Abstract
Spastic paraplegia 30 is a recently established autosomal recessive disease characterized by a complex form of spastic paraplegia associated with neuropathy. Homozygous mutations of KIF1A reportedly lead to hereditary spastic paraplegia or hereditary sensory and autonomic neuropathy type 2 (HSAN2), whereas heterozygous mutations can cause nonsyndromic and syndromic intellectual disability (MRD9). Here we report the case of a 37-year-old female who presented with gait disturbance complicated with moyamoya disease.
Results
The patient exhibited hypotonia during infancy, after which intellectual disability, epileptic fits, spastic paraplegia, and cerebellar atrophy occurred. Genetic analysis revealed a novel de novo mutation (c.254C > A, p.A85D) in the motor domain of KIF1A.
Keywords: KIF1A, Spastic paraplegia 30: SPG30, Moyamoya disease, Gene, HSAN2, Mental retardation, Autosomal dominant 9, MRD9
1. Introduction
Hereditary spastic paraplegias (HSPs) are a group of clinically and genetically heterogeneous neurodegenerative diseases characterized by progressive spasticity and lower limb weakness. To date, as many as 79 loci [spastic paraplegia (SPG) 1–79] have been identified. In 2011, hereditary SPG 30 caused by a homozygous mutation in KIF1A (Kinesin family member1A) was identified in a Palestinian family presenting early childhood onset HSP [1]. Recessively inherited cases of the KIF1A mutations were further identified in several families [2,3]. KIF1A is located on the long arm of chromosome 2 (2q37.3) and is responsible for hereditary sensory and autonomic neuropathy type 2 (HSAN2) [4]. Furthermore, a heterozygous KIF1A mutation has been described to cause nonsyndromic intellectual disability (mental retardation, autosomal dominant 9: MRD9) [5]. Cases exhibiting a diverse range of symptoms caused by heterozygous mutations in KIF1A, such as nonsyndromic and syndromic intellectual disability, cerebellar atrophy, spastic paraplegia, and axonal neuropathy, have recently been reported [6,7].
Here we report the case of a 37-year-old woman who exhibited hypotonia during infancy, epileptic fits and intellectual disability during childhood, and late development of SPG and cerebellar atrophy during adulthood, in whom a novel heterozygous mutation of KIF1A (c.254C > A,p.A85D) was detected via exome analysis.
2. Methods
2.1. Patient
The proband and her parents (Japanese origin) were examined. This study was approved by the Kindai University genetics ethics committee and the institutional review board of the University of Tokyo. Written informed consent was obtained from the participant and her parents.
2.2. Whole exome sequence analysis and confirmation of the mutations
Genomic DNA was extracted from peripheral blood leukocytes obtained from the proband and her parents, following standard procedures. Exome sequencing analysis was performed in the proband. In brief, exon sequences were enriched using the SureSelect Human All Exon v5 + UTRs kit (Agilent, Santa Clara, CA) and subjected to massively parallel sequence analysis employing an Illumina Hiseq2500 platform (Illumina, San Diego, CA), following manufacturer's instructions. Burrows Wheeler Aligner [8] and sequence alignment/map tools [9] were used with default settings for aligning raw reads and detecting variations. Variant confirmation was performed via direct nucleotide sequence analysis using the ABI 3730 Genetic Analyzer. The mutation in KIF1A and the variant in RNF213 were analyzed using the primer pairs described in the studies conducted by Riviere et al. [6] and Liu et al. [10], respectively.
2.3. Clinical course
The patient's family history was unremarkable. She was delivered normally, and no abnormalities were noted at her 3-month checkup. At 5 months of age, she was examined at a local doctor's clinic owing to hypotonia; however, no clear developmental abnormality was identified. Because she exhibited short stature from 5 to 6 years of age, she was examined at another hospital at 11 years of age and subsequently received growth hormone therapy. Epileptic fits were noted at 12 years of age, and she underwent electroencephalography at a local clinic. Hence, administration of antiepileptic agents was initiated. At this time, no abnormal findings were noted on magnetic resonance imaging (MRI) or computed tomography scans. Muscle weakness gradually became apparent, and the patient was admitted to our hospital at 29 years of age for the evaluation of her gait disorder. Magnetic resonance angiography performed during hospitalization revealed moyamoya disease by chance; however, the disease was considered to have not contributed to her gait disturbance.
