin-glycan links work together to form a lasting bond that can be competitively inhibited only by very high concentrations of soluble glycans, an effective mechanism that microbes may have evolved to strengthen their attachment to host surfaces.
Bacterial adhesins are frequently associated with large, non-flagellar surface structures, called pili or fimbriae, that extend away from the bacterial surface. Due to their length, these pili are likely the first bacterial proteins to come into contact with host surfaces [117]. Bacterial adhesins are themselves often glycosylated. The purpose of this glycosylation is not firmly established, but it may help maintain the physical shape of the adhesin, preventing folding and extending the protein away from the bacterial cell surface [118–120]. Oral microbes encounter a rich diversity of glycan structures in the human mouth. This diversity comes from the variety of mucous glycoproteins present in saliva [49, 50] (Table 1), food intake by the host, and the innumerable glycan motifs expressed by other members of the oral biofilm community [121, 122]. Considering this diversity of potential binding targets, the range of oral bacterial glycanbinding adhesins is likely infinitely greater than what is currently known.
A selection of well-characterized microbial glycan-binding adhesins is presented in Table 2. Below, we will provide a more detailed description of several representative, well characterized adhesin families including the serine rich repeat (SRR) protein adhesins found on oral streptococci (subsection 5.1) and the type 2 fimbrial lectin-like adhesins of oral actinomyces (subsection 5.2). For both of these adhesin families, ample and detailed structural knowledge exists for both the glycan-binding bacterial adhesins and the glycan motifs recognized by the corresponding adhesin glycan-binding pocket.
Table 2.
Oral microbial glycan-binding proteins
| Microbial Protein Adhesin1 | Microbial Species | Host or Microbial Glycans Recognized |
|---|---|---|
| Bacteria | ||
| Serine-rich repeat protein (SRR) adhesins (e.g. Hsa, GspB, SrpA) | Streptococcus gordonii, Streptococcus sanguinis | Sialic acid, Neu5Ac/Neu5Gcα2–3~ [137] |
| Glycan-binding protein A, GbpA/B/C/D | Streptococcus mutans | Glucose (dextran) [302] |
| Antigen I/II family, SpaP, SSP-5, SspA/B | Streptococcus gordonii, Streptococcus mutans, Streptococcus sanguinis | Sialic acid [303], fucose, lactose [304], N- acetylgalactosamine [305] |
| Type-2 fimbriae coaggregation factor A (CafA) | Actinomyces oris, Actinomyces naeslundii | Galß1–3GalNAc, GalNAcβ1–3Gal [156, 167] |
| NanH | Tannerella forsythia | Sialic acid [205, 306] |
| Fungi | ||
| Als1 | Candida albicans | Fuca1–2Galß1–4GlcNAc, Fucose [185] |
| Epa1/6/7 | Candida glabrata | Galß1–3~, Galβ1–4~ [188, 189] |
| Viruses | ||
| Capsid proteins | Hand-foot-and-mouth disease virus (FMDV) | Heparan sulfate [173] |
| Hemagglutinin (HA) | Influenza A and B | Neu5Aca2–3Gal, Neu5Acα2–6Gal [177, 178] |
| gB/C/D | Herpes simplex virus (HSV) | 3-O-sulfated heparan sulfate [174, 175] |
| Gp120 | Human immunodeficiency viurs (HIV) | Heparan sulfate [176] |
Table 2 lists only a select number of oral microbial glycan-binding proteins for which both the adhesin and the cognate glycan receptors have been identified and well characterized. Many more glycan-binding activities of oral microorganisms have been described or postulated but could not be listed here due to space limitations.
5.1. Streptococcal serine-rich repeat protein adhesins.
Viridans streptococci are prevalent among the earliest colonizers of the tooth surface [123]. Oral streptococci express multiple adhesins [124] which allow them to recognize a wide spectrum of adhesion substrates, including salivary glycoproteins present in the acquired enamel pellicle on the tooth surface. [125, 126]. The existence of a streptococcal lectin that binds host sialic acids on salivary mucins, was first discovered in the late 1970’s [28, 29, 127]. Later, a lectin-like adhesin expressed by S. gordonii DL1, named Hsa, was isolated based on its ability to bind sialic acid on red blood cells, and was found to be a high molecular weight glycoprotein associated with fibrillar structures on the bacterial surface [128]. This adhesin was cloned by Takahashi and coworkers and determined to be a streptococcal serine-rich repeat (SRR) protein [129]. An especially detailed description of the serine rich repeat family of adhesins is included in the following to illustrate the variation in glycan binding specificity possible within such a closely-related group of adhesins.
The SRR glycoproteins comprise a unique family of bacterial adhesins that bind a wide range of ligands, and have a significant impact on biofilm formation and virulence [130–136]. The overall domain organization of the SRR glycoproteins is conserved, with an atypical Nterminal signal peptide followed by a short SRR domain, a ligand binding region, a long SRR domain, and a C-terminal LPXTG cell wall anchoring motif. The long SRR domain is highly glycosylated. These cell wall-associated glycoproteins most likely facilitate the early colonization of the oropharynx by streptococci, and are associated with pathogenicity in the setting of an infective endocarditis model [130]. Although the ligands for the SRR glycoproteins are not known for most species, it is clear that they include both glycans and proteins. The binding regions vary considerably between species, which likely accounts for the range of ligands bound by this family of adhesins.
Through binding of whole bacteria and of their recombinant ligand binding domains to a sialoglycan array, it was found that each unique sialic acid binding SRR protein adhesin has a distinctive binding profile that is influenced by both the subtype of sialic acid, its linkage, and the underlying glycan chain [137–139]. For example, Hsa of S. gordonii DL1 showed broad specificity for a sialic acids α2–3-lined to a wide range of subterminal glycans, but GspB of S. gordonii M99 showed a narrower specificity, binding primarily sialyl-T antigen (Neu5Acα2–3Galβ1–3GalNAc) terminating glycans [137]. The binding patterns of purified SRR ligand binding domains closely matched the binding patterns of whole cells, suggesting that the SRR proteins are sufficient to explain streptococcal sialic acid binding. Furthermore, isogenic mutants lacking SRR genes did not bind to any sialoglycans on the array [137]. The binding regions of SRR proteins are stable modules that can be efficiently expressed in a standard bacterial host such as E. coli. This stability, combined with the selectivity for specific underlying glycan structures, makes SRR adhesins potentially useful tools for studying glycobiology, which will be discussed in the last section of this review (section 9). Interestingly, none of the SRR protein adhesins bound to α2–6-linked sialic acids, even though this sialic acid configuration is quite prevalent in the oral cavity and on salivary glycoproteins [49, 50, 72, 73, 140].
Among the best structurally characterized of the SRR glycoproteins is gordonii surface protein B (GspB) of Streptococcus gordonii strain M99. GspB mediates the binding of streptococci to human salivary glycoproteins and surface glycoproteins on platelets [141, 142]. Recent crystallographic studies have revealed that the binding region of GspB is comprised of three independently folded domains, with the second domain exhibiting the same topology as eukaryotic Siglecs [143]. Domain deletion studies indicate that only two of the domains, the Siglec and Unique domains, are required for sialic acid binding. The purpose of the third domain, CnaA, is unclear, but its structure resembles MSCRAMM-related peptide-binding domains, first identified within the S. aureus CNA protein adhesin [131, 138]. SrpA of S. sanguinis SK36 is another well-characterized, lectin-like SRR adhesins that binds sialic acids [129, 141, 142, 144–146]. Interestingly, Hsa, GspB, and SrpA mediate shear-enhanced binding to their cognate receptor glycans on salivary glycoproteins and on platelets [147, 148]. In that respect it may be of relevance that CnaA domains have been shown to be efficient dissipaters of mechanical forces [149].
5.2. Other bacterial glycan-binding proteins.
Similar to viridans streptococci, oral actinomyces are among the earliest colonizers of the tooth surface [150]. Unlike streptococci, oral actinomyces have also been associated with initiation of periodontal disease [151, 152]. Two types of fimbrial adhesin have been characterized in actinomyces. Type-1 fimbriae were found to be responsible for binding to salivary proline-rich proteins through recognition of an amino acid motif [112, 113]. Type-2 fimbriae were shown to carry a galactose-specific lectin [150] that mediates bacterial coaggregation with certain strains of streptococci [153, 154]. The receptor glycan motif for type-2 fimbriae on streptococcal partner cells lies within the so-called streptococcal receptor polysaccharides (see section 7) [121]. Further investigation into the glycan-binding specificity of the type-2 fimbriae showed a preference for Galβ1–3GalNAc, GalNAcβ1–3Gal, and to a lesser extent other glycans with a terminal Galβ1–3 linkage [155–158]. The glycan motif Galβ13GalNAc, also called the T antigen, is one of the most common O-glycan cores, but in human tissues it is typically cryptic, meaning other saccharides are attached to the terminal end of the glycan chain [159]. Type-2 fimbriated actinomyces cannot bind to sialyl-T antigen unless terminal sialic acid is removed by the action of sialidase [160, 161]. Notably, actinomyces express a sialidase [162] to gain access to their preferred glycan receptor (further discussed in sections 7 and 8). Type-2 fimbriated actinomyces bind to eukaryotic cell surfaces by recognizing cognate Gal/GalNAc-containing glycan motifs on epithelial cells [163] and phagocytes [164, 165]. Salivary glycoproteins including salivary mucin MUC7 [166] and secretory immunoglobulin A [88] are also bound by type-2 fimbriae. Molecular cloning of the fimbrial subunits revealed the fimbrial tip as the adhesin protein responsible for binding Gal/GalNAc-containing glycans, and was named coaggregation factor A (CafA) [167].
Streptococcus mutans is strongly implicated in the development of dental caries [168]. In consequence, the adhesins that allow it to attach to oral surfaces have been intensely studied for many years. Initial attachment of S. mutans to the tooth surface is most likely initiated by the SpaP protein, an adhesin of the antigen I/II family that has been shown to bind salivary agglutinin [96]. Similar to the SRR family, antigen I/II family adhesins are fibrillar proteins found in many species of oral streptococci that display a range of binding substrates including proteins and glycans [169–171]. After initial attachment, S. mutans glucosyltransferase (Gtf) enzymes produce an extracellular matrix of glucan polysaccharides that help establish a permanent biofilm [172]. S. mutans attaches to this glucan matrix via lectin-like glucan-binding proteins (Gbp) [170].
5.3. Viral and fungal glycan-binding proteins.
Oral viruses and fungi also express lectin-like adhesins that aid their attachment to host tissues. Viruses that enter though the oral cavity often bind heparan sulfate, a polysaccharide commonly found in all animal tissues. Viral proteins that attach to heparan sulfate include herpes simplex glycoproteins (gB, gC, and gD), enterovirus capsid proteins, and HIV gp120 [173] [174, 175] [176]. The viruses likely use heparan sulfate as part of a co-receptor system in which initial contact is made with heparan sulfate, followed by adhesion to a more specific cellular receptor. The hemagglutinin (HA) protein of the influenza virus is possibly the most widely known sialic acid binding protein [177, 178]. A shift in binding preference from α2–3-linked sialic acid to α2–6-linked sialic acid is believed to facilitate the transition from avian to human hosts [179]. The binding of HA to sialic acid on host cell surfaces is a crucial step in the influenza lifecycle, but salivary proteins decorated with sialic acid can intercept HA binding, leading to agglutination and clearance of the viral particles [180, 181]. The salivary mucin MUC5B in particular has been shown to have antiviral effects against influenza virus [182]. As many more pathogenic viruses use the oral cavity as a port of entry [183], likely a high number of viral glycan-binding interactions with salivary glycoproteins are yet to be discovered.
Candida albicans is a commensal yeast species that is a common part of the human oral flora but can cause oral candidiasis in immunocompromised patients or in individuals with a dry mouth [184]. It expresses agglutinin like sequence (Als) proteins that bind to a broad range of mammalian proteins, and likely also interact with bacteria. Als1 binds to fucose-containing glycans, and has been shown to bind fibronectin and laminin glycoproteins in vitro [185]. Binding of the N-terminal domain of the Als1 protein (Als1p-N) was tested on a glycan array. Als1p-N bound most readily to Fucα1–2Galβ1–4GlcNAc, but it also bound to many glycans with a terminal fucose, and even to fucose alone [185]. Candida albicans also binds to salivary mucin MUC7 [186] and salivary glycans inhibit binding of C. albicans to oral epithelial cells [187]. Thus, additional glycan-binding adhesins expressed by C. albicans likely play a role in binding to salivary glycoproteins. Candida glabrata is less common than C. albicans, but has been shown to also cause oral candidiasis. This species expresses several epithelial adhesin (Epa) proteins that bind to glycans [188]. Epa1 appears to be the main mediator of adherence to human epithelial cells. It binds to most glycans with a terminal galactose β-linked to galactose or glucose [188]. Deletion of Epa1 substantially reduced Candida adherence to epithelial cells [189].
6. Bacterial surface glycans.
In conjunction with their glycan binding, oral bacteria produce a wide variety of extracellular glycans of their own [121, 190]. This combination of glycan production and glycan attachment results in elaborate, interconnected patterns of interspecies coadhesion and interaction (Fig. 1B) [125]. For example, oral streptococci produce long extracellular glycans with a repeating pattern, called streptococcal receptor polysaccharides (RPS). There are six common variations in RPS structure, and all of them are bound by actinomyces type-2 fimbrial adhesins. Only four of the RPS structures are also bound by oral streptococci [121, 191]. Additional variations in streptococcal polysaccharide structure have been discovered that differ from the six common RPS types [190, 192, 193]. Based on analogy with the RPS glycan motifs, it can be predicted that unknown partner bacteria might exist in oral biofilms that express hitherto undiscovered adhesins recognizing these rare glycan motifs to engage in bacterial coadhesion.
Many bacterial genera present in oral biofilms, including Fusobacterium, Neisseria, Porphyromonas, Streptococcus, and Tannerella, express sialic acids or other nonulosonic acids on their surface [194–196]. Bacterial sialic acid may be synthesized de novo, scavenged from host glycans, or produced from host-derived intermediates [197, 198]. Since sialic acid is a very common human glycan structure, its presence on the bacterial surface may prevent attack by the immune system by serving as a type of molecular camouflage. For instance, H. influenzae gains a survival advantage in human serum from presenting sialic acid on lipopolysaccharide (LPS) [199]. Sialylation of Porphyromonas gingivalis LPS appeared to have no effect on its inflammatory potential [196]. The effects of bacterial sialylation may not always be protective when considering the variety of glycan structures underlying the sialic acid and the variety of sialic acid-binding Siglec receptors on eukaryotic cells [200].
7. Bacterial foraging on oral host glycans.
Scavenging glycans from host glycoproteins appears to be a common strategy of oral and intestinal bacteria [197, 201]. Strains of S. gordonii, S. oralis, S. mitis, and S. sanguinis are able to survive using saliva as a sole carbon source [202]. These strains show a preference for Nglycans linked to the proline-rich glycoprotein (PRG) [202]. For S. gordonii, this specificity for PRG is due to the sequential action of three glycoside hydrolases that are able to break down the most common glycan structure on PRG [203]. The ability to scavenge host sialic acids has also been observed in Haemophilus influenzae, Tannerella forsythia, and Vibrio cholerae. These species carry an orthologous N-acetylneuraminic acid lyase (nanA) gene, which is necessary in the first step of sialic acid catabolism [198, 204, 205]. The nanA gene has been found in other bacterial genera including Streptomyces, Streptococcus, Staphylococcus, Clostridium, Lactobacillus, Escherichia, Salmonella, and many more, suggesting that they may also be able to scavenge host sialic acids from their environment [204].
Bacteria that can metabolize sialic acid often express sialidase enzymes in order to liberate terminal sialic acids from glycans. NanH is an interesting sialidase expressed by T. forsythia and several species of Actinomyces [160, 161, 205]. The NanH enzyme is believed to play a role in bacterial attachment to human epithelial cells. It first acts as a lectin, anchoring the bacteria to the host cell, then it removes sialic acid from the host glycoprotein, providing a useful carbon source [205]. The removal of sialic acid from O-linked glycans may reveal the T antigen (Galβ1–3GalNAc) or other beta-galactoside structures for bacterial access [206]. This, for instance, allows binding of the actinomyces type-2 fimbrial adhesins to their receptor glycan motifs as described in section 5.2. Sialidase-dependent attachment of Actinomyces naeslundii and Actinomyces viscosus to human erythrocytes [207] and polymorphonuclear leukocytes [164, 206] has been described (Fig. 1F). Similar sialidase-dependent attachment has been observed for S. gordonii, S. oralis, and S. pneumoniae, but these species rely on different adhesins, which bind to β1–4-linked galactose [208–211]. Streptococcus gordonii does not produce its own sialidase, but instead relies on sialidases produced by other vicinal bacterial species in the biofilm environment [208]. Depending on their host species and specific site of colonization, oral bacteria should be expected to express sialidases that match the types of sialic acids prevalent in their surroundings. In this context, a sialidase was characterized from oral S. sanguinis that can release O-acetylated sialic acids which are resistant to cleavage by commonly used microbial sialidases [212]. The ability of bacteria to modify glycans in their environment, combined with their ability to bind the underlying, previously cryptic glycans, adds yet another level of complexity to interspecies coadhesion and interaction [125].
8. Role of glycans and glycan binding in oral biofilm formation.
Bacteria normally do not adhere directly to the mineralized tooth surface. In the mouth, tooth enamel is coated with the acquired enamel pellicle, a thin film of adsorbed salivary proteins and glycoproteins [61]. The interactions between lectin-like adhesins on bacteria and complementary glycan motifs on glycoproteins in the salivary pellicle play central roles in initial bacterial colonization [213] (Fig. 1A). Several human salivary proteins have been identified as counter-receptors for oral bacteria and their glycan-binding adhesins [88, 140, 166, 214, 215]. Major salivary glycoproteins such as the salivary mucin MUC7 serve as prominent targets for bacterial binding [166, 215]. Also systemic respiratory or gastrointestinal pathogens were found to bind to specific glycan motifs on MUC7 and on other salivary glycoproteins [69, 70, 73, 93, 216].
Lectin-glycan binding is also important for bacterial coadhesion and coaggregation, which lead to the formation of multispecies microbial biofilms that have the potential to become pathogenic communities causing dental, oral, and perhaps also systemic disease. Bacterial coaggregation is generally defined as the formation of bacterial aggregates due to bacterial cellcell recognition of genetically distinct partner strains [217]. Bacterial coadhesion is defined as the binding of a planktonic bacterium to an already surface-attached partner strain, as in the formation of early multispecies dental plaque biofilms [218] (Fig. 1B).
The spatial organization of oral biofilms plays an important role in their development and survival [219, 220]. Species with mutualistic symbiotic relationships benefit from close proximity, and beneficial interspecies coaggregation and coadhesion is often mediated by lectinglycan binding [154, 221, 222] (Fig. 1B). The enormous diversity of species commonly found in the oral environment [223–225], combined with the potentially infinite number of possible glycan motifs, [121, 122] results in an extremely complex ecosystem that is constantly being destroyed by chewing or brushing, and rebuilt by microbial coadhesion and coaggregation.
Despite the overwhelming number of species and interactions in the oral environment, progress is being made in investigating which microbial species are associated with oral health or disease, and in determining their mechanisms of attachment [226]. Also, numerous laboratory strains and clinical isolates have been tested for inter-bacterial binding in vitro by the conventional bacterial coaggregation assay [154, 227]. Thus far however, the mechanisms of inter-bacterial adhesion down to the molecular level have been studied for only a few of these interactions [121, 167]. The traditional in-suspension coaggregation assay has provided a wealth of information for interbacterial adhesion, but is not well suited to handle the multitude of microbial species present in oral biofilms. A method was recently developed that enables investigators to screen multi-species biofilms for coadhesion partners with the option to isolate and identify them [228]. This coadhesion assay, based on immobilization of one bacterial partner strain [229], allows the use of well-known reference strains as probes to efficiently screen mixed bacterial cultures for unknown co-adhering partner organisms that express complementary adhesins or glycan receptors [228]. Clearly, a better understanding is warranted of how oral biofilm bacteria act together in vivo. Imaging and functional approaches used in more recent insitu studies will likely provide further insight in glycan-mediated mechanisms of interbacterial interactions [230].
Dysbiosis in oral microbial biofilms can lead to the onset of dental caries and periodontal disease [4, 231]. If bacteria get dispersed in the blood circulation, they can even become causative agents of systemic disease [226, 232]. One example that is clearly based on glycan-recognition is infective endocarditis caused by streptococci of the oral viridans group that are normally harmless commensal inhabitants of the oral cavity. As mentioned further above, they are often among the first bacteria to attach to the saliva-coated surfaces of teeth [125, 233]. However, when oral streptococci gain access to the blood circulation through accidental microlesions in the oral epithelia or through invasive dental treatment, these organisms can become causative agents of infective endocarditis and sepsis [108, 234–236]. The binding of bacteria to platelets has been recognized as a central process in the pathogenesis of infective endocarditis [237, 238], and that binding is partially mediated by lectin-like bacterial adhesins. The Sigleclike binding regions of streptococcal SRR adhesins selectively target GPIbα on human platelets [137], where they adhere specifically to O-linked glycans on the mucin-like stalk of GPIbα [130, 131, 137, 141, 214]. This binding may cause a tropism of oral streptococci to platelets on injured heart valves. An alternative mechanism is suggested by a recent study which found that streptococci adhere preferentially to platelets in the context of whole blood, and may thus be carried by circulating platelets to the damaged endocardial surfaces [137]. The contribution of Siglec-like adhesins in the development of endocarditis has been demonstrated in vivo using a rat model [130, 239]. Recently, streptococcal recognition of sialoglycans on plasma glycoproteins was investigated and suggested to modulate the propensity of streptococci to establish endocardial infections [240].
9. Perspectives for use of glycans or glycan-binding molecules in prevention or diagnosis of oral disease.
Since initial attachment is a crucial first step in the development of pathogenic oral biofilms, many research groups have attempted to identify treatments that inhibit the binding ability of bacterial lectin-like adhesins. A long-sought potential solution to these problems is immunization against the bacterial adhesins [241, 242]. Specifically, the glucosyltransferase (Gtf) and glycan-binding protein B (GbpB) of S. mutans have been the target of vaccine development for many years [243]. Rodent studies have demonstrated that vaccines containing the Gtf or GbpB protein result in detectable levels of specific salivary IgA and serum IgG. Importantly, this immunization results in significantly less total caries [244–246]. It is believed that IgA and IgG present in saliva bind to Gtf or GbpB, thereby preventing bacterial adhesion to extracellular bacterial polysaccharides in dental plaque and leading to agglutination and clearance of the S. mutans organisms [246]. Immunization with dead or attenuated S. mutans cells was not pursued because antibodies against S. mutans whole cells showed crossreactions with human heart tissue [247–249].
Several human trials have tested vaccines based on Gtf, GbpB, and other S. mutans surface proteins. Immunization consistently resulted in detectable salivary IgA antibodies that target the desired antigen [250, 251]. Small-scale human trials suggest that vaccination using Gtf reduces the proportion of S. mutans in the oral cavity [252, 253]. Regrettably, no studies on the effectiveness of these vaccines in preventing dental caries have been published, and no large scale clinical trials have been initiated. Future attempts will have to consider the existence of multiple other cariogenic species besides S. mutans, the maintenance of protective levels of antibodies in saliva, and the public reluctance to yet another vaccine for pediatric use [254]. Passive immunization against S. mutans may circumvent some of these concerns, and has been tested in several animal models. Topically applied antibodies to S. mutans surface proteins, including GbpB, significantly reduced caries formation in rats and macaques [255, 256]. There is also evidence that passive immunization in humans can reduce the bacterial burden of S. mutans in the oral cavity [257–259], but thus far it was not proven that passive immunization can reduce the incidence or severity of dental caries in humans. The lack of clinical data showing efficacy of these vaccines in preventing dental caries is unfortunate, but could present a major opportunity for future dental research.