Her general physical examination was unremarkable upon admission. Neurological examination revealed mild proximal muscle weakness in the limbs, but tendon reflexes were normal. The Babinski sign was extensor on both sides. No abnormal limb coordination was noted. In the sensory system, vibration sensation was mildly diminished in both legs, and spastic gait was noted. The Hasegawa's dementia scale-revised (HDS-R) score was 18/30. Blood examination revealed no electrolyte, thyroid function, or adrenal gland function abnormalities. Cerebrospinal fluid examination revealed normal results. Head MRI scan indicated atrophic cerebellar vermis and mild cerebral atrophy. MR angiography revealed occlusion from the origin of the middle cerebral arteries on both sides accompanied by rich perforating branches and collateral pathways on the parietal side, consistent with the diagnosis of moyamoya disease. Leg MRI scans revealed diffuse atrophy of the muscle tissue, with prominent atrophy of the quadriceps femoris muscle. A mesh-patterned, high-signal intensity was noted, indicating fatty degeneration (Fig. 1A). Nerve conduction studies revealed decreased sensory nerve conduction velocity of the ulnar nerve and motor nerve conduction velocity of the posterior tibial nerve, indicating a mildly demyelinating pattern. Needle electromyography revealed low amplitude potential in the left biceps brachii, and myopathy was suspected. Muscle biopsy of the vastus muscle on the outer side of the left femur revealed differences in the muscle fiber sizes, with the same extent of atrophy of type I & type II fibers. Abnormal intermyofibrillar networks were observed within muscle fibers, with two to three muscle fibers with cytoplasmic bodies observed in the entire region. However, there were few specific findings, and only mild myopathy-like changes observed. Neurogenic changes were not apparent. Electroencephalography revealed the widespread appearance of a 1–2 Hz spike-and-slow-wave complex.
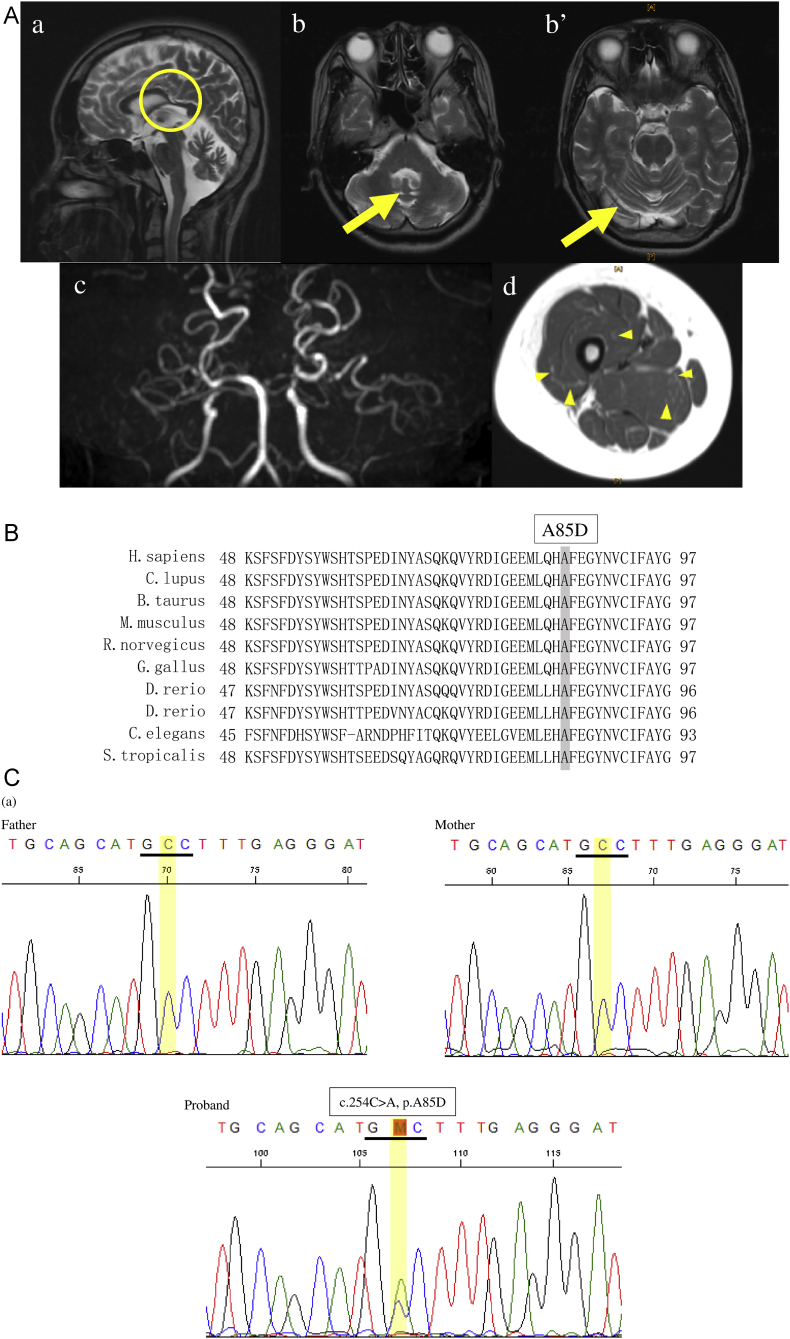
Fig. 1.