As mentioned before, the number of well-characterized bacterial lectins represents likely only a miniscule fraction of the total potential lectin diversity in the oral cavity. Further investigating the unidentified bacterial glycan-binding molecules will allow to better understand the role of glycans in microbe-host and microbe-microbe interactions in the human mouth and, ultimately, to study their relevance for oral disease, including the two major diseases in the oral cavity, dental caries and periodontal disease. Once the molecular basis of these glycan-mediated bacteria-host interactions is revealed, it might be possible to refine the use of custom-designed glycan or lectin analogues, and specifically target the adhesins of pathogenic microbes in the oral cavity. Similar glyco-therapeutic approaches are already under investigation for other pathogens [260–262].
Besides their role in human health, the extreme variety of lectins in the oral microbial community represents an untapped source of highly specific, affordable, and easy-to-use molecular glycan probes. The majority of the currently available glycan-binding molecules, i.e. the lectins, were traditionally discovered by their ability to agglutinate red blood cells. Because of this selection, there is a bias towards host glycans present on erythrocytes, which represent only a limited portion of the diversity of glycans found in nature. This approach has likely missed many existing natural lectins that bind to other kinds of glycan motifs, e.g. the polysaccharide motifs expressed on bacteria [121, 122]. Considering (a) the multitude of bacterial species identified in oral biofilms, (b) the rich diversity of species-typical sialoglycans found on salivary glycoproteins, and (c) the innumerable glycan motifs expressed on the surface of other bacteria, it is likely that many more unidentified lectin-like bacterial adhesins exist in the oral microbiome, beyond the few ones currently known. It seems only logical and timely to harvest these unidentified glycan-binding proteins already honed by evolution, and shape them into tools for glycan recognition.
The creation of a toolkit of novel, highly specific bacteria-derived glycan-binding probes will be groundbreaking not only for identification and study of glycans in oral biology, but also for use as glycan-tracking reagents for salivary diagnostics pertaining to oral and systemic disease. In a broader perspective, novel bacterial glycan-binding proteins will become useful tools to probe the glycomes of all types of cells and tissues, allowing for simple glycan analysis in most fields of vertebrate biology. Such glycan-binding probes, derived from oral bacteria and engineered to suit experimental needs, might be used with any medical or biological sample in a number of assays including array technology, flow cytometry, lectin histochemistry, lectin blotting, and lectin ELISA. Many more uses for lectin probes will likely become obvious as we continue to discover new classes of lectins and new examples of lectin-glycan interactions in nature.
HIGHLIGHTS.
Saliva plays an important role for host defense in the oral cavity.
Glycoproteins in saliva create a diverse glycan landscape in the mouth.
Oral bacterial adhesins bind to glycan motifs on salivary glycoproteins.
Oral bacterial glycan binding contributes to biofilm formation.
The majority of glycan-binding molecules in the mouth still remains unexplored.
ACKNOWLEDGEMENTS
Funding: This work was supported by the National Institutes of Health [R01 DE019807, R21 DE025826, and U01 CA221244]. BC was supported by NIDCR training grant T32DE023526.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Gaffen SL, Herzberg MC, Taubman MA, Van Dyke TE, Recent advances in host defense mechanisms/therapies against oral infectious diseases and consequences for systemic disease, Adv Dent Res, 26 (2014) 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sreebny LM, Saliva in health and disease: an appraisal and update, Int Dent J, 50 (2000) 140–161. [DOI] [PubMed] [Google Scholar]
- [3].Dawes C, Pedersen AM, Villa A, Ekstrom J, Proctor GB, Vissink A, Aframian D, McGowan R, Aliko A, Narayana N, Sia YW, Joshi RK, Jensen SB, Kerr AR, Wolff A, The functions of human saliva: A review sponsored by the World Workshop on Oral Medicine VI, Arch Oral Biol, 60 (2015) 863–874. [DOI] [PubMed] [Google Scholar]
- [4].Lamont RJ, Hajishengallis G, Polymicrobial synergy and dysbiosis in inflammatory disease, Trends Mol Med, 21 (2015) 172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Eichwald E, Beiträge zur Chemie der gewebbildenden Substanzen und ihrer Abkömmlinge. I. Über das Mucin, besonders der Weinbergschnecke, Annalen der Chemie und Pharmacie, 134 (1865) 177–211. [Google Scholar]
- [6].Obolensky S, Über das Mucin aus der Submaxillardrüse, Pflüger, Archiv für die Gesammte Physiologie des Menschen und der Thiere, 4 (1871) 336–346. [Google Scholar]
- [7].Hammarsten O, Ueber das Mucin der Submaxillardrüse, Zeitschrift für physiologische Chemie, 1888, pp. 163. [Google Scholar]
- [8].Hoppe-Seyler F, Zeitschrift für physiologische Chemie, I. Teil (1877) 94. [Google Scholar]
- [9].Blix G, Über die Kohlenhydratgruppen des Submaxillarismucins, Hoppe-Seyler´s Zeitschrift für physiologische Chemie, 240 (1936) 43. [Google Scholar]
- [10].Blix G, Svennerholm L, Werner I, The isolation of chondrosamine from gangliosides and from submaxillary mucin, Acta Chem Scand, 6 (1952) 358–362. [Google Scholar]
- [11].Klenk E, Faillard H, Zur Kenntnis der Kohlenhydratgruppen der Mucoproteide, Hoppe Seylers Z Physiol Chem, 298 (1954) 230. [PubMed] [Google Scholar]
- [12].Comb DG, Roseman S, Composition and enzymatic synthesis of N-acetylneuraminic acid (sialic acid), J Am Chem Soc, 80 (1958) 497–499. [Google Scholar]
- [13].Cornforth JW, Firth ME, Gottschalk A, The synthesis of N-acetylneuraminic acid, Biochem J, 68 (1958) 57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Klenk E, Neuraminsäure, das Spaltprodukt eines neuen Gehirnlipoids, Hoppe Seylers Z Physiol Chem, 268 (1941) 50. [Google Scholar]
- [15].Tanaka K, Bertolini M, Pigman W, Serine and threonine glycosidic linkages in bovine submaxillary mucin, Biochem Biophys Res Commun, 16 (1964) 404–409. [DOI] [PubMed] [Google Scholar]
- [16].Blix FG, Gottschalk A, Klenk E, Proposed nomenclature in the field of neuraminic and sialic acids, Nature, 179 (1957) 1088. [DOI] [PubMed] [Google Scholar]
- [17].Varki A, Loss of N-glycolylneuraminic acid in humans: Mechanisms, consequences, and implications for hominid evolution, Am J Phys Anthropol, Suppl 33 (2001) 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Blix G, Lindberg E, The sialic acids of bovine and equine submaxillary mucins, Acta Chem Scand, 14 (1960). [Google Scholar]
- [19].Blix G, Lindberg E, Odin L, Werner I, Sialic acids, Nature, 175 (1955) 340–341. [DOI] [PubMed] [Google Scholar]
- [20].Buscher HP, Casals-Stenzel J, Schaufer R, New sialic acids. Identification of N-glycoloyl-O-acetylneuraminic acids and N-acetyl-O-glycoloylneuraminic acids by improved methods for detection of N-acyl and O-acyl groups and by gas-liquid chromatography, Eur J Biochem, 50 (1974) 71–82. [DOI] [PubMed] [Google Scholar]
- [21].Schauer R, Characterization of sialic acids, Methods Enzymol, 50 (1978) 64–89. [DOI] [PubMed] [Google Scholar]
- [22].Schauer R, Chemistry, metabolism, and biological functions of sialic acids, Adv Carbohydr Chem Biochem, 40 (1982) 131–234. [DOI] [PubMed] [Google Scholar]
- [23].Tuppy H, Gottschalk A, Glycoproteins, their composition, structure and function, Gottschalk A, ed. BBA Library, Elsevier Publishing Co, Amsterdam, London, New York, 5 (1972) 403–449. [Google Scholar]
- [24].Gottschalk A, Chemistry and biology of sialic acids and related substances, Cambridge University Press: Cambridge: 1960. [Google Scholar]
- [25].Baer H, Kabat EA, Knaub V, Immuno-chemical studies on blood groups; the preparation of blood group A and B substances and an inactive substance from individual horse stomachs and of blood group B substance from human saliva, J Exp Med, 91 (1950) 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kabat EA, Bendich A, Bezer AE, Beiser SM, Immunochemical studies on blood groups : Iv. preparation of blood group A substances from human sources and a comparison of their chemical and immunochemical properties with those of the blood group A substance from hog stomach, J Exp Med, 85 (1947) 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gibbons RJ, Adherent interactions which may affect microbial ecology in the mouth, J Dent Res, 63 (1984) 378–385. [DOI] [PubMed] [Google Scholar]
- [28].Levine MJ, Herzberg MC, Levine MS, Ellison SA, Stinson MW, Li HC, van Dyke T, Specificity of salivary-bacterial interactions: role of terminal sialic acid residues in the interaction of salivary glycoproteins with Streptococcus sanguis and Streptococcus mutans, Infect Immun, 19 (1978) 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].McBride BC, Gisslow MT, Role of sialic acid in saliva-induced aggregation of Streptococcus sanguis, Infect Immun, 18 (1977) 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mandel ID, The functions of saliva, J Dent Res, 66 Spec No (1987) 623–627. [DOI] [PubMed] [Google Scholar]
- [31].Slomiany BL, Murty VL, Slomiany A, Salivary lipids in health and disease, Prog Lipid Res, 24 (1985) 311–324. [DOI] [PubMed] [Google Scholar]
- [32].Slomiany BL, Murty VL, Mandel ID, Sengupta S, Slomiany A, Effect of lipids on the lactic acid retardation capacity of tooth enamel and cementum pellicles formed in vitro from saliva of caries-resistant and caries-susceptible human adults, Arch Oral Biol, 35 (1990) 175–180. [DOI] [PubMed] [Google Scholar]
- [33].Larsson B, Olivecrona G, Ericson T, Lipids in human saliva, Arch Oral Biol, 41 (1996) 105–110. [DOI] [PubMed] [Google Scholar]
- [34].Lamkin MS, Oppenheim FG, Structural features of salivary function, Crit Rev Oral Biol Med, 4 (1993) 251–259. [DOI] [PubMed] [Google Scholar]
- [35].Tenovuo J, Jentsch H, Soukka T, Karhuvaara L, Antimicrobial factors of saliva in relation to dental caries and salivary levels of mutans streptococci, J Biol Buccale, 20 (1992) 85–90. [PubMed] [Google Scholar]
- [36].Tenovuo J, Antimicrobial agents in saliva--protection for the whole body, J Dent Res, 81 (2002) 807–809. [DOI] [PubMed] [Google Scholar]
- [37].Amerongen AV, Veerman EC, Saliva--the defender of the oral cavity, Oral Dis, 8 (2002) 12–22. [DOI] [PubMed] [Google Scholar]
- [38].Gorr SU, Antimicrobial peptides of the oral cavity, Periodontol 2000, 51 (2009) 152–180. [DOI] [PubMed] [Google Scholar]
- [39].Dale BA, Fredericks LP, Antimicrobial peptides in the oral environment: expression and function in health and disease, Curr Issues Mol Biol, 7 (2005) 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Van’t Hof W, Veerman EC, Nieuw Amerongen AV, Ligtenberg AJ, Antimicrobial defense systems in saliva, Monogr Oral Sci, 24 (2014) 40–51. [DOI] [PubMed] [Google Scholar]
- [41].Brown JW, Badahdah A, Iticovici M, Vickers TJ, Alvarado DM, Helmerhorst EJ, Oppenheim FG, Mills JC, Ciorba MA, Fleckenstein JM, Bullitt E, A role for salivary peptides in the innate defense against enterotoxigenic Escherichia coli, J Infect Dis, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zelles T, Purushotham KR, Macauley SP, Oxford GE, Humphreys-Beher MG, Saliva and growth factors: the fountain of youth resides in us all, J Dent Res, 74 (1995) 1826–1832. [DOI] [PubMed] [Google Scholar]