A. Brain and femoral magnetic resonance imaging data of the proband (a) The sagittal T2-weighted image shows thinning of corpus callosum (indicated by circle). (b, b') T2-weighted images show atrophy of cerebellar vermis and hemispheres (indicated by arrow). (c) Occlusions from the origin of the middle cerebral arteries on both sides are shown in the magnetic resonance angiography. Perforating branch and collateral pathway can be observed. (d) Diffuse atrophy of muscle tissues (particularly observed in the quadriceps femoris muscle), mesh-patterned high-signal intensity (arrow head) are shown in the T2-weighted image.
B. Amino acid sequence alignment of KIF1A. Positions 48–97 of KIF1A are highly conserved among species.
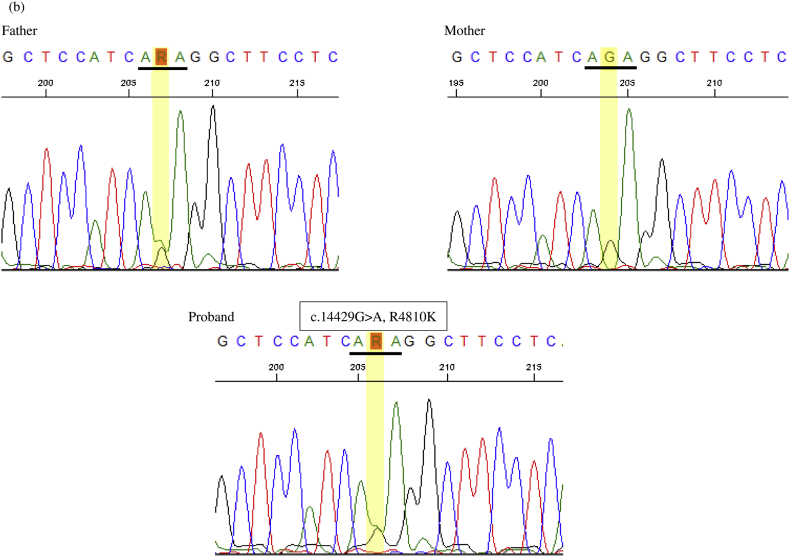
C. Results of the genetic analysis. (a) Electropherography revealed a missense mutation (c.254C > A, p.A85D) in exon 3 of KIF1A and (b) a high-risk variant (c.14429G > A, p.R4810K) in the RNF213 gene.
2.4. Genetic analysis
Because the patient exhibited decreased muscle strength, spastic gait, intellectual disability, and short stature, diseases including mitochondrial disease, chromosomal disorders, and hereditary spastic paraplegia of complex form were suspected for differential diagnosis. No mutations were identified at m.3243 or m.8344 in the mitochondrial DNA using Sanger sequencing. Furthermore, no chromosomal abnormalities were noted using G-banding.
Exome analysis revealed a novel heterozygous mutation (c.254C > A, p.A85D) in KIF1A, which is absent in the in-house database of 1261 control subjects or the public gnomAD database (http://gnomad.broadinstitute.org/). No other potential causative variants were detected in the genes known to cause HSP. The mutation was located in the motor domain of KIF1A, with A85 evolutionally conserved among species (Fig. 1B). Sanger sequencing revealed that this mutation was absent in the parents; therefore, the mutation in KIF1A was considered a de novo mutation. Sanger sequencing of seven rare variants located in different chromosomes identified via exome analysis of the proband were present in either of the biological parents. Moreover, a high-risk variant (rs112735431) for moyamoya disease in RNF213 was detected and found to be transmitted from the father (Fig. 1C).