- [43].Brand HS, Ligtenberg AJ, Veerman EC, Saliva and wound healing, Monogr Oral Sci, 24 (2014) 52–60. [DOI] [PubMed] [Google Scholar]
- [44].Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, Delahunty CM, Denny T, Dunsmore J, Faull KF, Gilligan J, Gonzalez-Begne M, Halgand F, Hall SC, Han X, Henson B, Hewel J, Hu S, Jeffrey S, Jiang J, Loo JA, Ogorzalek Loo RR, Malamud D, Melvin JE, Miroshnychenko O, Navazesh M, Niles R, Park SK, Prakobphol A, Ramachandran P, Richert M, Robinson S, Sondej M, Souda P, Sullivan MA, Takashima J, Than S, Wang J, Whitelegge JP, Witkowska HE, Wolinsky L, Xie Y, Xu T, Yu W, Ytterberg J, Wong DT, Yates JR 3rd, Fisher SJ, The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions, J Proteome Res, 7 (2008) 1994–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ruhl S, The scientific exploration of saliva in the post-proteomic era: from database back to basic function, Expert Rev Proteomics, 9 (2012) 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Walz A, Stühler K, Wattenberg A, Hawranke E, Meyer HE, Schmalz G, Blüggel M, Ruhl S, Proteome analysis of glandular parotid and submandibular-sublingual saliva in comparison to whole human saliva by two-dimensional gel electrophoresis, Proteomics, 6 (2006) 1631–1639. [DOI] [PubMed] [Google Scholar]
- [47].Helmerhorst EJ, Oppenheim FG, Saliva: a dynamic proteome, J Dent Res, 86 (2007) 680–693. [DOI] [PubMed] [Google Scholar]
- [48].Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ, Salivary proteome and its genetic polymorphisms, Ann N Y Acad Sci, 1098 (2007) 22–50. [DOI] [PubMed] [Google Scholar]
- [49].Karlsson NG, Thomsson KA, Salivary MUC7 is a major carrier of blood group I type O-linked oligosaccharides serving as the scaffold for sialyl Lewis x, Glycobiology, 19 (2009) 288–300. [DOI] [PubMed] [Google Scholar]
- [50].Thomsson KA, Prakobphol A, Leffler H, Reddy MS, Levine MJ, Fisher SJ, Hansson GC, The salivary mucin MG1 (MUC5B) carries a repertoire of unique oligosaccharides that is large and diverse, Glycobiology, 12 (2002) 1–14. [DOI] [PubMed] [Google Scholar]
- [51].Frenkel ES, Ribbeck K, Salivary mucins in host defense and disease prevention, J Oral Microbiol, 7 (2015) 29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tabak LA, In defense of the oral cavity: the protective role of the salivary secretions, Pediatric dentistry, 28 (2006) 110–117; discussion 192–118. [PubMed] [Google Scholar]
- [53].Tabak LA, In defense of the oral cavity: structure, biosynthesis, and function of salivary mucins, Annu Rev Physiol, 57 (1995) 547–564. [DOI] [PubMed] [Google Scholar]
- [54].Tabak LA, Structure and function of human salivary mucins, Crit Rev Oral Biol Med, 1 (1990) 229–234. [DOI] [PubMed] [Google Scholar]
- [55].Schauer R, Sialic acids: fascinating sugars in higher animals and man, Zoology (Jena), 107 (2004) 49–64. [DOI] [PubMed] [Google Scholar]
- [56].Dekker J, Rossen JW, Büller HA, Einerhand AW, The MUC family: an obituary, Trends Biochem Sci, 27 (2002) 126–131. [DOI] [PubMed] [Google Scholar]
- [57].Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J, Mucin-bacterial interactions in the human oral cavity and digestive tract, Gut Microbes, 1 (2010) 254–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Xu D, Pavlidis P, Taskent RO, Alachiotis N, Flanagan C, DeGiorgio M, Blekhman R, Ruhl S, Gokcumen O, Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation, Mol Biol Evol, 34 (2017) 2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xu D, Pavlidis P, Thamadilok S, Redwood E, Fox S, Blekhman R, Ruhl S, Gokcumen O, Recent evolution of the salivary mucin MUC7, Sci Rep, 6 (2016) 31791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Culp DJ, Robinson B, Cash MN, Bhattacharyya I, Stewart C, Cuadra-Saenz G, Salivary mucin 19 glycoproteins: innate immune functions in Streptococcus mutansinduced caries in mice and evidence for expression in human saliva, J Biol Chem, 290 (2015) 2993–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lendenmann U, Grogan J, Oppenheim FG, Saliva and dental pellicle - a review, Adv Dent Res, 14 (2000) 22–28. [DOI] [PubMed] [Google Scholar]
- [62].Siqueira WL, Zhang W, Helmerhorst EJ, Gygi SP, Oppenheim FG, Identification of protein components in in vivo human acquired enamel pellicle using LC-ESI-MS/MS, J Proteome Res, 6 (2007) 2152–2160. [DOI] [PubMed] [Google Scholar]
- [63].Nieuw Amerongen AV, Oderkerk CH, Driessen AA, Role of mucins from human whole saliva in the protection of tooth enamel against demineralization in vitro, Caries Res, 21 (1987) 297–309. [DOI] [PubMed] [Google Scholar]
- [64].Van Nieuw Amerongen A, Bolscher JG, Veerman EC, Salivary proteins: protective and diagnostic value in cariology?, Caries Res, 38 (2004) 247–253. [DOI] [PubMed] [Google Scholar]
- [65].Hwang G, Liu Y, Kim D, Sun V, Aviles-Reyes A, Kajfasz JK, Lemos JA, Koo H, Simultaneous spatiotemporal mapping of in situ pH and bacterial activity within an intact 3D microcolony structure, Sci Rep, 6 (2016) 32841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Prakobphol A, Thomsson KA, Hansson GC, Rosen SD, Singer MS, Phillips NJ, Medzihradszky KF, Burlingame AL, Leffler H, Fisher SJ, Human low-molecular-weight salivary mucin expresses the sialyl lewisx determinant and has L-selectin ligand activity, Biochemistry, 37 (1998) 4916–4927. [DOI] [PubMed] [Google Scholar]
- [67].Prakobphol A, Leffler H, Fisher SJ, The high-molecular-weight human mucin is the primary salivary carrier of ABH, Le(a), and Le(b) blood group antigens, Crit Rev Oral Biol Med, 4 (1993) 325–333. [DOI] [PubMed] [Google Scholar]
- [68].Albertolle ME, Hassis ME, Ng CJ, Cuison S, Williams K, Prakobphol A, Dykstra AB, Hall SC, Niles RK, Ewa Witkowska H, Fisher SJ, Mass spectrometry-based analyses showing the effects of secretor and blood group status on salivary Nglycosylation, Clin Proteomics, 12 (2015) 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Walz A, Odenbreit S, Mahdavi J, Borén T, Ruhl S, Identification and characterization of binding properties of Helicobacter pylori by glycoconjugate arrays, Glycobiology, 15 (2005) 700–708. [DOI] [PubMed] [Google Scholar]
- [70].Walz A, Odenbreit S, Stühler K, Wattenberg A, Meyer HE, Mahdavi J, Borén T, Ruhl S, Identification of glycoprotein receptors within the human salivary proteome for the lectin-like BabA and SabA adhesins of Helicobacter pylori by fluorescence-based 2-D bacterial overlay, Proteomics, 9 (2009) 1582–1592. [DOI] [PubMed] [Google Scholar]
- [71].Bürgers R, Schneider-Brachert W, Reischl U, Behr A, Hiller KA, Lehn N, Schmalz G, Ruhl S, Helicobacter pylori in human oral cavity and stomach, Eur J Oral Sci, 116 (2008) 297–304. [DOI] [PubMed] [Google Scholar]
- [72].Issa S, Moran AP, Ustinov SN, Lin JH, Ligtenberg AJ, Karlsson NG, O-linked oligosaccharides from salivary agglutinin: Helicobacter pylori binding sialyl-Lewis x and Lewis b are terminating moieties on hyperfucosylated oligo-N-acetyllactosamine, Glycobiology, 20 (2010) 1046–1057. [DOI] [PubMed] [Google Scholar]
- [73].Prakobphol A, Borén T, Ma W, Zhixiang P, Fisher SJ, Highly glycosylated human salivary molecules present oligosaccharides that mediate adhesion of leukocytes and Helicobacter pylori, Biochemistry, 44 (2005) 2216–2224. [DOI] [PubMed] [Google Scholar]
- [74].Dowsett SA, Kowolik MJ, Oral Helicobacter pylori: can we stomach it?, Crit Rev Oral Biol Med, 14 (2003) 226–233. [DOI] [PubMed] [Google Scholar]
- [75].Van den Brink GR, Tytgat KM, Van der Hulst RW, Van der Loos CM, Einerhand AW, Buller HA, Dekker J, H. pylori colocalises with MUC5AC in the human stomach, Gut, 46 (2000) 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Van de Bovenkamp JH, Mahdavi J, Korteland-Van Male AM, Buller HA, Einerhand AW, Boren T, Dekker J, The MUC5AC glycoprotein is the primary receptor for Helicobacter pylori in the human stomach, Helicobacter, 8 (2003) 521–532. [DOI] [PubMed] [Google Scholar]
- [77].Scannapieco FA, Saliva-bacterium interactions in oral microbial ecology, Crit Rev Oral Biol Med, 5 (1994) 203–248. [DOI] [PubMed] [Google Scholar]
- [78].Bobek LA, Liu J, Sait SN, Shows TB, Bobek YA, Levine MJ, Structure and chromosomal localization of the human salivary mucin gene, MUC7, Genomics, 31 (1996) 277–282. [DOI] [PubMed] [Google Scholar]
- [79].Ligtenberg AJ, Karlsson NG, Veerman EC, Deleted in malignant brain tumors-1 protein (DMBT1): a pattern recognition receptor with multiple binding sites, Int J Mol Sci, 11 (2010) 5212–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Madsen J, Mollenhauer J, Holmskov U, Review: Gp-340/DMBT1 in mucosal innate immunity, Innate Immun, 16 (2010) 160–167. [DOI] [PubMed] [Google Scholar]
- [81].Royle L, Roos A, Harvey DJ, Wormald MR, van Gijlswijk-Janssen D, Redwan el RM, Wilson IA, Daha MR, Dwek RA, Rudd PM, Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems, J Biol Chem, 278 (2003) 20140–20153. [DOI] [PubMed] [Google Scholar]
- [82].Baenziger JU, Structure of the oligosaccharide of human J chain, J Biol Chem, 254 (1979) 4063–4071. [PubMed] [Google Scholar]
- [83].Baenziger J, Kornfeld S, Structure of the carbohydrate units of IgA1 immunoglobulin. II. Structure of the O-glycosidically linked oligosaccharide units, J Biol Chem, 249 (1974) 7270–7281. [PubMed] [Google Scholar]
- [84].Baenziger J, Kornfeld S, Structure of the carbohydrate units of IgA1 immunoglobulin. I. Composition, glycopeptide isolation, and structure of the asparagine-linked oligosaccharide units, J Biol Chem, 249 (1974) 7260–7269. [PubMed] [Google Scholar]
- [85].Mizoguchi A, Mizuochi T, Kobata A, Structures of the carbohydrate moieties of secretory component purified from human milk, J Biol Chem, 257 (1982) 9612–9621. [PubMed] [Google Scholar]
- [86].Mestecky J, Saliva as a manifestation of the common mucosal immune system, Ann N Y Acad Sci, 694 (1993) 184–194. [DOI] [PubMed] [Google Scholar]
- [87].Kilian M, Mestecky J, Russell MW, Defense mechanisms involving Fc-dependent functions of immunoglobulin A and their subversion by bacterial immunoglobulin A proteases, Microbiol Rev, 52 (1988) 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ruhl S, Sandberg AL, Cole MF, Cisar JO, Recognition of immunoglobulin A1 by oral actinomyces and streptococcal lectins, Infect Immun, 64 (1996) 5421–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Itzek A, Chen Z, Merritt J, Kreth J, Effect of salivary agglutination on oral streptococcal clearance by human polymorphonuclear neutrophil granulocytes, Mol Oral Microbiol, 32 (2017) 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Rosan B, Appelbaum B, Golub E, Malamud D, Mandel ID, Enhanced saliva-mediated bacterial aggregation and decreased bacterial adhesion in caries-resistant versus caries-susceptible individuals, Infect Immun, 38 (1982) 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Emilson CG, Ciardi JE, Olsson J, Bowen WH, The influence of saliva on infection of the human mouth by mutans streptococci, Arch Oral Biol, 34 (1989) 335–340. [DOI] [PubMed] [Google Scholar]
- [92].Brack CM, Reynolds EC, Colonization of rat molar teeth by mutans streptococci with different salivary agglutination characteristics, Arch Oral Biol, 33 (1988) 695–699. [DOI] [PubMed] [Google Scholar]
- [93].Heo SM, Choi KS, Kazim LA, Reddy MS, Haase EM, Scannapieco FA, Ruhl S, Host defense proteins derived from human saliva bind to Staphylococcus aureus, Infect Immun, 81 (2013) 1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Heo SM, Ruhl S, Scannapieco FA, Implications of salivary protein binding to commensal and pathogenic bacteria, J Oral Biosci, 55 (2013) 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gibbons RJ, Cohen L, Hay DI, Strains of Streptococcus mutans and Streptococcus sobrinus attach to different pellicle receptors, Infect Immun, 52 (1986) 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Loimaranta V, Jakubovics NS, Hytonen J, Finne J, Jenkinson HF, Stromberg N, Fluidor surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties, Infect Immun, 73 (2005) 2245–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Springer SA, Gagneux P, Glycan evolution in response to collaboration, conflict, and constraint, J Biol Chem, 288 (2013) 6904–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Bishop JR, Gagneux P, Evolution of carbohydrate antigens--microbial forces shaping host glycomes?, Glycobiology, 17 (2007) 23R–34R. [DOI] [PubMed] [Google Scholar]
- [99].Varki A, Freeze HH, Gagneux P, Evolution of Glycan Diversity, in: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME (Eds.) Essentials of Glycobiology, Cold Spring Harbor (NY), 2009. [PubMed] [Google Scholar]
- [100].Varki A, Nothing in glycobiology makes sense, except in the light of evolution, Cell, 126 (2006) 841–845. [DOI] [PubMed] [Google Scholar]
- [101].Varki A, Multiple changes in sialic acid biology during human evolution, Glycoconj J, 26 (2009) 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cohen M, Varki A, Modulation of glycan recognition by clustered saccharide patches, Int Rev Cell Mol Biol, 308 (2014) 75–125. [DOI] [PubMed] [Google Scholar]
- [103].Polley S, Louzada S, Forni D, Sironi M, Balaskas T, Hains DS, Yang F, Hollox EJ, Evolution of the rapidly mutating human salivary agglutinin gene (DMBT1) and population subsistence strategy, Proc Natl Acad Sci U S A, 112 (2015) 5105–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lang T, Hansson GC, Samuelsson T, Gel-forming mucins appeared early in metazoan evolution, Proc Natl Acad Sci U S A, 104 (2007) 16209–16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Desseyn JL, Aubert JP, Porchet N, Laine A, Evolution of the large secreted gel-forming mucins, Mol Biol Evol, 17 (2000) 1175–1184. [DOI] [PubMed] [Google Scholar]
- [106].Gibbons RJ, Bacterial adhesion to oral tissues: a model for infectious diseases, J Dent Res, 68 (1989) 750–760. [DOI] [PubMed] [Google Scholar]
- [107].Cornejo OE, Lefebure T, Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ, Burne RA, Siepel A, Bustamante CD, Stanhope MJ, Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans, Mol Biol Evol, 30 (2013) 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Mitchell TJ, The pathogenesis of streptococcal infections: from tooth decay to meningitis, Nat Rev Microbiol, 1 (2003) 219–230. [DOI] [PubMed] [Google Scholar]
- [109].Busscher HJ, Norde W, van der Mei HC, Specific molecular recognition and nonspecific contributions to bacterial interaction forces, Appl Environ Microbiol, 74 (2008) 2559–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Jenkinson HF, Lamont RJ, Streptococcal adhesion and colonization, Crit Rev Oral Biol Med, 8 (1997) 175–200. [DOI] [PubMed] [Google Scholar]
- [111].Sato Y, Okamoto K, Kagami A, Yamamoto Y, Igarashi T, Kizaki H, Streptococcus mutans strains harboring collagen-binding adhesin, J Dent Res, 83 (2004) 534–539. [DOI] [PubMed] [Google Scholar]
- [112].Clark WB, Beem JE, Nesbitt WE, Cisar JO, Tseng CC, Levine MJ, Pellicle receptors for Actinomyces viscosus type 1 fimbriae in vitro, Infect Immun, 57 (1989) 3003–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Gibbons RJ, Hay DI, Schlesinger DH, Delineation of a segment of adsorbed salivary acidic proline-rich proteins which promotes adhesion of Streptococcus gordonii to apatitic surfaces, Infect Immun, 59 (1991) 2948–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lee RT, Lee YC, Affinity enhancement by multivalent lectin-carbohydrate interaction, Glycoconj J, 17 (2000) 543–551. [DOI] [PubMed] [Google Scholar]
- [115].Collins BE, Paulson JC, Cell surface biology mediated by low affinity multivalent protein-glycan interactions, Curr Opin Chem Biol, 8 (2004) 617–625. [DOI] [PubMed] [Google Scholar]
- [116].Dam TK, Brewer CF, Effects of clustered epitopes in multivalent ligand-receptor interactions, Biochemistry, 47 (2008) 8470–8476. [DOI] [PubMed] [Google Scholar]
- [117].Waksman G, Hultgren SJ, Structural biology of the chaperone-usher pathway of pilus biogenesis, Nat Rev Microbiol, 7 (2009) 765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Lizcano A, Sanchez CJ, Orihuela CJ, A role for glycosylated serine-rich repeat proteins in gram-positive bacterial pathogenesis, Mol Oral Microbiol, 27 (2012) 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Ramboarina S, Garnett JA, Zhou M, Li Y, Peng Z, Taylor JD, Lee WC, Bodey A, Murray JW, Alguel Y, Bergeron J, Bardiaux B, Sawyer E, Isaacson R, Tagliaferri C, Cota E, Nilges M, Simpson P, Ruiz T, Wu H, Matthews S, Structural insights into serine-rich fimbriae from Gram-positive bacteria, J Biol Chem, 285 (2010) 32446–32457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Zhou M, Wu H, Glycosylation and biogenesis of a family of serine-rich bacterial adhesins, Microbiology, 155 (2009) 317–327. [DOI] [PubMed] [Google Scholar]
- [121].Yoshida Y, Palmer RJ, Yang J, Kolenbrander PE, Cisar JO, Streptococcal receptor polysaccharides: recognition molecules for oral biofilm formation, BMC Oral Health, 6 Suppl 1 (2006) S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Stowell SR, Arthur CM, McBride R, Berger O, Razi N, Heimburg-Molinaro J, Rodrigues LC, Gourdine JP, Noll AJ, von Gunten S, Smith DF, Knirel YA, Paulson JC, Cummings RD, Microbial glycan microarrays define key features of host-microbial interactions, Nat Chem Biol, 10 (2014) 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Nyvad B, Kilian M, Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals, Caries Res, 24 (1990) 267–272. [DOI] [PubMed] [Google Scholar]
- [124].Hasty DL, Ofek I, Courtney HS, Doyle RJ, Multiple adhesins of streptococci, Infect Immun, 60 (1992) 2147–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Nobbs AH, Jenkinson HF, Jakubovics NS, Stick to your gums: mechanisms of oral microbial adherence, J Dent Res, 90 (2011) 1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Jenkinson HF, Lamont RJ, Oral microbial communities in sickness and in health, Trends Microbiol, 13 (2005) 589–595. [DOI] [PubMed] [Google Scholar]
- [127].Murray PA, Levine MJ, Tabak LA, Reddy MS, Specificity of salivary-bacterial interactions: II. Evidence for a lectin on Streptococcus sanguis with specificity for a NeuAc α2, 3Ga1 β1, 3Ga1NAc sequence, Biochem Biophys Res Comm, 106 (1982) 390–396. [DOI] [PubMed] [Google Scholar]
- [128].Takahashi Y, Sandberg AL, Ruhl S, Muller J, Cisar JO, A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to α2–3-linked sialic acid-containing receptors, Infect Immun, 65 (1997) 5042–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Takahashi Y, Konishi K, Cisar JO, Yoshikawa M, Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1, Infect Immun, 70 (2002) 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Xiong YQ, Bensing BA, Bayer AS, Chambers HF, Sullam PM, Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis, Microb Pathog, 45 (2008) 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Pyburn TM, Bensing BA, Xiong YQ, Melancon BJ, Tomasiak TM, Ward NJ, Yankovskaya V, Oliver KM, Cecchini G, Sulikowski GA, Tyska MJ, Sullam PM, Iverson TM, A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors, PLoS Pathog, 7 (2011) e1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Siboo IR, Chambers HF, Sullam PM, Role of SraP, a Serine-Rich Surface Protein of Staphylococcus aureus, in binding to human platelets, Infect Immun, 73 (2005) 2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].van Sorge NM, Quach D, Gurney MA, Sullam PM, Nizet V, Doran KS, The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier, J Infect Dis, 199 (2009) 1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Seo HS, Mu R, Kim BJ, Doran KS, Sullam PM, Binding of glycoprotein Srr1 of Streptococcus agalactiae to fibrinogen promotes attachment to brain endothelium and the development of meningitis, PLoS Pathog, 8 (2012) e1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Rose L, Shivshankar P, Hinojosa E, Rodriguez A, Sanchez CJ, Orihuela CJ, Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge, J Infect Dis, 198 (2008) 375–383. [DOI] [PubMed] [Google Scholar]
- [136].Shivshankar P, Sanchez C, Rose LF, Orihuela CJ, The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells, Mol Microbiol, 73 (2009) 663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Deng L, Bensing BA, Thamadilok S, Yu H, Lau K, Chen X, Ruhl S, Sullam PM, Varki A, Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood, PLoS Pathog, 10 (2014) e1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Bensing BA, Khedri Z, Deng L, Yu H, Prakobphol A, Fisher SJ, Chen X, Iverson TM, Varki A, Sullam PM, Novel aspects of sialoglycan recognition by the Sigleclike domains of streptococcal SRR glycoproteins, Glycobiology, 26 (2016) 1222–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Loukachevitch LV, Bensing BA, Yu H, Zeng J, Chen X, Sullam PM, Iverson TM, Structures of the Streptococcus sanguinis SrpA binding region with human sialoglycans suggest features of the physiological ligand, Biochemistry, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Gillece-Castro BL, Prakobphol A, Burlingame AL, Leffler H, Fisher SJ, Structure and bacterial receptor activity of a human salivary proline-rich glycoprotein, J Biol Chem, 266 (1991) 17358–17368. [PubMed] [Google Scholar]
- [141].Bensing BA, Lopez JA, Sullam PM, The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibalpha, Infect Immun, 72 (2004) 6528–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Takamatsu D, Bensing BA, Cheng H, Jarvis GA, Siboo IR, Lopez JA, Griffiss JM, Sullam PM, Binding of the Streptococcus gordonii surface glycoproteins GspB and Hsa to specific carbohydrate structures on platelet membrane glycoprotein Ibα, Mol Microbiol, 58 (2005) 380–392. [DOI] [PubMed] [Google Scholar]
- [143].Zhuravleva MA, Trandem K, Sun PD, Structural implications of Siglec-5-mediated sialoglycan recognition, J Mol Biol, 375 (2008) 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Jakubovics NS, Kerrigan SW, Nobbs AH, Stromberg N, van Dolleweerd CJ, Cox DM, Kelly CG, Jenkinson HF, Functions of cell surface-anchored antigen I/II family and Hsa polypeptides in interactions of Streptococcus gordonii with host receptors, Infect Immun, 73 (2005) 6629–6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Plummer C, Wu H, Kerrigan SW, Meade G, Cox D, Ian Douglas CW, A serine-rich glycoprotein of Streptococcus sanguis mediates adhesion to platelets via GPIb, Br J Haematol, 129 (2005) 101–109. [DOI] [PubMed] [Google Scholar]
- [146].Yajima A, Takahashi Y, Konishi K, Identification of platelet receptors for the Streptococcus gordonii DL1 sialic acid-binding adhesin, Microbiol Immunol, 49 (2005) 795–800. [DOI] [PubMed] [Google Scholar]