3. Discussion
While homozygous mutations in KIF1A cause hereditary sensory and autonomic neuropathy type 2C and HSP type 30, heterozygous mutations in the motor domain of KIF1A reportedly cause pure SPG, nonsyndromic intellectual disability, or syndromic intellectual disability with SPG. KIFs comprise a 40-kDa molecular weight spherical domain called the motor domain (MD). KIF1A comprises 1791 amino acids, and the MD is made up of amino acids 1–361 at the amino-terminus. Previously reported heterozygous KIF1A mutations are located in the MD [ref]. The mutation identified in our study (c.254C > A, p.A85D) affects a highly conserved amino acid in the MD. The mutation was absent in the in-house database of 1261 control subjects as well as in the public Genome Aggregation Database (gnomAD). De novo occurrence of the mutation was confirmed based on the trio analysis. These findings strongly support the pathogenicity of the novel mutation identified in the patient. Moreover, the results of the pathogenicity prediction database (PROVEAN, Polyphen2) indicate strong pathogenicity due to protein structural changes. Because intellectual disability, cerebellar atrophy, epileptic fits, and axonal neuropathy are the common clinical characteristics of patients with heterozygous KIF1A mutations, we conclude that these findings in the proband are attributable to the KIF1A mutation.
Although the patient had not experienced any clinical symptoms attributable to cerebral ischemia, magnetic resonance angiography performed at admission revealed cerebral vessel abnormalities consistent with that observed in moyamoya disease. Because moyamoya disease has been found to be highly associated with a SNP (rs112735431) of RNF213 in Asian populations [10,11], we examined it when performing exome sequencing. The data showed that the patient was a heterozygous carrier of the high-risk variant rs112735431. Posey et al. reported that the frequency of patients with two or more disease loci was approximately 4.9% (101 of 2076) and that among those patients, 97 received dual molecular diagnoses. Dual molecular diagnosis could be divided into two categories based on the exhibited features: distinct clinical features or similar phenotypic features [12]. Our case diagnosis can be categorized as that with distinct clinical features. However, it is difficult to clarify the complexity of the blending phenotype in a single patient with two molecular variants. Because this case did not present transient motor disturbances as referred to in the OMIM database, it seems to have a distinct phenotype. Regarding the modes of inheritance, variants in autosomal dominant disease genes were the most common [12], in line with the inheritance pattern of both variants in KIF1A and RNF213 noted here. Going forward, with the accumulation of genetic testing, many genetic variants will be discovered and detailed data from more cases will be investigated, eventually leading to easier and faster interpretation of cases with dual molecular diagnosis.
Acknowledgments
Acknowledgments
This study was supported in part by Ministry of Health, Labor, and Welfare of Japan and the Ministry of Education, Culture, Sports, Science, and Technology of Japan. This study was also supported by grants (Nos. 16kk0205001h0001 and 17kk0205001h0002) from the Japan Agency for Medical Research and Development (AMED) to S.T.
Declaration of interest
None.
References
- 1.Erlich Y., Edvardson S., Hodges E. Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res. 2011;21:658–664. doi: 10.1101/gr.117143.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klebe S., Azzedine H., Durr A. Autosomal recessive spastic paraplegia (SPG30) with mild ataxia and sensory neuropathy maps to chromosome 2q37.3. Brain. 2006;129:1456–1462. doi: 10.1093/brain/awl012. [DOI] [PubMed] [Google Scholar]
- 3.Klebe S., Lossos A., Azzedine H. KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur. J. Hum. Genet. 2012;20:645–6499. doi: 10.1038/ejhg.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riviere J.B., Ramalingam S., Lavastre V. KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am. J. Hum. Genet. 2011;89:219–230. doi: 10.1016/j.ajhg.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamdan F.F., Gauthier J., Araki Y. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am. J. Hum. Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J.R., Srour M., Kim D. De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy, and cerebellar atrophy. Hum. Mutat. 2015;36:69–78. doi: 10.1002/humu.22709. [DOI] [PubMed] [Google Scholar]
- 7.Ohba C., Haginoya K., Osaka H. De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. J. Hum. Genet. 2015;60:739–742. doi: 10.1038/jhg.2015.108. [DOI] [PubMed] [Google Scholar]
- 8.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H., Handsaker B., Wysoker A. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W., Morito D., Takashima S. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE. 2011;6:e22542. doi: 10.1371/journal.pone.0022542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamada F., Aoki Y., Narisawa A. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J. Hum. Genet. 2011;56:34–40. doi: 10.1038/jhg.2010.132. [DOI] [PubMed] [Google Scholar]
- 12.Posey J.E., Harel T., Liu P. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]