- [147].Yakovenko O, Nunez J, Bensing B, Yu H, Mount J, Zeng J, Hawkins J, Chen X, Sullam PM, Thomas W, Serine-rich repeat adhesins mediate shear-enhanced streptococcal binding to platelets, Infect Immun, 86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Ding AM, Palmer RJ Jr., Cisar JO, Kolenbrander PE, Shear-enhanced oral microbial adhesion, Appl Environ Microbiol, 76 (2010) 1294–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Echelman DJ, Alegre-Cebollada J, Badilla CL, Chang C, Ton-That H, Fernandez JM, CnaA domains in bacterial pili are efficient dissipaters of large mechanical shocks, Proc Natl Acad Sci U S A, 113 (2016) 2490–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Cisar JO, David VA, Curl SH, Vatter AE, Exclusive presence of lactose-sensitive fimbriae on a typical strain (WVU45) of Actinomyces naeslundii, Infect Immun, 46 (1984) 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Ellen RP, Establishment and distribution of Actinomyces viscosus and Actinomyces naeslundii in the human oral cavity, Infect Immun, 14 (1976) 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Liljemark WF, Bloomquist CG, Bandt CL, Pihlstrom BL, Hinrichs JE, Wolff LF, Comparison of the distribution of Actinomyces in dental plaque on inserted enamel and natural tooth surfaces in periodontal health and disease, Oral Microbiol Immunol, 8 (1993) 5–15. [DOI] [PubMed] [Google Scholar]
- [153].McIntire FC, Crosby LK, Vatter AE, Cisar JO, McNeil MR, Bush CA, Tjoa SS, Fennessey PV, A polysaccharide from Streptococcus sanguis 34 that inhibits coaggregation of S. sanguis 34 with Actinomyces viscosus T14V, J Bacteriol, 170 (1988) 2229–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Cisar JO, Kolenbrander PE, McIntire FC, Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii, Infect Immun, 24 (1979) 742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Strömberg N, Karlsson KA, Characterization of the binding of Actinomyces naeslundii (ATCC 12104) and Actinomyces viscosus (ATCC 19246) to glycosphingolipids, using a solid-phase overlay approach, J Biol Chem, 265 (1990) 11251–11258. [PubMed] [Google Scholar]
- [156].McIntire FC, Crosby LK, Vatter AE, Inhibitors of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34: beta-galactosides, related sugars, and anionic amphipathic compounds, Infect Immun, 36 (1982) 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Cisar JO, Sandberg AL, Abeygunawardana C, Reddy GP, Bush CA, Lectin recognition of host-like saccharide motifs in streptococcal cell wall polysaccharides, Glycobiology, 5 (1995) 655–662. [DOI] [PubMed] [Google Scholar]
- [158].Mishra A, Wu C, Yang J, Cisar JO, Das A, Ton-That H, The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development, Mol Microbiol, 77 (2010) 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Brockhausen I, Stanley P, O-GalNAc Glycans, in: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH (Eds.) Essentials of Glycobiology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor (NY), 2015, pp. 113–123. [Google Scholar]
- [160].Yeung MK, Molecular and genetic analyses of Actinomyces spp., Crit Rev Oral Biol Med, 10 (1999) 120–138. [DOI] [PubMed] [Google Scholar]
- [161].Do T, Henssge U, Gilbert SC, Clark D, Beighton D, Evidence for recombination between a sialidase (nanH) of Actinomyces naeslundii and Actinomyces oris, previously named ‘Actinomyces naeslundii genospecies 1 and 2’, FEMS Microbiol Lett, 288 (2008) 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Moncla BJ, Braham P, Detection of sialidase (neuraminidase) activity in Actinomyces species by using 2’-(4-methylumbelliferyl)alpha-D-N-acetylneuraminic acid in a filter paper spot test, J Clin Microbiol, 27 (1989) 182–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Brennan MJ, Cisar JO, Vatter AE, Sandberg AL, Lectin-dependent attachment of Actinomyces naeslundii to receptors on epithelial cells, Infect Immun, 46 (1984) 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Ruhl S, Cisar JO, Sandberg AL, Identification of polymorphonuclear leukocyte and HL-60 cell receptors for adhesins of Streptococcus gordonii and Actinomyces naeslundii, Infect Immun, 68 (2000) 6346–6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Sandberg AL, Ruhl S, Joralmon RA, Brennan MJ, Sutphin MJ, Cisar JO, Putative glycoprotein and glycolipid polymorphonuclear leukocyte receptors for the Actinomyces naeslundii WVU45 fimbrial lectin, Infect Immun, 63 (1995) 2625–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Ruhl S, Sandberg AL, Cisar JO, Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral actinomyces and streptococci, J Dent Res, 83 (2004) 505–510. [DOI] [PubMed] [Google Scholar]
- [167].Reardon-Robinson ME, Wu C, Mishra A, Chang C, Bier N, Das A, Ton-That H, Pilus hijacking by a bacterial coaggregation factor critical for oral biofilm development, Proc Natl Acad Sci U S A, 111 (2014) 3835–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Tanzer JM, Livingston J, Thompson AM, The microbiology of primary dental caries in humans, J Dent Educ, 65 (2001) 1028–1037. [PubMed] [Google Scholar]
- [169].Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, Jenkinson HF, The changing faces of Streptococcus antigen I/II polypeptide family adhesins, Mol Microbiol, 77 (2010) 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Banas JA, Vickerman MM, Glucan-binding proteins of the oral streptococci, Crit Rev Oral Biol Med, 14 (2003) 89–99. [DOI] [PubMed] [Google Scholar]
- [171].Yang C, Scoffield J, Wu R, Deivanayagam C, Zou J, Wu H, Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans, Mol Oral Microbiol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Yamashita Y, Bowen WH, Burne RA, Kuramitsu HK, Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model, Infect Immun, 61 (1993) 3811–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Fry EE, Lea SM, Jackson T, Newman JW, Ellard FM, Blakemore WE, Abu-Ghazaleh R, Samuel A, King AM, Stuart DI, The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex, EMBO J, 18 (1999) 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Herold BC, WuDunn D, Soltys N, Spear PG, Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity, J Virol, 65 (1991) 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG, A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry, Cell, 99 (1999) 13–22. [DOI] [PubMed] [Google Scholar]
- [176].Patel M, Yanagishita M, Roderiquez G, Bou-Habib DC, Oravecz T, Hascall VC, Norcross MA, Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of Tcell lines, AIDS Res Hum Retroviruses, 9 (1993) 167–174. [DOI] [PubMed] [Google Scholar]
- [177].Carroll SM, Higa HH, Paulson JC, Different cell-surface receptor determinants of antigenically similar influenza virus hemagglutinins, J Biol Chem, 256 (1981) 8357–8363. [PubMed] [Google Scholar]
- [178].Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC, Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid, Nature, 333 (1988) 426–431. [DOI] [PubMed] [Google Scholar]
- [179].Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R, Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin, Nat Biotechnol, 26 (2008) 107–113. [DOI] [PubMed] [Google Scholar]
- [180].Limsuwat N, Suptawiwat O, Boonarkart C, Puthavathana P, Wiriyarat W, Auewarakul P, Sialic acid content in human saliva and anti-influenza activity against human and avian influenza viruses, Arch Virol, 161 (2016) 649–656. [DOI] [PubMed] [Google Scholar]
- [181].Qin Y, Zhong Y, Zhu M, Dang L, Yu H, Chen Z, Chen W, Wang X, Zhang H, Li Z, Age- and sex-associated differences in the glycopatterns of human salivary glycoproteins and their roles against influenza A virus, J Proteome Res, 12 (2013) 2742–2754. [DOI] [PubMed] [Google Scholar]
- [182].White MR, Helmerhorst EJ, Ligtenberg A, Karpel M, Tecle T, Siqueira WL, Oppenheim FG, Hartshorn KL, Multiple components contribute to ability of saliva to inhibit influenza viruses, Oral Microbiol Immunol, 24 (2009) 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Corstjens PL, Abrams WR, Malamud D, Saliva and viral infections, Periodontol 2000, 70 (2016) 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Cannon RD, Chaffin WL, Oral colonization by Candida albicans, Crit Rev Oral Biol Med, 10 (1999) 359–383. [DOI] [PubMed] [Google Scholar]
- [185].Donohue DS, Ielasi FS, Goossens KV, Willaert RG, The N-terminal part of Als1 protein from Candida albicans specifically binds fucose-containing glycans, Mol Microbiol, 80 (2011) 1667–1679. [DOI] [PubMed] [Google Scholar]
- [186].Hoffman MP, Haidaris CG, Analysis of Candida albicans adhesion to salivary mucin, Infect Immun, 61 (1993) 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Everest-Dass AV, Jin D, Thaysen-Andersen M, Nevalainen H, Kolarich D, Packer NH, Comparative structural analysis of the glycosylation of salivary and buccal cell proteins: innate protection against infection by Candida albicans, Glycobiology, 22 (2012) 1465–1479. [DOI] [PubMed] [Google Scholar]
- [188].Zupancic ML, Frieman M, Smith D, Alvarez RA, Cummings RD, Cormack BP, Glycan microarray analysis of Candida glabrata adhesin ligand specificity, Mol Microbiol, 68 (2008) 547–559. [DOI] [PubMed] [Google Scholar]
- [189].Cormack BP, Ghori N, Falkow S, An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells, Science, 285 (1999) 578–582. [DOI] [PubMed] [Google Scholar]
- [190].Skov Sorensen UB, Yao K, Yang Y, Tettelin H, Kilian M, Capsular polysaccharide expression in commensal Streptococcus species: genetic and antigenic similarities to Streptococcus pneumoniae, MBio, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Xu DQ, Thompson J, Cisar JO, Genetic loci for coaggregation receptor polysaccharide biosynthesis in Streptococcus gordonii 38, J Bacteriol, 185 (2003) 5419–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Glushka J, Cassels FJ, Carlson RW, van Halbeek H, Complete structure of the adhesin receptor polysaccharide of Streptococcus oralis ATCC 55229 (Streptococcus sanguis H1), Biochemistry, 31 (1992) 10741–10746. [DOI] [PubMed] [Google Scholar]
- [193].Bergström N, Jansson PE, Kilian M, Skov Sorensen UB, Structures of two cell wall-associated polysaccharides of a Streptococcus mitis biovar 1 strain. A unique teichoic acid-like polysaccharide and the group O antigen which is a C-polysaccharide in common with pneumococci, Eur J Biochem, 267 (2000) 7147–7157. [DOI] [PubMed] [Google Scholar]
- [194].Stafford G, Roy S, Honma K, Sharma A, Sialic acid, periodontal pathogens and Tannerella forsythia: stick around and enjoy the feast!, Mol Oral Microbiol, 27 (2012) 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Horie T, Inomata M, Into T, Hasegawa Y, Kitai N, Yoshimura F, Murakami Y, Identification of OmpA-like protein of Tannerella forsythia as an O-linked glycoprotein and its binding capability to lectins, PLoS One, 11 (2016) e0163974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Zaric SS, Lappin MJ, Fulton CR, Lundy FT, Coulter WA, Irwin CR, Sialylation of Porphyromonas gingivalis LPS and its effect on bacterial-host interactions, Innate Immun, 23 (2017) 319–326. [DOI] [PubMed] [Google Scholar]
- [197].Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM, Diversity of microbial sialic acid metabolism, Microbiol Mol Biol Rev, 68 (2004) 132–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Severi E, Hood DW, Thomas GH, Sialic acid utilization by bacterial pathogens, Microbiology, 153 (2007) 2817–2822. [DOI] [PubMed] [Google Scholar]
- [199].Severi E, Randle G, Kivlin P, Whitfield K, Young R, Moxon R, Kelly D, Hood D, Thomas GH, Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter, Mol Microbiol, 58 (2005) 1173–1185. [DOI] [PubMed] [Google Scholar]
- [200].Schwarz F, Landig CS, Siddiqui S, Secundino I, Olson J, Varki N, Nizet V, Varki A, Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen, EMBO J, 36 (2017) 751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Van der Hoeven JS, Camp PJ, Synergistic degradation of mucin by Streptococcus oralis and Streptococcus sanguis in mixed chemostat cultures, J Dent Res, 70 (1991) 1041–1044. [DOI] [PubMed] [Google Scholar]
- [202].Zhou Y, Yang J, Zhang L, Zhou X, Cisar JO, Palmer RJ Jr., Differential utilization of basic proline-rich glycoproteins during growth of oral bacteria in saliva, Appl Environ Microbiol, 82 (2016) 5249–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Yang J, Zhou Y, Zhang L, Shah N, Jin C, Palmer RJ Jr., Cisar JO, Cell surface glycoside hydrolases of Streptococcus gordonii promote growth in saliva, Appl Environ Microbiol, 82 (2016) 5278–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].McDonald ND, Lubin JB, Chowdhury N, Boyd EF, Host-derived sialic acids are an important nutrient source required for optimal bacterial fitness in vivo, MBio, 7 (2016) e02237–02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Honma K, Mishima E, Sharma A, Role of Tannerella forsythia NanH sialidase in epithelial cell attachment, Infect Immun, 79 (2011) 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Sandberg AL, Mudrick LL, Cisar JO, Brennan MJ, Mergenhagen SE, Vatter AE, Type 2 fimbrial lectin-mediated phagocytosis of oral Actinomyces spp. by polymorphonuclear leukocytes, Infect Immun, 54 (1986) 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Costello AH, Cisar JO, Kolenbrander PE, Gabriel O, Neuraminidase-dependent hamagglutination of human erythrocytes by human strains of Actinomyces viscosus and Actinomyces naeslundii, Infect Immun, 26 (1979) 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Wong A, Grau MA, Singh AK, Woodiga SA, King SJ, Role of neuraminidaseproducing bacteria in exposing cryptic carbohydrate receptors for Streptococcus gordonii adherence, Infect Immun, 86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Singh AK, Woodiga SA, Grau MA, King SJ, Streptococcus oralis neuraminidase modulates adherence to multiple carbohydrates on platelets, Infect Immun, 85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Limoli DH, Sladek JA, Fuller LA, Singh AK, King SJ, BgaA acts as an adhesin to mediate attachment of some pneumococcal strains to human epithelial cells, Microbiology, 157 (2011) 2369–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Singh AK, Pluvinage B, Higgins MA, Dalia AB, Woodiga SA, Flynn M, Lloyd AR, Weiser JN, Stubbs KA, Boraston AB, King SJ, Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA, PLoS Pathog, 10 (2014) e1004364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Varki A, Diaz S, A neuraminidase from Streptococcus sanguis that can release Oacetylated sialic acids, J Biol Chem, 258 (1983) 12465–12471. [PubMed] [Google Scholar]
- [213].Nobbs AH, Lamont RJ, Jenkinson HF, Streptococcus adherence and colonization, Microbiol Mol Biol Rev, 73 (2009) 407–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Takamatsu D, Bensing BA, Prakobphol A, Fisher SJ, Sullam PM, Binding of the streptococcal surface glycoproteins GspB and Hsa to human salivary proteins, Infect Immun, 74 (2006) 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Murray PA, Prakobphol A, Lee T, Hoover CI, Fisher SJ, Adherence of oral streptococci to salivary glycoproteins, Infect Immun, 60 (1992) 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Thamadilok S, Roche-Håkansson H, Håkansson AP, Ruhl S, Absence of capsule reveals glycan-mediated binding and recognition of salivary mucin MUC7 by Streptococcus pneumoniae, Mol Oral Microbiol, 31 (2016) 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Kolenbrander PE, Coaggregation of human oral bacteria: potential role in the accretion of dental plaque, J Appl Bacteriol, 74 Suppl (1993) 79S–86S. [DOI] [PubMed] [Google Scholar]
- [218].Kolenbrander PE, Ganeshkumar N, Cassels FJ, Hughes CV, Coaggregation: specific adherence among human oral plaque bacteria, FASEB J, 7 (1993) 406–413. [DOI] [PubMed] [Google Scholar]
- [219].Jenkinson HF, Beyond the oral microbiome, Environ Microbiol, 13 (2011) 3077–3087. [DOI] [PubMed] [Google Scholar]
- [220].Listgarten MA, The structure of dental plaque, Periodontol 2000, 5 (1994) 52–65. [DOI] [PubMed] [Google Scholar]
- [221].Wright CJ, Burns LH, Jack AA, Back CR, Dutton LC, Nobbs AH, Lamont RJ, Jenkinson HF, Microbial interactions in building of communities, Mol Oral Microbiol, 28 (2013) 83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Palmer RJ Jr., Gordon SM, Cisar JO, Kolenbrander PE, Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque, J Bacteriol, 185 (2003) 3400–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].T.H.M.P. Consortium, Structure, function and diversity of the healthy human microbiome, Nature, 486 (2012) 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG, The human oral microbiome, J Bacteriol, 192 (2010) 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG, Biogeography of a human oral microbiome at the micron scale, Proc Natl Acad Sci U S A, 113 (2016) E791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Wade WG, The oral microbiome in health and disease, Pharmacol Res, 69 (2013) 137–143. [DOI] [PubMed] [Google Scholar]
- [227].Kolenbrander PE, Palmer RJ Jr., Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI, Bacterial interactions and successions during plaque development, Periodontol 2000, 42 (2006) 47–79. [DOI] [PubMed] [Google Scholar]
- [228].Ruhl S, Eidt A, Melzl H, Reischl U, Cisar JO, Probing of microbial biofilm communities for coadhesion partners, Appl Environ Microbiol, 80 (2014) 6583–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Lamont RJ, Rosan B, Adherence of mutans streptococci to other oral bacteria, Infect Immun, 58 (1990) 1738–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Palmer RJ Jr., Shah N, Valm A, Paster B, Dewhirst F, Inui T, Cisar JO, Interbacterial adhesion networks within early oral biofilms of single human hosts, Appl Environ Microbiol, 83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Hajishengallis G, Lamont RJ, Dancing with the stars: How choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts, Trends Microbiol, 24 (2016) 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Garcia RI, Henshaw MM, Krall EA, Relationship between periodontal disease and systemic health, Periodontol 2000, 25 (2001) 21–36. [DOI] [PubMed] [Google Scholar]
- [233].Frandsen EV, Pedrazzoli V, Kilian M, Ecology of viridans streptococci in the oral cavity and pharynx, Oral Microbiol Immunol, 6 (1991) 129–133. [DOI] [PubMed] [Google Scholar]
- [234].Duval X, Leport C, Prophylaxis of infective endocarditis: current tendencies, continuing controversies, Lancet Infect Dis, 8 (2008) 225–232. [DOI] [PubMed] [Google Scholar]
- [235].Parahitiyawa NB, Jin LJ, Leung WK, Yam WC, Samaranayake LP, Microbiology of odontogenic bacteremia: beyond endocarditis, Clin Microbiol Rev, 22 (2009) 46–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Wilson W, Taubert KA, Gewitz M, Lockhart PB, Baddour LM, Levison M, Bolger A, Cabell CH, Takahashi M, Baltimore RS, Newburger JW, Strom BL, Tani LY, Gerber M, Bonow RO, Pallasch T, Shulman ST, Rowley AH, Burns JC, Ferrieri P, Gardner T, Goff D, Durack DT, Prevention of infective endocarditis: guidelines from the American Heart Association: a guideline from the American Heart Association Rheumatic Fever, Endocarditis and Kawasaki Disease Committee, Council on Cardiovascular Disease in the Young, and the Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and the Quality of Care and Outcomes Research Interdisciplinary Working Group, J Am Dent Assoc, 139 Suppl (2008) 3S–24S. [DOI] [PubMed] [Google Scholar]
- [237].Thiene G, Basso C, Pathology and pathogenesis of infective endocarditis in native heart valves, Cardiovasc Pathol, 15 (2006) 256–263. [DOI] [PubMed] [Google Scholar]
- [238].Fitzgerald JR, Foster TJ, Cox D, The interaction of bacterial pathogens with platelets, Nat Rev Microbiol, 4 (2006) 445–457. [DOI] [PubMed] [Google Scholar]
- [239].Takahashi Y, Takashima E, Shimazu K, Yagishita H, Aoba T, Konishi K, Contribution of sialic acid-binding adhesin to pathogenesis of experimental endocarditis caused by Streptococcus gordonii DL1, Infect Immun, 74 (2006) 740–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Bensing BA, Li Q, Park D, Lebrilla CB, Sullam PM, Streptococcal Siglec-like adhesins recognize different subsets of human plasma glycoproteins: implications for infective endocarditis, Glycobiology, 28 (2018) 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Russell MW, Childers NK, Michalek SM, Smith DJ, Taubman MA, A Caries Vaccine? The state of the science of immunization against dental caries, Caries Res, 38 (2004) 230–235. [DOI] [PubMed] [Google Scholar]
- [242].Taubman MA, Nash DA, The scientific and public-health imperative for a vaccine against dental caries, Nat Rev Immunol, 6 (2006) 555–563. [DOI] [PubMed] [Google Scholar]
- [243].Bowen WH, Vaccine against dental caries--a personal view, J Dent Res, 75 (1996) 1530–1533. [DOI] [PubMed] [Google Scholar]
- [244].Cao XX, Li YH, Ye QL, Hu X, Wang TF, Fan MW, Self-assembling anticaries mucosal vaccine containing ferritin cage nanostructure and glucan-binding region of S. mutans glucosyltransferase effectively prevents caries formation in rodents, Hum Vaccin Immunother, 13 (2017) 2332–2340. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [245].Kt S, Kmk M, B. N, Jimson S, S. R, Dental caries vaccine - a possible option?, J Clin Diagn Res, 7 (2013) 1250–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Smith DJ, Taubman MA, Experimental immunization of rats with a Streptococcus mutans 59-kilodalton glucan-binding protein protects against dental caries, Infect Immun, 64 (1996) 3069–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Ferretti JJ, Shea C, Humphrey MW, Cross-reactivity of Streptococcus mutans antigens and human heart tissue, Infect Immun, 30 (1980) 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Van de Rijn I, Bleiweis AS, Zabriskie JB, Antigens in Streptococcus mutans cross reactive with human heart muscle, J Dent Res, 55 Spec No (1976) C59–64. [DOI] [PubMed] [Google Scholar]
- [249].Wu HY, Russell MW, Immunological cross-reactivity between Streptococcus mutans and human heart tissue examined by cross-immunization experiments, Infect Immun, 58 (1990) 3545–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Childers NK, Li F, Dasanayake AP, Li Y, Kirk K, Michalek SM, Immune response in humans to a nasal boost with Streptococcus mutans antigens, Oral Microbiol Immunol, 21 (2006) 309–313. [DOI] [PubMed] [Google Scholar]
- [251].Childers NK, Zhang SS, Michalek SM, Oral immunization of humans with dehydrated liposomes containing Streptococcus mutans glucosyltransferase induces salivary immunoglobulin A2 antibody responses, Oral Microbiol Immunol, 9 (1994) 146–153. [DOI] [PubMed] [Google Scholar]
- [252].Smith DJ, Taubman MA, Oral immunization of humans with Streptococcus sobrinus glucosyltransferase, Infect Immun, 55 (1987) 2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Smith DJ, Taubman MA, Effect of local deposition of antigen on salivary immune responses and reaccumulation of mutans streptococci, J Clin Immunol, 10 (1990) 273–281. [DOI] [PubMed] [Google Scholar]
- [254].Mattos-Graner RO, Klein MI, Smith DJ, Lessons Learned from Clinical Studies: Roles of Mutans Streptococci in the Pathogenesis of Dental Caries, Curr Oral Health Rep, 1 (2013) 70–78. [Google Scholar]
- [255].Lehner T, Ma JK, Kelly CG, A mechanism of passive immunization with monoclonal antibodies to a 185,000 M(r) streptococcal antigen, Adv Exp Med Biol, 327 (1992) 151–163. [DOI] [PubMed] [Google Scholar]
- [256].Smith DJ, King WF, Godiska R, Passive transfer of immunoglobulin Y antibody to Streptococcus mutans glucan binding protein B can confer protection against experimental dental caries, Infect Immun, 69 (2001) 3135–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Robinette RA, Oli MW, McArthur WP, Brady LJ, A therapeutic anti-Streptococcus mutans monoclonal antibody used in human passive protection trials influences the adaptive immune response, Vaccine, 29 (2011) 6292–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Weintraub JA, Hilton JF, White JM, Hoover CI, Wycoff KL, Yu L, Larrick JW, Featherstone JD, Clinical trial of a plant-derived antibody on recolonization of mutans streptococci, Caries Res, 39 (2005) 241–250. [DOI] [PubMed] [Google Scholar]
- [259].Ma JK, Hikmat BY, Wycoff K, Vine ND, Chargelegue D, Yu L, Hein MB, Lehner T, Characterization of a recombinant plant monoclonal secretory antibody and preventive immunotherapy in humans, Nat Med, 4 (1998) 601–606. [DOI] [PubMed] [Google Scholar]
- [260].Jung HJ, Pamer EG, Bacterial pathogens: A spoonful of sugar could be the medicine, Nature, 546 (2017) 479–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Ernst B, Magnani JL, From carbohydrate leads to glycomimetic drugs, Nat Rev Drug Discov, 8 (2009) 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].von Itzstein M, Disease-associated carbohydrate-recognising proteins and structure-based inhibitor design, Curr Opin Struct Biol, 18 (2008) 558–566. [DOI] [PubMed] [Google Scholar]
- [263].Holmskov U, Mollenhauer J, Madsen J, Vitved L, Gronlund J, Tornoe I, Kliem A, Reid KB, Poustka A, Skjodt K, Cloning of gp-340, a putative opsonin receptor for lung surfactant protein D, Proc Natl Acad Sci U S A, 96 (1999) 10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Holmskov U, Lawson P, Teisner B, Tornoe I, Willis AC, Morgan C, Koch C, Reid KB, Isolation and characterization of a new member of the scavenger receptor superfamily, glycoprotein-340 (gp-340), as a lung surfactant protein-D binding molecule, J Biol Chem, 272 (1997) 13743–13749. [DOI] [PubMed] [Google Scholar]
- [265].Klein A, Carnoy C, Wieruszeski JM, Strecker G, Strang AM, van Halbeek H, Roussel P, Lamblin G, The broad diversity of neutral and sialylated oligosaccharides derived from human salivary mucins, Biochemistry, 31 (1992) 6152–6165. [DOI] [PubMed] [Google Scholar]
- [266].Cohen RE, Levine MJ, Salivary glycoproteins, in: Tenovuo JO (Ed.) Human saliva: clinical chemistry and microbiology, CRC Press, Inc., Boca Raton, Florida, 1989, pp. 101–130. [Google Scholar]
- [267].Oho T, Yu H, Yamashita Y, Koga T, Binding of salivary glycoprotein-secretory immunoglobulin A complex to the surface protein antigen of Streptococcus mutans, Infect Immun, 66 (1998) 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Mestecky J, Russell MW, IgA subclasses, Monogr Allergy, 19 (1986) 277–301. [PubMed] [Google Scholar]
- [269].Brandtzaeg P, Salivary Immunoglobulins, in: Tenovuo JO (Ed.) Human saliva: clinical chemistry and microbiology, CRC Press, Inc., Boca Raton, Florida, 1989, pp. 1–54. [Google Scholar]
- [270].Woof JM, Mestecky J, Mucosal Immunoglobulins, Mucosal Immunology, Academic Press, Boston, 2015, pp. 287–324. [Google Scholar]
- [271].Ericson T, Rundegren J, Characterization of a salivary agglutinin reacting with a serotype c strain of Streptococcus mutans, Eur J Biochem, 133 (1983) 255–261. [DOI] [PubMed] [Google Scholar]
- [272].Reddy MS, Levine MJ, Prakobphol A, Oligosaccharide structures of the low-molecularweight salivary mucin from a normal individual and one with cystic fibrosis, J Dent Res, 64 (1985) 33–36. [DOI] [PubMed] [Google Scholar]
- [273].Shin K, Hayasawa H, Lonnerdal B, Purification and quantification of lactoperoxidase in human milk with use of immunoadsorbents with antibodies against recombinant human lactoperoxidase, Am J Clin Nutr, 73 (2001) 984–989. [DOI] [PubMed] [Google Scholar]
- [274].Fine DH, Lactoferrin: A Roadmap to the Borderland between Caries and Periodontal Disease, J Dent Res, 94 (2015) 768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Spik G, Strecker G, Fournet B, Bouquelet S, Montreuil J, Dorland L, van Halbeek H, Vliegenthart JF, Primary structure of the glycans from human lactotransferrin, Eur J Biochem, 121 (1982) 413–419. [DOI] [PubMed] [Google Scholar]
- [276].Maurer MA, Meyer L, Bianchi M, Turner HL, Le NPL, Steck M, Wyrzucki A, Orlowski V, Ward AB, Crispin M, Hangartner L, Glycosylation of human IgA directly inhibits influenza A and other sialic-acid-binding viruses, Cell Rep, 23 (2018) 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Torano A, Tsuzukida Y, Liu YS, Putnam FW, Location and structural significance of the oligosaccharides in human Ig-A1 and IgA2 immunoglobulins, Proc Natl Acad Sci U S A, 74 (1977) 2301–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Rogers JD, Palmer RJ Jr., Kolenbrander PE, Scannapieco FA, Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation, Infect Immun, 69 (2001) 7046–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Kilian M, Nyvad B, Ability to bind salivary alpha-amylase discriminates certain viridans group streptococcal species, J Clin Microbiol, 28 (1990) 2576–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Douglas CW, Pease AA, Whiley RA, Amylase-binding as a discriminator among oral streptococci, FEMS Microbiol Lett, 54 (1990) 193–197. [DOI] [PubMed] [Google Scholar]
- [281].Yamashita K, Tachibana Y, Nakayama T, Kitamura M, Endo Y, Kobata A, Structural studies of the sugar chains of human parotid alpha-amylase, J Biol Chem, 255 (1980) 5635–5642. [PubMed] [Google Scholar]
- [282].Leinonen J, Kivelä J, Parkkila S, Parkkila AK, Rajaniemi H, Salivary carbonic anhydrase isoenzyme VI is located in the human enamel pellicle, Caries Res, 33 (1999) 185–190. [DOI] [PubMed] [Google Scholar]
- [283].Murakami H, Sly WS, Purification and characterization of human salivary carbonic anhydrase, J Biol Chem, 262 (1987) 1382–1388. [PubMed] [Google Scholar]
- [284].van de Bovenkamp FS, Hafkenscheid L, Rispens T, Rombouts Y, The emerging importance of IgG Fab glycosylation in immunity, J Immunol, 196 (2016) 1435–1441. [DOI] [PubMed] [Google Scholar]
- [285].Zheng K, Bantog C, Bayer R, The impact of glycosylation on monoclonal antibody conformation and stability, mAbs, 3 (2011) 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Hassan MI, Waheed A, Yadav S, Singh TP, Ahmad F, Zinc alpha 2-glycoprotein: a multidisciplinary protein, Mol Cancer Res, 6 (2008) 892–906. [DOI] [PubMed] [Google Scholar]
- [287].Sanchez LM, Chirino AJ, Bjorkman P, Crystal structure of human ZAG, a fat-depleting factor related to MHC molecules, Science, 283 (1999) 1914–1919. [DOI] [PubMed] [Google Scholar]
- [288].Sakamoto W, Yoshikawa K, Uehara S, Nishikaze O, Characterization of human salivary kallikrein: reactivities to human plasma kininogens and proteinase inhibitors, J Biochem, 93 (1983) 833–838. [DOI] [PubMed] [Google Scholar]
- [289].Reddy MS, Levine MJ, Tabak LA, Structure of the carbohydrate chains of the proline-rich glycoprotein from human parotid saliva, Biochem Biophys Res Commun, 104 (1982) 882–888. [DOI] [PubMed] [Google Scholar]
- [290].Abdolhosseini M, Sotsky JB, Shelar AP, Joyce PB, Gorr SU, Human parotid secretory protein is a lipopolysaccharide-binding protein: identification of an anti-inflammatory peptide domain, Mol Cell Biochem, 359 (2012) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Geetha C, Venkatesh SG, Dunn BH, Gorr SU, Expression and anti-bacterial activity of human parotid secretory protein (PSP), Biochem Soc Trans, 31 (2003) 815–818. [DOI] [PubMed] [Google Scholar]
- [292].Bingle L, Barnes FA, Lunn H, Musa M, Webster S, Douglas CW, Cross SS, High AS, Bingle CD, Characterisation and expression of SPLUNC2, the human orthologue of rodent parotid secretory protein, Histochem Cell Biol, 132 (2009) 339–349. [DOI] [PubMed] [Google Scholar]
- [293].Prokopovic V, Popovic M, Andjelkovic U, Marsavelski A, Raskovic B, Gavrovic-Jankulovic M, Polovic N, Isolation, biochemical characterization and anti-bacterial activity of BPIFA2 protein, Arch Oral Biol, 59 (2014) 302–309. [DOI] [PubMed] [Google Scholar]
- [294].Schaller J, Akiyama K, Kimura H, Hess D, Affolter M, Rickli EE, Primary structure of a new actin-binding protein from human seminal plasma, Eur J Biochem, 196 (1991) 743–750. [DOI] [PubMed] [Google Scholar]
- [295].Caputo E, Manco G, Mandrich L, Guardiola J, A novel aspartyl proteinase from apocrine epithelia and breast tumors, J Biol Chem, 275 (2000) 7935–7941. [DOI] [PubMed] [Google Scholar]
- [296].Gussow D, Rein R, Ginjaar I, Hochstenbach F, Seemann G, Kottman A, Ploegh HL, The human beta 2-microglobulin gene. Primary structure and definition of the transcriptional unit, J Immunol, 139 (1987) 3132–3138. [PubMed] [Google Scholar]
- [297].Manconi B, Castagnola M, Cabras T, Olianas A, Vitali A, Desiderio C, Sanna MT, Messana I, The intriguing heterogeneity of human salivary proline-rich proteins, J Proteomics, 134 (2016) 47–56. [DOI] [PubMed] [Google Scholar]
- [298].Manconi B, Cabras T, Sanna M, Piras V, Liori B, Pisano E, Iavarone F, Vincenzoni F, Cordaro M, Faa G, Castagnola M, Messana I, N- and O-linked glycosylation site profiling of the human basic salivary proline-rich protein 3M, J Sep Sci, 39 (2016) 1987–1997. [DOI] [PubMed] [Google Scholar]
- [299].Lu Y, Bennick A, Interaction of tannin with human salivary proline-rich proteins, Arch Oral Biol, 43 (1998) 717–728. [DOI] [PubMed] [Google Scholar]
- [300].Stubbs M, Chan J, Kwan A, So J, Barchynsky U, Rassouli-Rahsti M, Robinson R, Bennick A, Encoding of human basic and glycosylated proline-rich proteins by the PRB gene complex and proteolytic processing of their precursor proteins, Arch Oral Biol, 43 (1998) 753–770. [DOI] [PubMed] [Google Scholar]
- [301].Carlson DM, Salivary proline-rich proteins: biochemistry, molecular biology, and regulation of expression, Crit Rev Oral Biol Med, 4 (1993) 495–502. [DOI] [PubMed] [Google Scholar]
- [302].Lynch DJ, Fountain TL, Mazurkiewicz JE, Banas JA, Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture, FEMS Microbiol Lett, 268 (2007) 158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Demuth DR, Golub EE, Malamud D, Streptococcal-host interactions. Structural and functional analysis of a Streptococcus sanguis receptor for a human salivary glycoprotein, J Biol Chem, 265 (1990) 7120–7126. [PubMed] [Google Scholar]
- [304].Demuth DR, Lammey MS, Huck M, Lally ET, Malamud D, Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin, Microb Pathog, 9 (1990) 199–211. [DOI] [PubMed] [Google Scholar]
- [305].Clemans DL, Kolenbrander PE, Isolation and characterization of coaggregation-defective (Cog-) mutants of Streptococcus gordonii DL1 (Challis), J Ind Microbiol, 15 (1995) 193–197. [DOI] [PubMed] [Google Scholar]
- [306].Frey AM, Satur MJ, Phansopa C, Parker JL, Bradshaw D, Pratten J, Stafford GP, Evidence for a carbohydrate-binding module (CBM) of Tannerella forsythia NanH sialidase, key to interactions at the host-pathogen interface, Biochem J, 475 (2018) 1159–1176. [DOI] [PubMed] [Google Scholar]